This study describes new yeast expression systems for each subunit of the heterotrimeric epithelial sodium channel (ENaC). We found that a significant amount of each subunit resides in the ER and is destroyed via ERAD. We also found that the chaperone requirements for ENaC subunit degradation were unlike any other ERAD substrate examined.
Abstract
The epithelial sodium channel (ENaC) is composed of a single copy of an α-, β-, and γ-subunit and plays an essential role in water and salt balance. Because ENaC assembles inefficiently after its insertion into the ER, a substantial percentage of each subunit is targeted for ER-associated degradation (ERAD). To define how the ENaC subunits are selected for degradation, we developed novel yeast expression systems for each ENaC subunit. Data from this analysis suggested that ENaC subunits display folding defects in more than one compartment and that subunit turnover might require a unique group of factors. Consistent with this hypothesis, yeast lacking the lumenal Hsp40s, Jem1 and Scj1, exhibited defects in ENaC degradation, whereas BiP function was dispensable. We also discovered that Jem1 and Scj1 assist in ENaC ubiquitination, and overexpression of ERdj3 and ERdj4, two lumenal mammalian Hsp40s, increased the proteasome-mediated degradation of ENaC in vertebrate cells. Our data indicate that Hsp40s can act independently of Hsp70 to select substrates for ERAD.
INTRODUCTION
Proteins that transit the secretory pathway are translocated into the endoplasmic reticulum (ER) during or soon after synthesis, and as this diverse family of macromolecules enter the ER protein folding and posttranslational modifications commence. The efficacy of the posttranslational modification and folding processes are closely monitored by the ER quality control system, which is responsible for recognizing immature secreted proteins (Ellgaard and Helenius, 2003). Key components of the quality system are molecular chaperones, which bind and maintain the solubility of peptides with overall hydrophobic character (Flynn et al., 1991; Blond-Elguindi et al., 1993; Rudiger et al., 1997, 2001). Through the action of chaperones and chaperone-like lectins, misfolded proteins in the ER can be subjected to repeated rounds of assisted folding (Hebert and Molinari, 2007). Nevertheless, terminally misfolded proteins or improperly modified proteins may be targeted for degradation by the cytoplasmic proteasome. This process has been termed ER-associated degradation (ERAD; McCracken and Brodsky, 1996) and can be loosely subdivided into three steps: substrate recognition, retrotranslocation from the ER to the cytosol, and ubiquitination and degradation by the 26S proteasome (Meusser et al., 2005; Ismail and Ng, 2006; Vembar and Brodsky, 2008). Because an ever-increasing number ERAD substrates have been identified that are associated with disease, including the cystic fibrosis transmembrane conductance regulator (CFTR; cystic fibrosis), α1-antitrypsin (antitrypsin deficiency), aquaporin-2 (nephrogenic diabetes insipidus), and HMG-CoA reductase (atherosclerosis), a better definition of the requirements for the ERAD of these and other substrates may lead to novel opportunities for therapeutic intervention.
ERAD substrate recognition is carried out primarily by molecular chaperones that reside within the ER lumen and on the cytosolic face of the ER membrane. One class of chaperones, the heat-shock protein (Hsp) 70s, includes an ER lumenal Hsp70, BiP (also known as Kar2 in yeast), and cytosolic Hsp70s. Hsp70s promote protein folding by maintaining substrate solubility, but these ATP-dependent chaperones have also been intimately linked to ERAD substrate selection in both yeast (Plemper et al., 1997; Brodsky et al., 1999; Hill and Cooper, 2000; Zhang et al., 2001; Kabani et al., 2003) and mammalian cells (Knittler et al., 1995; Schmitz et al., 1995; Beggah et al., 1996; Meerovitch et al., 1998; Skowronek et al., 1998; Meacham et al., 2001; Molinari et al., 2002; Goldfarb et al., 2006; Okuda-Shimizu and Hendershot, 2007). Another class of chaperones, the Hsp40s, are Hsp70 cochaperones. Hsp40s contain a J-domain that binds and stimulates Hsp70's ATPase activity, which is required for high-affinity substrate interaction (Walsh et al., 2004; Craig et al., 2006). In addition, Hsp40 homologues bind peptides and may “hand-off” unfolded proteins to Hsp70s. In all cases thus far examined (see for example Meacham et al., 1999; Nishikawa et al., 2001; Huyer et al., 2004; Youker et al., 2004; Dong et al., 2008), the contributions of Hsp40s during ERAD have been shown to require their interaction with cognate Hsp70s.
In the yeast ER lumen, there are two Hsp40 homologues that partner with BiP, Jem1 and Scj1, and in the yeast cytosol there are two Hsp40s, Hlj1 and Ydj1, that partner with a yeast Hsp70, known as Ssa1. Genetic analysis indicates that the lumenal and cytosolic Hsp40s function redundantly during ERAD (Nishikawa et al., 2001; Youker et al., 2004). Other chaperones and chaperone-like proteins also contribute to ERAD substrate selection and may act in concert with the Hsp70/Hsp40s. These factors include lectins such as EDEM, Yos9/Os-9, and XTP-3B; the protein disulfide isomerases; and Grp94 (Gillece et al., 1999; Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005; Buck et al., 2007; Christianson et al., 2008; Hosokawa et al., 2008; Quan et al., 2008; Clerc et al., 2009; Cormier et al., 2009). Current evidence indicates that a large repertoire of chaperones may simultaneously associate with a given substrate, or groups of chaperones may bind substrates in a hierarchical manner that mediate the “decision” between folding and degradation (Wang et al., 2006). Nevertheless, it remains impossible to predict which chaperones might be required for the folding and/or ERAD of a given substrate.
After selection, an ERAD substrate is retrotranslocated to the cytoplasm. The identity of this retrotranslocation channel is still unclear, but in most cases retrotranslocation and substrate ubiquitination and degradation are tightly coupled (Meusser et al., 2005; Ismail and Ng, 2006; Vembar and Brodsky, 2008). Consistent with these data, multiprotein complexes have been identified in the ER membrane that house putative “retrotranslocons,” E3 ubiquitin ligases, components required for substrate selection, and/or a cytoplasmic ATPase (p97, or Cdc48 in yeast), which helps drive substrate extraction from the ER (Lilley and Ploegh, 2005; Neuber et al., 2005; Ye et al., 2005; Carvalho et al., 2006; DeLaBarre et al., 2006; Denic et al., 2006; Gauss et al., 2006; Oda et al., 2006). In fact, the yeast E3s required for ERAD, Hrd1 and Doa10, might select substrates and moonlight as retrotranslocation channels (Ravid et al., 2006).
ERAD substrates have been classified according to the sites of their folding lesions. ER membrane proteins with lesions in the cytoplasm (ERAD-C substrates) require the Doa10 ubiquitin ligase, whereas proteins with lesions in the lumen (ERAD-L substrates) are ubiquitinated by Hrd1. Proteins with folding lesions within the transmembrane domains, termed ERAD-M substrates, are also ubiquitinated by Hrd1 (Vashist and Ng, 2004; Carvalho et al., 2006; Denic et al., 2006; Hirsch et al., 2009; Sato et al., 2009).
A membrane protein of significant medical importance is the epithelial sodium channel (ENaC). ENaC is a heterotrimeric channel composed of α-, β-, and γ-subunits, and each subunit possesses two membrane-spanning domains: ∼75% of the mass of each subunit resides in the ER lumen and membrane and ∼25% of the mass resides in the cytoplasm (Canessa et al., 1994a; Renard et al., 1994; Snyder et al., 1994). After assembling in the ER, the channel traffics through the secretory pathway to the plasma membrane and helps maintain salt and water balance across several epithelia, including kidney and lung. Mutations in ENaC are responsible for inherited forms of both hyper- and hypotension. Liddle's syndrome is a gain of function mutation in ENaC that prevents channel endocytosis and leads to hypertension. Loss of function mutations result in pseudohypoaldosteronism type I, which is characterized by salt wasting and hypotension (Bhalla and Hallows, 2008). ENaC function is also tied to the pathogenesis associated with cystic fibrosis (Mall et al., 2004). Given its importance, ENaC levels and activity are highly regulated. For example, in the kidney the β- and γ-subunits are constitutively expressed at much higher levels than the α-subunit (Staub et al., 1997; Valentijn et al., 1998; Snyder, 2005). In the absence of the α-subunit, the β- and γ-subunits are degraded by the proteasome (Staub et al., 1997; Valentijn et al., 1998). The aldosterone-induced expression of the α-subunit leads to ENaC assembly and thus helps stabilize the β- and γ-subunits (Asher et al., 1996; Masilamani et al., 1999; Snyder, 2005), but even when all three subunits are expressed, a significant percentage of each subunit is targeted for proteasome-mediated degradation (Staub et al., 1997; Valentijn et al., 1998). This observation suggests that ENaC, like some other epithelial channels (Cheng et al., 1990; Li et al., 2000; Weisz et al., 2000; Yan et al., 2005), assembles inefficiently in the ER.
