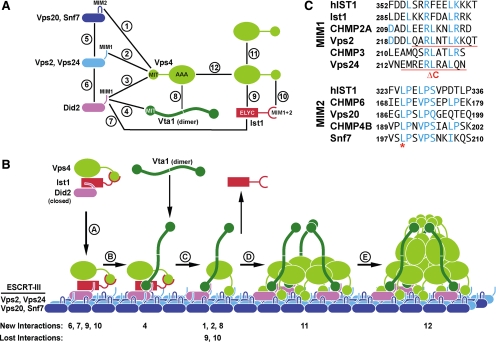
A complex network of interactions mediates the recruitment of Vps4 to ESCRT-III and its subsequent assembly, two key steps in the ESCRT-dependent vesicle formation at the endosome. A model is presented depicting the order of events that lead to active, ESCRT-III–associated Vps4.
Abstract
Vps4 is a key enzyme that functions in endosomal protein trafficking, cytokinesis, and retroviral budding. Vps4 activity is regulated by its recruitment from the cytoplasm to ESCRT-III, where the protein oligomerizes into an active ATPase. The recruitment and oligomerization steps are mediated by a complex network of at least 12 distinct interactions between Vps4, ESCRT-III, Ist1, Vta1, and Did2. The order of events leading to active, ESCRT-III–associated Vps4 is poorly understood. In this study we present a systematic in vivo analysis of the Vps4 interaction network. The data demonstrated a high degree of redundancy in the network. Although no single interaction was found to be essential for the localization or activity of Vps4, certain interactions proved more important than others. The most significant among these were the binding of Vps4 to Vta1 and to the ESCRT-III subunits Vps2 and Snf7. In our model we propose the formation of a recruitment complex in the cytoplasm that is composed of Did2-Ist1-Vps4, which upon binding to ESCRT-III recruits Vta1. Vta1 in turn is predicted to cause a rearrangement of the Vps4 interactions that initiates the assembly of the active Vps4 oligomer.
INTRODUCTION
Plasma membrane proteins are continuously endocytosed and either sorted into the multivesicular body (MVB) pathway for eventual degradation in the lysosome (vacuole in yeast) or recycled to the plasma membrane. Thus, modulation of surface protein trafficking is an important regulatory element for numerous cellular responses, such as growth factor receptor function, nutrient uptake, cell–cell communication and the immune response (for review, see Piper and Katzmann, 2007; Davies et al., 2009; Raiborg and Stenmark, 2009; Saksena and Emr, 2009). Ubiquitination of endosomal cargo proteins initiates their sorting into the MVB pathway where they are sequestered into vesicles formed when the outer/limiting endosomal membrane invaginates into the lumen of the compartment, giving the structure a multivesicular appearance. MVBs then fuse with lysosomes, delivering the vesicles into the lumen of the hydrolytic compartment for degradation. The proper recognition and sorting of ubiquitinated cargoes requires the coordinated interaction of the Vps4 ATPase and the ESCRT (endosomal-sorting complex required for transport) protein complexes that work in sequence to mediate MVB protein sorting and vesicle formation events (for review, see Babst, 2005; Hurley and Emr, 2006; Williams and Urbe, 2007). Current models suggest that an initial cargo-sorting event occurs when the ubiquitinated cargo is recognized by the ESCRT-0 and ESCRT-I complexes on the cytoplasmic side of the MVB membrane. ESCRT-I subsequently activates ESCRT-II, which, in turn, initiates the formation of ESCRT-III. This latter step is thought to concentrate cargo and recruit additional factors, including Vps4. Vps4 is an AAA (ATPase associated with various cellular activities)-type ATPase that releases ESCRT-III from the MVB membrane for additional sorting events, which is the final discernable step in the MVB-sorting process. In the absence of Vps4 function, the ESCRT machinery accumulates on the endosome, and vesicle formation is inhibited.