Even though the ENaC endocytic, recycling, and lysosome-mediated degradation pathways are well defined (Snyder, 2005), little is known about the requirements underlying channel quality control in the ER. Given its complex topology and proposed quaternary structure (Jasti et al., 2007; Gonzales et al., 2009), the ENaC subunits might require a unique cadre of molecular chaperones for degradation. Moreover, the E3 requirements for the turnover of the individual subunits are unknown, and thus it is unclear whether they are targeted to the ERAD-L, -C, or -M pathway. Finally, studies on ENaC function, trafficking, and regulation have uncovered differences between the subunits when they are examined individually (Weisz et al., 2000; Bhalla and Hallows, 2008) even though they are 30–40% identical. Therefore, the selection and degradation of each subunit might exhibit distinct requirements. To begin to address these questions, we have used a yeast ENaC expression system in which the factors that mediate substrate-specific ERAD can be readily defined. Using this model system, we show that Hsp40s can function independently of a cognate Hsp70 to facilitate ERAD substrate targeting and that the turnover of each ENaC subunit requires the E3 ubiquitin ligases required for both ERAD-L/M and ERAD-C pathways.
MATERIALS AND METHODS
Yeast Strains, Growth Conditions, and Plasmids
Yeast strains were propagated at 26°C, and standard methods for growth, media preparation, and transformation were used unless indicated otherwise (Adams et al., 1997a). A complete list of the strains used for this study is presented in Supplemental Table S1.
Expression Plasmids
To create the pRS426GPD and pRS423GPD ENaC-HA constructs, which represent URA and HIS-marked vectors, respectively, the sequences encoding ENaC subunits were PCR amplified from murine ENaC cDNA constructs (Hughey et al., 2003) and inserted into the pRS426GPD or pRS423GPD expression vectors (Mumberg et al., 1995) between the following restriction sites: α (ClaI and EcoRI), β (BamHI and EcoRI), and γ (SpeI and HindIII). In each case the primer corresponding to the 3′ end of the gene included sequences allowing for the insertion of an HA epitope immediately 5′ to a translational stop site. The integrity of each insert was confirmed by DNA sequence analysis. Primer sequences are available upon request.
To monitor the degradation of CPY* (see below), we obtained pRS316CPY*-3HA as a gift from the Weissman lab (University of California, San Francisco) (Bhamidipati et al., 2005). For some studies, we used pRS313CPY*-3HA, which was constructed by PCR amplification of pRS316CPY*-3HA and insertion into the ClaI and SpeI sites of pRS313 (Sikorski and Hieter, 1989).
The expression of epitope-tagged forms of Jem1 utilizes plasmids pRS316JEM1–3HA and pRS316JEM1H566Q-3HA, which were generous gifts from the Nishikawa lab (Nagoya University) (Nishikawa and Endo, 1997).
The analysis of the unfolded protein response (UPR) in yeast utilized plasmid pJC104, which was obtained from the Walter laboratory (University of California, San Francisco). UPR assays were performed as previously reported (Kabani et al., 2003).
Indirect Immunofluorescence Microscopy
Indirect immunofluorescence microscopy was performed based on a previously reported protocol (Coughlan et al., 2004) with several modifications. Wild-type yeast (HRD1/DOA10) (Pagant et al., 2007) containing either the pRS426GPD-αENaC-HA, pRS426GPD-βENaC-HA, or pRS426GPD-γENaC-HA expression vectors were grown to an OD600 ∼0.5 and fixed for 30 min with 3.7% formaldehyde. Cells were harvested by pelleting in a clinical centrifuge at 3000 rpm for 3 min and washed two times with 3 ml of solution A (2 M sorbitol, 0.5 M KPO4 pH 7). Cells were then resuspended in 500 μl solution A, 60 ng/ml 100T zymolase (MP Biomedicals, Solon, OH), and 25 mM 2-mercaptoethanol and incubated at 37°C for 30 min. Cells were pelleted at 3000 rpm for 3 min, washed with solution A twice, resuspended in 800 μl of solution A before adding 30 μl of the cell suspension to a microscope slide (pretreated with 1 mg/ml poly-lysine) and incubated at room temperature for 30 min. The cell suspension was aspirated and the wells were washed once with PBS/0.1% bovine serum albumin (BSA) and twice with PBS/0.1% BSA/0.1% NP40, and then incubated with the following primary antibodies over night at 4°C: anti-HA (Roche, Indianapolis, IN) at 1:250, and anti-Kar2 (Brodsky et al., 1993) at 1:500. The slides were washed once with PBS/0.1% BSA, once with PBS/0.1%BSA/0.1% NP40, and once with PBS/0.1% BSA and were then incubated with the appropriate secondary antibodies (Alexa Fluor 568 goat anti-rabbit 1:500 and Alexa Fluor 488 goat anti-mouse, Invitrogen, Carlsbad, CA) in PBS/0.1% BSA for 1 h at room temperature. The slides were then washed as above, and coverslips were mounted using ProLong Gold Antifade mounting media (Invitrogen). Images were captured on an Olympus BX60 microscope (Olympus, Tokyo, Japan) fitted with a Hamamatsu C4742–95 digital camera (Hamamatsu, Bridgewater, NJ), and analyzed using QED Imaging software (Media Cybernetics, Silver Spring, MD).
Assays to Measure the Degradation of ERAD Substrates
To assess the degradation of ENaC subunits, overnight cultures of yeast at 26°C containing the indicated ENaC expression vector and in the appropriate selective medium were diluted into the same medium and then grown to midlog phase (OD600 = 0.3–0.8). Cultures with an OD600 >1 were not used because ENaC protein expression was minimal under these conditions (our unpublished data). Next, protein synthesis was stopped by the addition cycloheximide to a final concentration of 50 μg/ml, and the cultures were shifted to 37°C and incubated with vigorous shaking. At the indicated time points, 1 ml of cells was harvested and pelleted, and the yeast were washed once with ice-cold water and then flash-frozen in liquid nitrogen. Total cellular protein was precipitated as described (Zhang et al., 2001) and was immediately resolved by SDS-PAGE before Western blot analysis. The ENaC subunits were detected using either anti-hemagglutinin (HA)-horseradish peroxidase (HRP; clone 3F10; Roche) or anti-V5-HRP (Invitrogen) antibody, as indicated. In addition, immunoblots were probed with either anti-Sec61 (Stirling et al., 1992) or anti-glucose-6-phosphate dehydrogenase (Sigma-Aldrich, St. Louis, MO) antisera, which served as a loading control. The primary antibodies were decorated with donkey HRP-conjugated anti-rabbit IgG secondary (GE Healthcare, Waukesha, WI), and the signal was detected using enhanced chemiluminescence (Pierce, Rockford, IL) and visualized on a Kodak 440CF Image Station and quantified with the associated Kodak 1D software (Eastman Kodak, Rochester, NY).
The degradation of CPY* was assessed as described above except that an expression vector for CPY*-3HA (see above) was used.
In Vitro and In Vivo Ubiquitination Assays
The in vitro ubiquitination assay (Nakatsukasa et al., 2008) was adapted to measure the extent of ubiquitinated ENaC in ER-derived microsomes prepared from wild-type and mutant yeast strains expressing the indicated HA-tagged ENaC subunit. Microsomes and cytosol from transformed strains grown in selective medium were prepared as described (McCracken and Brodsky, 1996), and 20-μl ubiquitination reactions contained ∼20 μg of microsomes, an ATP-regenerating system, and 2 mg/ml cytosol in B88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 5 mM MgOAc, and protease inhibitor cocktail [2 μg/ml leupeptin, 2 μg/ml aprotinin, 10 μg/ml trypsin inhibitor, 1 μg/ml E64, 10 μg/ml TPCK, 1 mM PMSF, 0.1 μg/ml pepstatin A]). Reactions were assembled on ice with protease inhibitor cocktail and were then prewarmed to 23°C for 10 min before 2 μl of 125I-labeled ubiquitin was added (∼200,000 cpm/μl). At the indicated time point, the reaction was quenched with 1.25% SDS containing a protease inhibitor cocktail and 10 mM N-ethylmaleimide (NEM). ENaC subunits were immunoprecipitated with anti-HA conjugated resin (Roche) with protease inhibitors. The precipitated samples were split and half of the material was used for a Western blot analysis to detect the precipitated ENaC subunit and the other half was used for phosphorimager analysis. Data were analyzed and quantified using Image Gauge Software (v3.45; Fuji Film Science Lab).
The detection of ubiquitinated ENaC in yeast was performed essentially as described (Ahner et al., 2007). In brief, cells expressing the indicated HA-tagged ENaC subunit were grown to log phase in 30 ml of selective medium. Next, the cells were disrupted with glass beads in lysis buffer (150 mM NaCl, 50 mM Tris, pH7.5, 0.1% NP40, 10 mM NEM, and a protease inhibitor cocktail) by agitation on a Vortex mixer 10 times for 30 s with 30-s incubations on ice between each cycle. Cell debris was removed by centrifugation, and the protein concentration was estimated by measuring the absorbance at 280 nm. ENaC was immunoprecipitated from equal amounts of lysate with anti-HA–conjugated resin (Roche), and the precipitated proteins were resolved by SDS-PAGE. Half of each sample was used to detect ENaC, and the other half was used to detect polyubiquitinated ENaC. ENaC was detected with anti-HA antibody (Roche), and ubiquitinated ENaC was detected with anti-ubiquitin antiserum (obtained from the laboratory of C. Pickart, Johns Hopkins University School of Medicine [deceased]) after boiling the nitrocellulose membrane for 30 min in water. Data were obtained and quantified as described above.
Functional Analysis of ENaC Channel Activity in Xenopus Oocytes
Functional analysis of ENaC was performed in Xenopus oocytes using two-electrode voltage clamp (TEV) and by measuring surface expression (Zerangue et al., 1999; Kashlan et al., 2007). The cDNAs encoding the murine ENaC subunits (α, β, γ) were inserted into pBluescript SK− (Stratagene, La Jolla, CA; Ahn et al., 1999), and plasmids containing cDNA encoding the human Hsp40 homologues, ERdj3 and ERdj4, were generously provided by the Weaver laboratory (University of Cincinnati College of Medicine) (Dong et al., 2008). For both ENaC and the Hsp40s, the cRNAs were prepared from linearized DNA templates using T3 RNA polymerase according to the manufacturer's instructions (Ambion, Austin, TX). Stage V and VI oocytes were injected with cRNAs encoding α-, β-, γ-ENaC (1 ng of each subunit) and between 0.2 and 5 ng of either ERdj3 or ERdj4 cRNA. The oocytes were then maintained at 18°C in modified Barth's saline [MBS; 15 mM HEPES, pH 7.4, 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.3 mM Ca(NO3)2, 0.41 mM CaCl2, 0.82 mM MgSO4, 10 μg/ml sodium penicillin, 10 μg/ml streptomycin sulfate, and 100 μg/ml gentamicin sulfate]. TEV was performed 24 h after injection using a DigiData 1320A interface and a GeneClamp 500B voltage clamp amplifier (Axon Instruments, Foster City, CA). Data acquisition and analyses were performed using pClamp software, v. 8.2 (Axon Instruments). Pipettes were pulled from borosilicate glass capillaries (World Precision Instruments, Sarasota, CA) with a Micropipette Puller (Sutter Instrument, Novato, CA) and had a resistance of 0.3–5 MΩ when filled with 3 M KCl and inserted into the bath solution. Oocytes were maintained in a recording chamber (AutoMate Scientific, Berkeley, CA) and perfused continuously at a flow rate of 3 ml/min with bath solution (10 mM HEPES, pH 7.4, 110 mM NaCl, 2 mM KCl, 2 mM CaCl2). For experiments in the presence of a proteasome inhibitor, oocytes were incubated for 3 h before TEV in MBS supplemented with 6 μM of MG132 (Calbiochem, La Jolla, CA) or an equivalent volume of DMSO.
To measure ENaC surface expression, oocytes were injected with 1 ng of the cRNAs encoding α-, γ-, and FLAG epitope-tagged β-ENaC (βF). Where indicated, oocytes were also injected with 5 ng of the cRNAs encoding ERdj3 or ERdj4. As a negative control, a cRNA encoding an untagged form of β was injected. All subsequent blocking and washing methods were performed at 4°C. Approximately 48 h after injection, oocytes were blocked for 30 min in MBS supplemented with 10 mg/ml (MBS/BSA) and then incubated for 1 h with MBS/BSA containing 1 μg/ml mouse monoclonal anti-FLAG antibody (M2; Sigma-Aldrich). Oocytes were then washed for 1 h in MBS/BSA and incubated with MBS/BSA supplemented with 1 μg/ml HRP-coupled secondary antibody for 1 h [peroxidase-conjugated F(ab′2), goat anti-mouse IgG; Jackson ImmunoResearch, West Grove, PA]. The cells were washed 12 times over a 2 h period and transferred into MBS lacking BSA. Individual oocytes were placed in 100 μl of SuperSignal Elisa Femto maximum sensitivity substrate (Pierce) and incubated for 1 min. Chemiluminescence was quantified in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA).
RESULTS
The ERAD of the ENaC α-, β-, and γ-Subunits Requires Both the Hrd1 and Doa10 Ubiquitin Ligases
At the plasma membrane, each of the three ENaC subunits has a large extracellular loop that accounts for the bulk of the protein's mass. After subunit synthesis, this loop resides within the ER (Figure 1A) and folds into its native structure when the associated subunits are present. Therefore, we predicted that the ERAD-L pathway would be required to degrade individual ENaC subunits and consequently that the Hrd1 but not Doa10 ubiquitin ligase would catalyze subunit ubiquitination and degradation. A Hrd1 degradation requirement might also reflect the fact that the transmembrane helices in each subunit fail to pack unless the subunits have oligomerized. These exposed helices might then be recognized as folding lesions within the membrane; thus, the ERAD-M E3 ligase, which is also Hrd1, would be expected to select ENaC for degradation (Carvalho et al., 2006; Sato et al., 2009). We further envisioned that ER lumenal Hsp70 and Hsp40 chaperones would be required to facilitate ERAD because ENaC's lumenal segment includes a folded domain (Snyder et al., 2000; Rossier, 2003; Jasti et al., 2007; Gonzales et al., 2009). In the absence of an associated subunit, these domains might mis-fold and would have to be retained in a retrotranslocation-competent conformation. Conversely, the deposition of some subunit mass in the cytoplasmic space might lead to the limited requirement for components of the ERAD-C machinery. By analogy, the cytoplasmic portion of ENaC subunits is recognized by an E3 ligase that triggers channel endocytosis from the plasma membrane (Snyder, 2005).
Figure 1.
ENaC is ER localized in yeast. (A) Schematic representation of the ENaC subunits demonstrating topology and N-linked glycosylation sites (triangles), which initially reside in the ER lumen. (B) Cell lysates from wild-type yeast expressing C-terminally HA-tagged α-, β-, or γ-ENaC treated with endoglycosidase H (Endo H). A small amount of glycosylated β-ENaC was evident due to incomplete conversion. In all cases, the fastest migrating species corresponds to the predicted size for each unglycosylated subunit. (C) Localization of ENaC by immunofluorescence. Panels left to right show the same fluorescent field for DAPI nuclear staining, Kar2 (yeast BiP), used as an ER marker, and HA antibody for ENaC, respectively.
To begin to test these hypotheses, we expressed each of the ENaC subunits in yeast. The subunits were tagged with an HA epitope at the C-terminus because epitopes at this position have no effect on channel function when examined in oocyte expression systems and in Madin-Darby canine kidney cells (Adams et al., 1997b; Hanwell et al., 2002). Also, because we wanted to assess chaperone-dependent effects on ERAD, we chose to constitutively express each subunit to minimize stress responses that might arise from using an inducible system. Initial pilot studies expressing α-ENaC using a variety of promoters (Mumberg et al., 1995) established that driving ENaC subunit expression from the GPD promoter in a 2μ vector yielded adequate levels of expression without significant effects on cell growth (our unpublished data).