The formation of MVB vesicles requires membrane deformation and fusion steps that utilize a reversed topology compared with other vesicle formation events in the cell (e.g., clathrin- and COP-mediated vesicle formation). Interestingly, the ESCRT machinery has been implicated in two other membrane fusion events with reverse topology. Retroviruses such as HIV-1 form new virus particles at the plasma membrane by an ESCRT-dependent budding event, and the final step in cytokinesis requires the ESCRT-dependent abscission of the plasma membrane in order to form two separate cells (Garrus et al., 2001; VerPlank et al., 2001; Spitzer et al., 2006; Carlton and Martin-Serrano, 2007; Morita et al., 2007; McDonald and Martin-Serrano, 2009).
The assembly of ESCRT-III on endosomal membranes has been suggested to both concentrate cargo and deform the membrane, two essential steps in formation of MVB vesicles (Babst et al., 2002; Hanson et al., 2008; Wollert et al., 2009). Yeast ESCRT-III is composed of four subunits (Vps2, Vps20, Vps24, Snf7), each of which has at least one homologue in mammalian cells. These four subunits are predicted to have similar three-dimensional structures (Muziol et al., 2006). In the cytoplasm the ESCRT-III subunits are in a “closed” inactive conformation that inhibits complex formation. On the endosomal membrane, however, the ESCRT-III subunits seem to switch to an “open” conformation that promotes the formation of ESCRT-III and allows interactions with other ESCRT-III–associated factors (Shim et al., 2007; Lata et al., 2008; Bajorek et al., 2009b). Within ESCRT-III the four subunits form two functionally distinct subcomplexes (Babst et al., 2002). The Vps20–Snf7 subcomplex interacts with the endosomal membrane and with ESCRT-II, the complex that initiates ESCRT-III formation. The second subcomplex, Vps2–Vps24, is recruited by Vps20–Snf7 and thus functions downstream of the Vps20–Snf7 subcomplex. Although recent studies provided new insights into the arrangement of the subunits within ESCRT-III (Saksena et al., 2009), the overall structure of ESCRT-III remains to be determined.
Vps4 belongs to the large protein family of AAA-type ATPases (for review, see Lupas and Martin, 2002). These proteins function as mechano-enzymes that, in most studied cases, use the energy of ATP hydrolysis to induce conformational changes in the bound substrate. Vps4 is composed of an N-terminal substrate-binding domain, called the MIT (microtubule interacting and transport) domain, one central AAA domain (hallmark of type-1 AAA ATPases), and a C-terminal region that is involved in Vps4 dimerization (Babst et al., 1998; Scott et al., 2005a; Gonciarz et al., 2008; Vajjhala et al., 2008). Vps4 uses energy from ATP hydrolysis to disassemble ESCRT-III, thereby recycling the ESCRT-III subunits for additional rounds of MVB cargo sorting. The disassembly reaction is initiated by the recruitment of Vps4 monomers or dimers from the cytoplasm to ESCRT-III, where they assemble into the active oligomeric ATPase (Babst et al., 1998). Because of its dynamic nature the structural analysis of the Vps4 oligomer has been problematic and controversial. However recent studies indicate that the Vps4 oligomer is composed of 12 subunits that assemble into two hexameric rings in a tail-to-tail (antiparallel) orientation (Yu et al., 2008; Landsberg et al., 2009). Both the recruitment of Vps4 and the consequent disassembly reaction require the interaction of the Vps4 MIT domain with the ESCRT-III substrate. This interaction is mediated by two distinct motifs, termed MIMs (MIT-interacting motifs), found in the ESCRT-III subunits. MIM1 motifs are located in the very C-terminus of subunits Vps2 and Vps24, whereas MIM2 motifs are found in the C-terminal regions of the Vps20 and Snf7 subunits (Obita et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008; Shim et al., 2008). These two motifs bind to distinct surfaces of the MIT domain, allowing both types of interactions to occur simultaneously. Phenotypic analyses have indicated that both types of MIM motifs are important for proper Vps4 activity, but it remains unknown if all four potential yeast ESCRT-III MIM sites are functional.