As shown in Figure 1B, each subunit acquired N-linked glycosylation when expressed in yeast, as evidenced by the multiple bands that were apparent after Western blot analysis and by the fact that these bands collapsed mostly into a single species after treatment with endoglycosidase H. Indirect immunofluorescence was then used to uncover the steady-state residence of each subunit in yeast, and strong colocalization with BiP was apparent (Figure 1C). These data indicate that the α-, β-, and γ-subunits reside primarily within the ER. The results are consistent with studies in mammalian cells and in Xenopus oocyte expression systems, indicating that most of the α-, β-, and γ-subunits are degraded in the ER and that little if any of the β- and γ-ENaC subunits can traffic to the plasma membrane (see for example, Valentijn et al., 1998; Hanwell et al., 2002; Mohan et al., 2004). Of interest, the more diffuse signal corresponding to the α-subunit might reflect data indicating that some fraction of this subunit resides at the cell surface in other systems (Harris et al., 2008).
In a recent report, we obtained preliminary data using the yeast system to establish that the turnover of a doubly tagged version of α-ENaC was proteasome-dependent, as observed in higher cell types (Kashlan et al., 2007). Degradation was also slowed in yeast containing a loss-of-function allele in UFD1, which encodes a member of the Cdc48 complex that extracts ubiquitinated substrates from the ER (Jentsch and Rumpf, 2007). Therefore, to determine which E3 ligase ubiquitinates the α-subunit, the protein was expressed in yeast lacking Hrd1, Doa10, or both Hrd1 and Doa10, and cycloheximide chase analyses were performed. Interestingly, we found that the degree of subunit stabilization was similar regardless of which mutant strain was used (Figure 2). We also developed expression systems for the β- and γ-subunits, and like the α-subunit expression system the proteins contained C-terminal HA tags. We found that the β- and γ-subunits were stabilized to a somewhat greater degree in the hrd1Δdoa10Δ mutant strain, but as observed with the α-subunit the degradation of the β- and γ-subunits was again similar in either the hrd1Δ or doa10Δ mutant.
Figure 2.
The ERAD of ENaC subunits is dependent on both the Hrd1 and Doa10 ubiquitin ligases. Cycloheximide chase reactions were performed as described in Materials and Methods in HRD1/DOA10 (●), hrd1Δ (▴), doa10Δ (▵), or hrd1Δdoa10Δ (○) yeast strains (Pagant et al., 2007) expressing C-terminally HA-tagged α-, β-, or γ-ENaC. Chase reactions were performed at 37°C, lysates were immunoblotted with anti-HA (ENaC) or with anti-Sec61 (as a loading control). Data represent the means of 4–6 experiments, ±SEM.
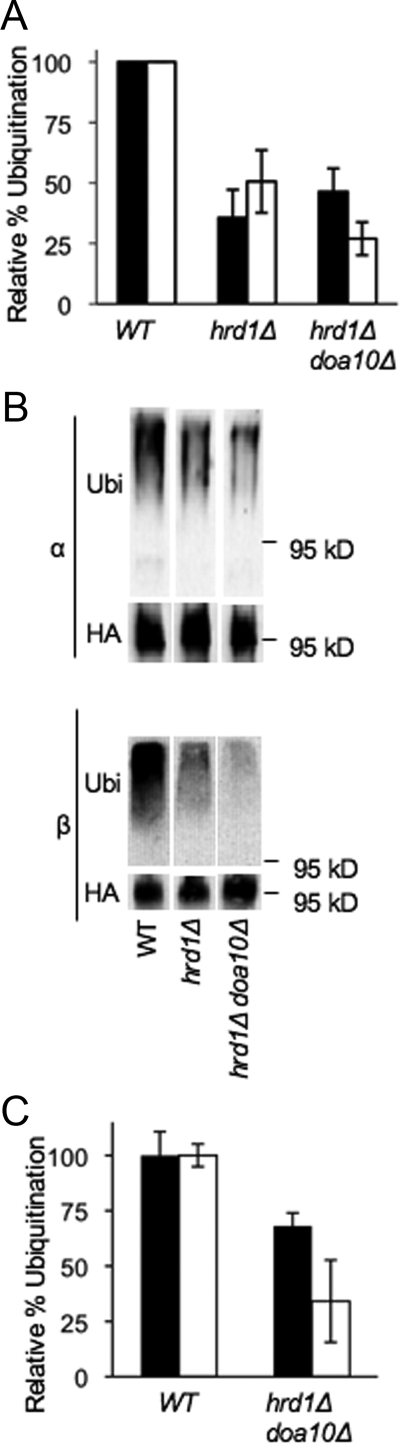
To assay whether the extent of stabilization correlated with the degree of ubiquitination—and thus that the effect of deleting the ligases was direct—we examined α- and β-subunit ubiquitination in vivo. To this end, the α- and β-subunits were immunoprecipitated from wild type and from hrd1Δ and hrd1Δdoa10Δ mutant yeast, and an anti-ubiquitin antibody was used in a subsequent Western blot analysis. The results presented in Figure 3, A and B, indicate that subunit ubiquitination was significantly decreased in yeast lacking Hrd1 or both Hrd1 and Doa10. These data are consistent with a requirement for either Hrd1 or Doa10, as observed in Figure 2, and indicate that the effects of deleting the ligases are direct.
Figure 3.
ENaC ubiquitination is impaired in yeast strains lacking the E3 ubiquitin ligases Hrd1 and Doa10. (A) Wild type (WT) or the indicated mutant yeast strains expressing either the α- (■) or β- (□) ENaC subunit were processed as described in Materials and Methods, and the level of ubiquitination was assessed. The bar graph represents the means of 5–7 determinations, ±SEM, and the data in the mutant strains were standardized to the amount in the wild-type cells. A typical experimental result is shown in B. (C) Microsomes from wild type (HRD1/DOA10) or the hrd1Δdoa10Δ mutant strain expressing either α- or β-ENaC were prepared and subjected to the in vitro ubiquitination assay as described in Figure S1. The bar graphs represent the means of three (β) or eight (α) determinations, ±SEM.
To confirm these data, we next developed an assay in which the degree of ENaC subunit ubiquitination could be assessed in vitro. This assay was based on a recently reported system in which the conjugation of 125I-ubiquitin onto membrane-integrated ERAD substrates could be monitored after their expression in yeast (Nakatsukasa et al., 2008). We therefore prepared ER-derived microsomes from α-ENaC-expressing yeast and delineated the time, temperature, and cytosol dependence on ENaC subunit ubiquitination in vitro (Figure S1). We next prepared microsomes from either wild-type yeast or hrd1Δdoa10Δ cells that expressed the α- or β-subunit and used the optimized conditions to assess subunit ubiquitination in vitro. We again found that subunit ubiquitination decreased in the ligase mutant strain (Figure 3C), whereas polyubiquitination was more modestly affected in the single mutants (our unpublished data). Together, and contrary to our expectations, these results suggest that Hrd1 and Doa10 perform distinct functions during ENaC subunit degradation. Thus, the ENaC subunits cannot be easily classified as ERAD-M/L or -C substrates. Based on the decreased stability of the α-subunit compared with the β- and γ-subunits in the hrd1Δdoa10Δ strain, our results also suggest that the mechanism by which the α-subunit is destroyed might possess unique attributes (see Discussion).
The ER Lumenal Hsp40s, Jem1 and Scj1, Facilitate the Ubiquitination and Degradation of ENaC Subunits
After synthesis, a significant portion of ENaC's mass is deposited into the ER lumen, including a folded domain that regulates channel gating (Snyder et al., 2000; Rossier, 2003; Jasti et al., 2007; Gonzales et al., 2009). Thus, we reasoned that the turnover of ENaC subunits would require lumenal Hsp70 and Hsp40 chaperones.
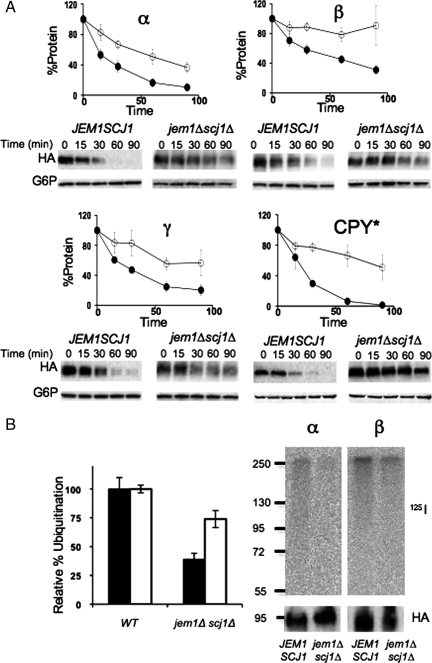
Jem1 is one of two Hsp40 homologues within the yeast ER (the other being Scj1) that function redundantly to prevent the aggregation of soluble, lumenal ERAD substrates before degradation (Nishikawa et al., 2001). Consequently, we examined whether strains lacking Jem1 and Scj1 proficiently degraded the ENaC subunits. As shown in Figure 4A, we noted a significant reduction in the rate and extent of α-, β-, and γ-subunit degradation in the mutant strains relative to the wild-type control. We believe that this was a direct effect of a reduction in ERAD efficiency because ENaC subunit aggregation was not evident in this or any other experiment (our unpublished data). Deletion of JEM1 or SCJ1 individually had no effect on subunit turnover (our unpublished data).