Both recruitment and assembly of Vps4 are aided by additional factors that seem to ensure proper localization and timing of the disassembly reaction. Ist1 and Did2 have been implicated in the recruitment of Vps4 to ESCRT-III, whereas Vta1 has been shown to support the assembly and ATPase activity of Vps4 (Yeo et al., 2003; Shiflett et al., 2004; Azmi et al., 2006; Lottridge et al., 2006; Nickerson et al., 2006; Azmi et al., 2008; Dimaano et al., 2008; Rue et al., 2008). A fourth factor, Vps60, seems to function together with Vta1 at late stage of Vps4 activation; however, its precise role remains unknown (Ward et al., 2005; Azmi et al., 2008; Rue et al., 2008; Shim et al., 2008). Together, Vps4, ESCRT-III, Did2, Ist1, and Vta1 form a complex network of interactions that leads to the formation of an active ATPase complex and the disassembly of ESCRT-III. Although much is known about each of these interactions individually, the temporal arrangement of the interactions and how they work together to achieve the disassembly reaction is poorly understood. We present a detailed in vivo analysis of the interactions known to be involved in the recruitment and assembly steps of Vps4.
MATERIALS AND METHODS
Antibodies
The anti-HA (hemagglutinin) mAb used for immunoprecipitations and Western blotting was purchased from Covance (Princeton, NJ). The antisera against Vps4, Snf7, and Vps24 were previously described (Babst et al., 1998). The antiserum against Vps20 was a gift from Scott D. Emr (Cornell University, Ithaca, NY).
Strains and Media
Saccharomyces cerevisiae strains used in this work are listed in Table 1. To maintain plasmids, yeast strains were grown in corresponding complete synthetic dropout medium (Sherman et al., 1979). Wild-type, integrated, and knockout strains were grown in rich YPD medium (yeast extract-peptone-dextrose). Yeast gene knockouts were constructed as previously described (Baudin et al., 1993).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptive name | Genotype or description | Reference or source |
|---|---|---|---|
| Straina | |||
| SEY6210 | WT | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| BHY10 | WT CPY-I | SEY6210, CPY-INVERTASE::LEU2 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Horazdovsky et al. (1994) |
| MBY2 | vps4Δ CPY-I | BHY10, VPS4::TRP1 | Babst et al. (1997) |
| MBY3 | vps4Δ | SEY6210, VPS4::TRP1 | Babst et al. (1997) |
| MBY16 | vps4Δ vps36Δ | SEY62010.1, VPS4::TRP1, VPS36::HIS3 | Babst et al. (2002) |
| MBY28 | vps2Δ | SEY6210, VPS2::HIS3 | Babst et al. (2002) |
| BWY102 | vps24Δ | SEY6210, VPS24::HIS3 | Babst et al. (2002) |
| MBY37 | vps4Δ vps20Δ | MBY3, VPS20::HIS3 | Babst et al. (2002) |
| JPY50 | vps4Δ vta1Δ | MBY4, VTA1::HIS3 | Azmi et al. (2006) |