Figure 4.
The ERAD of ENaC subunits depends on the ER lumenal Hsp40s, Jem1 and Scj1. (A) Cycloheximide chase reactions were performed as described in Materials and Methods in JEM1/SCJ1 (●) and jem1Δscj1Δ (○) yeast strains expressing C-terminally HA-tagged α-, β-, or γ-ENaC or CPY*-3HA. Chase reactions were performed at 37°C and lysates were immunoblotted with anti-HA (ENaC) or with anti-glucose-6-phosphate dehydrogenase (as a loading control) antibodies. Data represent the means of 4–6 experiments, ±SEM. (B) Microsomes from wild type (JEM1/SCJ1) or the jem1Δscj1Δ mutant strain expressing either α- or β-ENaC were prepared and subjected to the in vitro ubiquitination assay as described in Materials and Methods. The bar graphs represent the means of six determinations, ±SEM.
The inefficient degradation of the ENaC subunits in the scj1Δjem1Δ strain might either result from an inability of the protein to be delivered or recognized by E3 ligases or result from an inability to transfer the ubiquitinated species to the proteasome. To differentiate between these scenarios, we assessed subunit ubiquitination in vitro, as described above, in a wild-type strain and in cells lacking Scj1 and Jem1 (Figure 4B). The relative percentage of ubiquitinated protein decreased significantly for both the α- and β-subunit, consistent with the Hsp40s playing a role before ubiquitination in the ERAD process.
Hsp40s partner with Hsp70s, and in every case examined Hsp40 function requires the activity of its cognate Hsp70 during ERAD. Previous work established that Jem1 and Scj1 cooperate with BiP, the lone Hsp70 in the ER lumen, to support nuclear membrane fusion and protein folding, respectively (Schlenstedt et al., 1995; Nishikawa and Endo, 1997; Silberstein et al., 1998). As a result, it seemed reasonable to ask whether yeast containing an ERAD-specific mutant allele in the gene encoding yeast BiP, KAR2 (Kabani et al., 2003), would exhibit a defect in the degradation of the α-, β-, and γ-ENaC subunits. To our surprise, we found that the destruction of each subunit was unaffected in the kar2 mutant (Figure 5). The kar2 mutant (kar2-1) is a strain in which the ERAD of Scj1-Jem1–dependent substrates is significantly reduced and in which the off-rate of substrates from the Kar2-1 protein results in ERAD substrate aggregation (Kabani et al., 2003). Nevertheless, to confirm that the mutant was ERAD-deficient, the wild-type and kar2-1 strains were transformed with a vector that drove the expression of CPY*, a well-characterized substrate that was previously demonstrated to require BiP (Plemper et al., 1997). As anticipated, CPY* degradation was significantly slowed in kar2-1 mutant yeast relative to the wild-type strain, as it was in the Hsp40 mutant (Figure 4). Combined with the data presented in Figure 4, these results implicate the ER lumenal Hsp40s acting independently of an Hsp70 as mediators of ENaC subunit quality control.
Figure 5.
The ERAD of ENaC is BiP-independent. Cycloheximide chase reactions were performed as described in Materials and Methods in KAR2 (●) and kar2-1 (○) yeast strains expressing C-terminally HA-tagged α-, β-, or γ-ENaC or CPY*-3HA. Chase reactions were performed at 37°C, and lysates were immunoblotted with anti-HA (ENaC) or with anti-glucose-6-phosphate dehydrogenase (as a loading control) antibodies. Data represent the means of 4–6 experiments, ±SEM.
Even though BiP was dispensable for the degradation of ENaC, the chaperone might assist during subunit folding. BiP plays both a direct and contributory role in the UPR (Kimata et al., 2007). If BiP is recruited from Ire1, the UPR sensor (Cox et al., 1993), and helps fold the unassembled lumenal domain of ENaC, then ENaC subunit expression might induce the UPR. Alternatively, ENaC expression might titrate other ER chaperones (e.g., Jem1 and Scj1), which would compromise general protein folding and induce the UPR. To test these hypotheses, yeast were transformed with the α-ENaC expression vector or with a vector control, as well as with a UPR reporter (see Materials and Methods). We then assessed the extent of UPR induction, as described (Kabani et al., 2003). As a control for these assays, cells were treated with 5 mM dithiothreitol (DTT) for 1 h, which increased the UPR 2.5-fold in wild-type yeast (Table S2). As an additional control the UPR response was examined in ire1Δ cells, which are unable to trigger the UPR (Cox et al., 1993; Mori et al., 1993). We found that α- and β-subunit expression induced the UPR and that the level of induction did not rise further with DTT treatment. These data suggest that the lumenal portion of ENaC interacts with and possibly titrates away BiP and/or other chaperones that are important for maintaining ER homeostasis. As discussed above, one of these chaperones may be Scj1, and of note, the deletion of SCJ1 is known to induce the UPR (Silberstein et al., 1998).
To further verify that Scj1 and Jem1 play a direct role during the ERAD of ENaC subunits, and that degradation is BiP-independent, we reasoned that ENaC should be proficiently degraded in scj1Δjem1Δ yeast expressing a plasmid-borne copy of Jem1 with a mutation in the HPD motif, which interferes with BiP association (Nishikawa and Endo, 1997). In contrast, these cells should exhibit an ERAD defect for a substrate that is Scj1, Jem1, and BiP dependent, such as CPY* (Nishikawa et al., 2001). We also transformed a wild-type copy of Jem1 into the mutant cells. As expected, only the wild-type Jem1 protein rescued the temperature-sensitive phenotype of scj1Δjem1Δ yeast (Figure S2A). Next, the α-ENaC or CPY* expression vector was transformed into each strain, and ERAD was assessed. Even though the signal-to-noise ratio was low in these experiments, the data shown in Figure S2, B and C, are consistent with our hypothesis: Expression of either wild type or the HPD mutant Jem1 resulted in identical levels of α-ENaC degradation, whereas there was a statistically significant attenuation of subunit turnover in the vector control; in contrast, CPY* degradation was slowed the most in the HPD mutant Jem1-expressing strain.
The Proteolysis of ENaC Is Unaffected in Strains Mutated for Cytoplasmic Hsp40 Chaperones That Facilitate the Degradation of Other ERAD Substrates
Based on previously derived models (Snyder et al., 1994) and on the recent structural determination of an ENaC family member (Figure 1; Jasti et al., 2007; Gonzales et al., 2009), only ∼25% of the total mass of ENaC resides in the cytoplasm. Thus, the Doa10 dependence of ENaC degradation was unexpected, but suggested that ENaC's cytoplasmic motifs might also represent ERAD recognition sites. In other words, the subunits might exhibit ERAD-C-like features. The degradation of some ERAD-C substrates requires the action of cytoplasmic Hsp70 and Hsp40s and are therefore stabilized in temperature-sensitive Hsp70 (ssa1-45) and Hsp40 (hlj1Δydj1-151) mutant strains (Zhang et al., 2001; Huyer et al., 2004; Youker et al., 2004). We therefore examined the turnover of each of the ENaC subunits in the chaperone mutant strains. First, we noted that there were variable levels of stabilization of each subunit in the ssa1-45 strain (Figure S3). However, the degradation of each of the three subunits was unaffected in hlj1Δydj1-151 yeast relative to the wild-type control (Figure S4). Interestingly, Rubenstein and colleagues (Goldfarb et al., 2006) reported that the vertebrate homolog of Ydj1, Hdj2, also had no effect on ENaC biogenesis. At this point, we are unable to conclude whether the variable Ssa1 dependence for some subunits but not others reflects distinct subunit recognition or arises from a secondary effect. However, consistent with a secondary effect, we found that α-subunit expression in a bona fide wild-type strain induces a heat-shock response (our unpublished data), which significantly increases SSA1, SSA3, and SSA4 levels. In the ssa1-45 mutant, this compensatory response might be absent, because of the ssa1 mutant allele and because of the absence of the SSA2, SSA3, and SSA4 genes (Becker et al., 1996). Of note, contributions of the heat-shock response on ERAD efficiency have previously been reported in yeast (Liu and Chang, 2008).
The Overexpression of ER-resident Mammalian Hsp40s in Xenopus Oocytes Accelerates the Proteasome-mediated Degradation of ENaC
The human Hsp40s most similar to Scj1 and Jem1 in both domain architecture and cellular localization are, respectively, ERdj3 and ERdj4 (Nishikawa and Endo, 1997; Shen et al., 2002; Walsh et al., 2004; Hennessy et al., 2005; Shen and Hendershot, 2005). Scj1 and ERdj3 are type I Hsp40s that possess an N-terminal J-domain, a glycine/phenylalanine-rich domain, and a cysteine-rich domain. Jem1 and ERdj4 are both membrane-associated J-domain–containing proteins. ERdj3's role in biological processes has yet to be determined, but the chaperone resides in BiP-containing complexes, associates with unfolded BiP substrates, and can bind unfolded proteins in the absence of BiP (Yu et al., 2000; Meunier et al., 2002; Shen and Hendershot, 2005; Yu and Haslam, 2005; Jin et al., 2009). ERdj4 also binds unfolded proteins and was recently established to play an Hsp70-dependent role in the ERAD of mutant surfactant protein C (Shen et al., 2002; Dong et al., 2008). On the basis of these data, we predicted that ERdj3 and ERdj4 might facilitate the ERAD of ENaC in higher cell types.
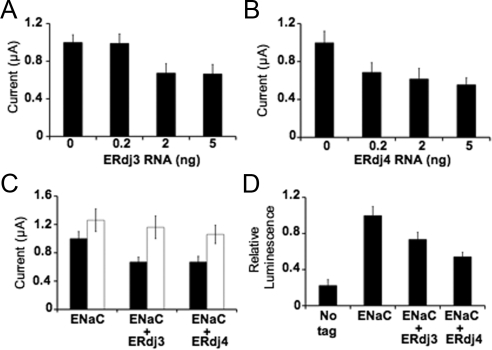
To test this hypothesis, we used a Xenopus oocytes expression system in which exogenous factors that impact ENaC function and trafficking can be readily examined (Adams et al., 1997b; Valentijn et al., 1998; Goldfarb et al., 2006; Kashlan et al., 2007). First, increasing amounts of ERdj3 or ERdj4 cRNA were coinjected into oocytes in the presence or absence of cRNAs encoding the three ENaC subunits. Next, the residence of active ENaC at the plasma membrane was detected by measuring the amiloride-sensitive Na+ current (see Materials and Methods). The result of this analysis indicated a dose-dependent decrease in ENaC current when the cRNA for either of the Hsp40s was coinjected (Figure 6, A and B). To determine whether the reduction in current was ERAD-dependent, the proteasome inhibitor MG132 was added 21 h after the cRNAs encoding either ERdj3 or ERdj4 were coinjected with cRNAs for the ENaC subunits. The amiloride-sensitive current was then measured after a 3 h incubation. As displayed in Figure 6C, we noted a small rise in ENaC current even in the absence of ERdj3 or ERdj4 coinjection, consistent with the known instability of ENaC in the ER (see Introduction). The ERdj3 and ERdj4 dependent decrease in ENaC activity was again observed in this independent experiment, but in the presence of MG132 the current (∼1.2 nA) was similar to that obtained in the absence of Hsp40 overexpression and in the presence of MG132. These data indicate that the proteasome is responsible for the ERdj3 and ERdj4 mediated decrease in ENaC activity.
Figure 6.
ENaC current is reduced in a proteasome dependent manner in oocytes coinjected with the human ER lumenal Hsp40s, ERdj3 and ERdj4. (A–C) Amiloride sensitive current was measured 24 h after injection by TEV. Oocytes were injected with α-, β-, and γ-ENaC cRNA (1 ng each) with increasing amounts of either ERdj3 (A) or ERdj4 (B) cRNA. (C) Oocytes were injected with ENaC cRNA as in A and B with either no additional cRNA or 5 ng ERdj3 or ERdj4 cRNA in either the presence (□) or absence (■) of 6 μM MG132. Average baseline current for oocytes expressing ENaC with no additional cRNA was −2.5 μA. (D) ENaC surface expression was assessed as described in Materials and Methods. Oocytes were injected with 1 ng α-, γ-, and β-FLAG ENaC and 5 ng ERdj3 or ERdj4 cRNA. The “No tag” control includes the same amounts of subunits injected but the β-subunit lacks the FLAG tag. In each panel, data are expressed as the means of 17 oocytes from three frogs. Data in D are the means of >35 oocytes from two frogs, ±SEM.
Another formal interpretation of these data is that MG132 inactivates ENaC that resides at the plasma membrane. To address this possibility, the level of ENaC surface expression was assessed. Of note, the population of amiloride-sensitive ENaC at the plasma membrane reflects the active protein, as measured in parts A–C. This experiment requires the coinjection of cRNAs for the α- and γ-subunit along with the message for an ectopic, FLAG-tagged β-subunit; the addition of an enzyme-linked antibody reports on the surface residence of the assembled channel (Kashlan et al., 2007). As a negative control for this experiment, cRNAs encoding the α- and γ-subunits were coinjected along with the cRNA for an untagged β-subunit, and under this condition, a low amount of luminescence was detected. In the presence of the tagged β-subunit, the signal rose by about fivefold. In contrast, the coinjection of cRNAs corresponding either to ERdj3 or ERdj4 significantly decreased the surface expression of ENaC (Figure 6D). We also established that the coexpression of ERdj3 or ERdj4 had no effect on the rates of channel endocytosis or exocytosis (our unpublished data). Overall, these data support our conclusion that ER lumenal Hsp40s act as mediators of ER protein quality control during ENaC biogenesis, and indicate that this function is conserved between yeast and vertebrates.
DISCUSSION
The data reported in this manuscript contain two novel findings. This is the first example in which lumenal Hsp40s have been shown to play a role in the ERAD of a membrane protein in yeast. To date, the ERAD of these substrates has been found to be independent of ER chaperone function (Hill and Cooper, 2000; Zhang et al., 2001; Huyer et al., 2004). This phenomenon has most likely been observed because the examined integral membrane proteins deposit only a limited amount of their total mass in the ER lumen. Thus, the folding “problem” in these substrates is primarily confined to the cytoplasmic space. As expected, then, a cytosolic Hsp70 (Ssa1) and Hsp40s (Hlj1 and Ydj1) play an important role in the disposal of ERAD-C substrates. In contrast, the majority of ENaC resides within the ER lumen, and consistent with what has been noted for soluble ERAD-L substrates (Nishikawa et al., 2001), we discovered that maximal rates of ENaC subunit turnover require two ER lumenal Hsp40s, Scj1 and Jem1.
The second novel finding is that ENaC is the first ERAD substrate for which Hsp40s have been found to facilitate degradation independent of Hsp70 function. Hsp40s function as Hsp70 cofactors, but these chaperones also bind directly to peptide substrates, which in nearly all cases is followed by the transfer of the bound substrates to an Hsp70 (Hennessy et al., 2005; Qiu et al., 2006; Buck et al., 2007). Therefore, it was surprising that the function of the Scj1-Jem1 Hsp70 cognate, BiP, was dispensable for ENaC subunit degradation. This suggests that the substrate-binding activities of Scj1 and Jem1 directly target ENaC subunits for ERAD. But, another formal possibility is that the residual BiP activity in the kar2-1 strain, which is unable to mediate the efficient ERAD of other Scj1-Jem1–dependent substrates, is still sufficient to promote ENaC subunit degradation.
The degradation and ubiquitination of the ENaC subunits is inhibited when the gene encoding either one of two ER resident E3 ubiquitin ligases, Hrd1 and Doa10, is deleted. This suggests that the activities of both ligases are required to append a critical level of ubiquitin to effectively target the substrate for proteasome-mediated degradation. Consistent with this view, we noted that the extent of Hrd1-Doa10 stabilization of the α- and β-subunits (Figure 2) mirrored their proteasome-dependent degradation (Figure S5). In contrast, the degradation of most other ERAD substrates requires either Hrd1 or Doa10 (Vembar and Brodsky, 2008), although exceptions to this rule have emerged (Gnann et al., 2004; Huyer et al., 2004; Kota et al., 2007; Nakatsukasa et al., 2008). One interpretation of these data is that folding lesions in the individual ENaC subunits reside both in the ER membrane/lumen and in the cytoplasm. Based on the topology of the ENaC subunits and the crystal structure of an assembled, homotrimeric ENaC relative, ASIC (Jasti et al., 2007; Gonzales et al., 2009), it may not be surprising that Hrd1 plays a role in ENaC ubiquitination. Approximately 75% of the mass of each ENaC subunit is either in the ER lumen or membrane and there are abundant intrasubunit contacts that would be absent if the subunits fail to assemble (Jasti et al., 2007; Gonzales et al., 2009). It was initially more surprising that Doa10 plays a role in the degradation of the ENaC subunits. The cytoplasmic segments of ENaC might be recognized by several cytoplasmic chaperones, perhaps including Ssa1, which then transfers ENaC to Doa10, as suggested for ERAD-C substrates (Han et al., 2007; Nakatsukasa et al., 2008). It is also possible that Doa10 directly recognizes and ubiquitinates ENaC subunits. Consistent with this notion, Pca1, a Doa10-requiring ERAD substrate does not show a significant dependence on cytoplasmic Hsp70 for its disposal (Adle et al., 2009).
The modest stabilization of the α- and γ-ENaC subunits observed in the ssa1 mutant strain is consistent with a role for cytoplasmic Hsp70 in transferring the substrate to Doa10, and we note that Hsp70 and Hsc70 have been found to facilitate, respectively, the folding and degradation of ENaC and CFTR in other systems (Rubenstein and Zeitlin, 2000; Goldfarb et al., 2006). However, the relatively subtle effect on ENaC degradation in the ssa1 strain—compared with that observed for other Ssa1-dependent substrates (Zhang et al., 2001)—suggests that different cytoplasmic chaperones act in place of this abundant Hsp70. As an initial test of this hypothesis, we examined ENaC subunit degradation in yeast that contained a temperature-sensitive mutation in Hsp90 but observed that degradation was unaffected (our unpublished data). Nevertheless, we cannot exclude the possibility that the Ssa1 dependence arises from an indirect effect, which was discussed above. Of note, ENaC was cleaved when ssa1 strains reached the late-log phase, which necessitated the use of cultures at significantly lower ODs than used in other experiments (our unpublished data). This observation again suggests that compensatory, stress-induced phenomena may be triggered to provide an alternate mechanism to dispose of ENaC subunits.
Based on the data presented in Figure 2 and Figure S5, the Hrd1-Doa10 and proteasome dependence for the ERAD of α is somewhat different from the other subunits. One interpretation of this result is that the α-subunit might utilize another E3 or that α-ENaC might be turned over in an ERAD-independent manner. To begin to address the second hypothesis, we found that the degradation of the α-subunit was unaffected in a pep4 mutant (our unpublished data), suggesting that the protein is not destroyed after its delivery to the vacuole by the secretory or autophagic pathway. However, we note that the α-subunit can function—albeit quite inefficiently—as a sodium channel on its own; in contrast, the β- and γ-subunits fail to form active channels (Canessa et al., 1994b). In fact, the expression of an α-β chimera in yeast leads to a salt-sensitive growth phenotype (Gupta and Canessa, 2000); therefore, it is possible that the expression of the α-subunit triggers an osmotic stress response that leads to a fraction of the protein being degraded in a Hrd1 and Doa10-independent manner. Moreover, only the α-subunit possesses an ER export sequence (Mueller et al., 2007). Even though each of the subunits resided in the ER under steady-state conditions (Figure 1C), a fraction of the population of α-subunits might migrate to an ER subfraction where the subunits are destroyed through an alternate mechanism. Indeed, the turnover of CFTR in yeast has been proposed to require localization to an ER subdomain (Fu and Sztul, 2003). Future efforts may lead to a better understanding of the nature of the Hrd1 and Doa10-independent degradation pathway.
In sum, we propose that a continued characterization of the factors required for ENaC degradation will yield additional insights into the varied pathways by which ERAD substrates are targeted for degradation. In addition, it will be interesting to determine if other ERAD substrates are targeted for degradation in an Hsp40-dependent but Hsp70-independent manner, and to define what features distinguish this apparently rare class of secreted proteins. Finally, given links between ENaC function and a number of human maladies, these future efforts might uncover previously ill-defined, new therapeutic targets.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Elizabeth Miller (Columbia University), Timothy Weaver (University of Cincinnati), Shuh-ichi Nishikawa (Nagoya University), Jonathan Weissman (University of California, San Francisco), Gunhild Mueller (University of Pittsburgh), Osama Kashlan (University of Pittsburgh), Shaohu Sheng (University of Pittsburgh), Lindsay Plavchak (University of Pittsburgh), Joseph Clay (University of Pittsburgh), Rebecca Hughey (University of Pittsburgh), Peter Walter (University of California, San Francisco), and Shruthi Vembar (University of Pittsburgh) for technical assistance, advice, and/or reagents. This work was supported by National Institutes of Health Grants GM75061 to J.L.B., DK65161 to T.R.K., and DK79307 (the University of Pittsburgh George O'Brien Kidney Research Core Center) to J.L.B. and T.R.K. T.M.B. is supported by a National Institutes of Health T32 training grant (DK61296) and a National Research Service Award (GM83540) and A.R.K. received support from American Heart Association Grant 09PRE2050048.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0944) on January 28, 2010.
REFERENCES
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997a. [Google Scholar]
- Adams C. M., Snyder P. M., Welsh M. J. Interactions between subunits of the human epithelial sodium channel. J. Biol. Chem. 1997b;272:27295–27300. doi: 10.1074/jbc.272.43.27295. [DOI] [PubMed] [Google Scholar]
- Adle D. J., Wei W., Smith N., Bies J. J., Lee J. Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proc. Natl. Acad. Sci. USA. 2009;106:10189–10194. doi: 10.1073/pnas.0812114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y. J., Brooker D. R., Kosari F., Harte B. J., Li J., Mackler S. A., Kleyman T. R. Cloning and functional expression of the mouse epithelial sodium channel. Am J. Physiol. 1999;277:F121–F129. doi: 10.1152/ajprenal.1999.277.1.F121. [DOI] [PubMed] [Google Scholar]
- Ahner A., Nakatsukasa K., Zhang H., Frizzell R. A., Brodsky J. L. Small heat-shock proteins select ΔF508-CFTR for endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 2007;18:806–814. doi: 10.1091/mbc.E06-05-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher C., Wald H., Rossier B. C., Garty H. Aldosterone-induced increase in the abundance of Na+ channel subunits. Am J. Physiol. 1996;271:C605–C611. doi: 10.1152/ajpcell.1996.271.2.C605. [DOI] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E. A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggah A., Mathews P., Beguin P., Geering K. Degradation and endoplasmic reticulum retention of unassembled alpha- and beta-subunits of Na,K-ATPase correlate with interaction of BiP. J. Biol. Chem. 1996;271:20895–20902. doi: 10.1074/jbc.271.34.20895. [DOI] [PubMed] [Google Scholar]
- Bhalla V., Hallows K. R. Mechanisms of ENaC regulation and clinical implications. J. Am Soc. Nephrol. 2008;19:1845–1854. doi: 10.1681/ASN.2008020225. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A., Denic V., Quan E. M., Weissman J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Hamamoto S., Feldheim D., Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J. Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Buck T. M., Wright C. M., Brodsky J. L. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev. Biol. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa C. M., Merillat A. M., Rossier B. C. Membrane topology of the epithelial sodium channel in intact cells. Am J. Physiol. 1994a;267:C1682–C1690. doi: 10.1152/ajpcell.1994.267.6.C1682. [DOI] [PubMed] [Google Scholar]
- Canessa C. M., Schild L., Buell G., Thorens B., Gautschi I., Horisberger J. D., Rossier B. C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994b;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier J. H., Tamura T., Sunryd J. C., Hebert D. N. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan C. M., Walker J. L., Cochran J. C., Wittrup K. D., Brodsky J. L. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 2004;279:15289–15297. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Craig E. A., Huang P., Aron R., Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dong M., Bridges J. P., Apsley K., Xu Y., Weaver T. E. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Flynn G. C., Pohl J., Flocco M. T., Rothman J. E. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Fu L., Sztul E. Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 2003;160:157–163. doi: 10.1083/jcb.200210086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece P., Luz J. M., Lennarz W. J., de La Cruz F. J., Romisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol. 1999;147:1443–1456. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnann A., Riordan J. R., Wolf D. H. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 complex in yeast. Mol. Biol. Cell. 2004;15:4125–4135. doi: 10.1091/mbc.E04-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb S. B., Kashlan O. B., Watkins J. N., Suaud L., Yan W., Kleyman T. R., Rubenstein R. C. Differential effects of Hsc70 and Hsp70 on the intracellular trafficking and functional expression of epithelial sodium channels. Proc. Natl. Acad. Sci. USA. 2006;103:5817–5822. doi: 10.1073/pnas.0507903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales E. B., Kawate T., Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. S., Canessa C. M. Heterologous expression of a mammalian epithelial sodium channel in yeast. FEBS Lett. 2000;481:77–80. doi: 10.1016/s0014-5793(00)01977-3. [DOI] [PubMed] [Google Scholar]
- Han S., Liu Y., Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Hanwell D., Ishikawa T., Saleki R., Rotin D. Trafficking and cell surface stability of the epithelial Na+ channel expressed in epithelial Madin-Darby canine kidney cells. J. Biol. Chem. 2002;277:9772–9779. doi: 10.1074/jbc.M110904200. [DOI] [PubMed] [Google Scholar]
- Harris M., Garcia-Caballero A., Stutts M. J., Firsov D., Rossier B. C. Preferential assembly of epithelial sodium channel (ENaC) subunits in Xenopus oocytes: role of furin-mediated endogenous proteolysis. J. Biol. Chem. 2008;283:7455–7463. doi: 10.1074/jbc.M707399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert D. N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Hennessy F., Nicoll W. S., Zimmermann R., Cheetham M. E., Blatch G. L. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Cooper A. A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. Maturation of the epithelial Na+ channel involves proteolytic processing of the alpha- and gamma-subunits. J. Biol. Chem. 2003;278:37073–37082. doi: 10.1074/jbc.M307003200. [DOI] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Ismail N., Ng D. T. Have you HRD? Understanding ERAD is DOAble! Cell. 2006;126:237–239. doi: 10.1016/j.cell.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Jasti J., Furukawa H., Gonzales E. B., Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Jin Y., Zhuang M., Hendershot L. M. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–49. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan O. B., Mueller G. M., Qamar M. Z., Poland P. A., Ahner A., Rubenstein R. C., Hughey R. P., Brodsky J. L., Kleyman T. R. Small heat shock protein alphaA-crystallin regulates epithelial sodium channel expression. J. Biol. Chem. 2007;282:28149–28156. doi: 10.1074/jbc.M703409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Spear E. D., Ng D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Kimata Y., Ishiwata-Kimata Y., Ito T., Hirata A., Suzuki T., Oikawa D., Takeuchi M., Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler M. R., Dirks S., Haas I. G. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J., Gilstring C. F., Ljungdahl P. O. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J. Cell Biol. 2007;176:617–628. doi: 10.1083/jcb.200612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Quilty J., Popov M., Reithmeier R. A. Processing of N-linked oligosaccharide depends on its location in the anion exchanger, AE1, membrane glycoprotein. Biochem. J. 2000;349:51–57. doi: 10.1042/0264-6021:3490051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M., Grubb B. R., Harkema J. R., O'Neal W. K., Boucher R. C. Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat. Med. 2004;10:487–493. doi: 10.1038/nm1028. [DOI] [PubMed] [Google Scholar]
- Masilamani S., Kim G. H., Mitchell C., Wade J. B., Knepper M. A. Aldosterone-mediated regulation of ENaC alpha, beta, and gamma subunit proteins in rat kidney. J. Clin Invest. 1999;104:R19–23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Meerovitch K., Wing S., Goltzman D. Proparathyroid hormone-related protein is associated with the chaperone protein BiP and undergoes proteasome-mediated degradation. J. Biol. Chem. 1998;273:21025–21030. doi: 10.1074/jbc.273.33.21025. [DOI] [PubMed] [Google Scholar]
- Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mohan S., Bruns J. R., Weixel K. M., Edinger R. S., Bruns J. B., Kleyman T. R., Johnson J. P., Weisz O. A. Differential current decay profiles of epithelial sodium channel subunit combinations in polarized renal epithelial cells. J. Biol. Chem. 2004;279:32071–32078. doi: 10.1074/jbc.M405091200. [DOI] [PubMed] [Google Scholar]
- Molinari M., Galli C., Piccaluga V., Pieren M., Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J. Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M. J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mueller G. M., Kashlan O. B., Bruns J. B., Maarouf A. B., Aridor M., Kleyman T. R., Hughey R. P. Epithelial sodium channel exit from the endoplasmic reticulum is regulated by a signal within the carboxyl cytoplasmic domain of the alpha subunit. J. Biol. Chem. 2007;282:33475–33483. doi: 10.1074/jbc.M707339200. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L. M. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S., Kung L., Dorrington M., Lee M. C., Miller E. A. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol. Biol. Cell. 2007;18:3398–3413. doi: 10.1091/mbc.E07-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R. K., Bohmler S., Bordallo J., Sommer T., Wolf D. H. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Qiu X. B., Shao Y. M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T., Kreft S. G., Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard S., Lingueglia E., Voilley N., Lazdunski M., Barbry P. Biochemical analysis of the membrane topology of the amiloride-sensitive Na+ channel. J. Biol. Chem. 1994;269:12981–12986. [PubMed] [Google Scholar]
- Rossier B. C. The epithelial sodium channel (ENaC): new insights into ENaC gating. Pfluegers Arch. 2003;446:314–316. doi: 10.1007/s00424-003-1056-5. [DOI] [PubMed] [Google Scholar]
- Rubenstein R. C., Zeitlin P. L. Sodium 4-phenylbutyrate downregulates Hsc 70, implications for intracellular trafficking of DeltaF508-CFTR. Am J. Physiol. Cell Physiol. 2000;278:C259–C267. doi: 10.1152/ajpcell.2000.278.2.C259. [DOI] [PubMed] [Google Scholar]
- Rudiger S., Germeroth L., Schneider-Mergener J., Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S., Schneider-Mergener J., Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato B. K., Schulz D., Do P. H., Hampton R. Y. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol. Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G., Harris S., Risse B., Lill R., Silver P. A. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Maintz M., Kehle T., Herzog V. In vivo iodination of a misfolded proinsulin reveals co-localized signals for Bip binding and for degradation in the ER. EMBO J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Hendershot L. M. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Meunier L., Hendershot L. M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S., Schlenstedt G., Silver P. A., Gilmore R. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek M. H., Hendershot L. M., Haas I. G. The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc. Natl. Acad. Sci. USA. 1998;95:1574–1578. doi: 10.1073/pnas.95.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder P. M. Minireview: regulation of epithelial Na+ channel trafficking. Endocrinology. 2005;146:5079–5085. doi: 10.1210/en.2005-0894. [DOI] [PubMed] [Google Scholar]
- Snyder P. M., Bucher D. B., Olson D. R. Gating induces a conformational change in the outer vestibule of ENaC. J. Gen. Physiol. 2000;116:781–790. doi: 10.1085/jgp.116.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder P. M., McDonald F. J., Stokes J. B., Welsh M. J. Membrane topology of the amiloride-sensitive epithelial sodium channel. J. Biol. Chem. 1994;269:24379–24383. [PubMed] [Google Scholar]
- Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R., Bielmann R., Nita-Lazar M., Burda P., Jakob C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Valentijn J. A., Fyfe G. K., Canessa C. M. Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J. Biol. Chem. 1998;273:30344–30351. doi: 10.1074/jbc.273.46.30344. [DOI] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Weisz O. A., Wang J. M., Edinger R. S., Johnson J. P. Non-coordinate regulation of endogenous epithelial sodium channel (ENaC) subunit expression at the apical membrane of A6 cells in response to various transporting conditions. J. Biol. Chem. 2000;275:39886–39893. doi: 10.1074/jbc.M003822200. [DOI] [PubMed] [Google Scholar]
- Yan F. F., Lin C. W., Cartier E. A., Shyng S. L. Role of ubiquitin-proteasome degradation pathway in biogenesis efficiency of β-cell ATP-sensitive potassium channels. Am J. Physiol. Cell Physiol. 2005;289:C1351–C1359. doi: 10.1152/ajpcell.00240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. Inaugural Article: recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Haslam D. B. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect Immun. 2005;73:2524–2532. doi: 10.1128/IAI.73.4.2524-2532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Haslam R. H., Haslam D. B. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., Brodsky J. L. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.