| MCY3 | vps4Δ ist1Δ | MBY3, IST1::HIS3 | Dimaano et al. (2008) |
| EEY1-3 | vps20Δ CPY-I | BHY10, VPS20::HIS3 | This study |
| EEY2-1 | vps20Δ | 6210, VPS20::HIS3 | Babst et al. (2002) |
| EEY8 | snf7Δ CPY-I | BHY10, SNF7::HIS3 | This study |
| EEY9 | snf7Δ | SEY6210, SNF7::HIS3 | Babst et al. (2002) |
| EEY12 | vps4Δ snf7Δ | MBY3, SNF7::HIS3 | Babst et al. (2002) |
| EEY26-1 | vps4Δ did2Δ | MBY3, DID2::HIS3 | Dimaano et al. (2008) |
| ASY4 | vps4Δ vps2Δ vps24 | 6210.1, VPS4::TRP1, VPS2::HIS3, VPS24::HIS3 | This study |
| ASY5 | vps2Δ vps24Δ | SEY6210, VPS24::HIS3, VPS2::HIS3 | This study |
| ASY8 | vps2Δ CPY-I | BHY10, VPS2::G418 | This study |
| ASY9 | vps24Δ CPY-I | BHY10, VPS24::G418 | This study |
| ASY12 | vps2Δ vps24Δ CPY-I | ASY8, VPS24::HIS3 | This study |
| ASY16 | vps4Δ vps20Δ snf7Δ | MBY37, SNF7::G418 | This study |
| ASY19 | snf7Δ vps20Δ CPY-I | EEY1-3, SNF7::G418 | This study |
| ASY20 | vps4Δ vps20Δ snf7Δ ist1Δ | ASY16, IST1::URA3 | This study |
| E. coli: XL1-blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(tetr)] | Stratagene(La Jolla, CA) | |
| Plasmids | |||
| pAS22 | vps2(ΔC)-HA | URA3 (pRS416) vps2(ΔC)-HA | This study |
| pAS23 | vps24(ΔC)-HA | URA3 (pRS416) vps24(ΔC)-HA | This study |
| pAS28 | vps2(ΔC)-HA | LEU2 (pRS415) vps2(ΔC)-HA | This study |
| pAS42 | vps2(ΔC)-HA, vps24(ΔC)-HA | TRP1 (pRS414) vps2(ΔC)-HA, vps24(ΔC)-HA | This study |
| pAS44 | MIT-GFP | URA3 (pRS426) vps4(MIT)-GFP | This study |
| pAS47 | vps2(ΔC)-HA, vps24(ΔC)-HA | URA3 (pRS416) vps2(ΔC)-HA, vps24(ΔC)-HA | This study |
| pAS51 | GFP-vps4(ΔMIT(E233Q, S377A)) | URA3 (pRS416) GFP-vps4(ΔMIT(E233Q, S377A)) | This study |
| pAS58 | vps2(ΔC)-HA, vps24(ΔC)-HA | LEU2 (pRS415) vps2(ΔC)-HA, vps24(ΔC)-HA | This study |
| pAS59 | MIT(L64D)-GFP | URA3 (pRS426) vps4(MIT (L64D))-GFP | This study |
| pAS62 | vps4 (ΔMIT(E233Q)) | URA3 (pRS416) vps4(ΔMIT(E233Q)) | This study |
| pAS72 | snf7(L199D) | LEU2 (pRS415) snf(L199D) | This study |
| pAS74 | vps20(L188D) | LEU2 (pRS415) vps20(L188D) | This study |
| pAS76 | vps20(L188D)-HA | URA3 (pRS416) vps20(L188D)-HA | This study |
| pAS79 | vps4(S377A) | URA3 (pRS416) vps4(S377A) | This study |
| pMB4 | VPS4 | HIS3 (pRS413) VPS4 | Babst et al. (1997) |
| pMB66 | vps4(E233Q) | HIS3 (pRS413) vps4(E233Q) | Babst et al. (1998) |
| pMB168 | VPS20-HA | URA3 (pRS416) VPS20-HA | Babst et al. (2002) |
| pMB341 | GFP-vps4(ΔMIT(E233Q)) | URA3 (pRS416) P(CPY)-GFP-vps4(ΔMIT(E233Q)) | This study |
| pMB343 | vps4(E233Q)–GFP | URA3 (pRS416) vps4(E233Q)-HA-GFP | This study |
| pMB370 | vps4(L64D, E233Q)-GFP | URA3 (pRS416) vps4(L64D, E233Q)-HA-GFP | This study |
| pMB380 | vps4(I18D, E233Q)-GFP | URA3 (pRS416) vps4(I18D, E233Q)-HA-GFP | This study |
| pMB393 | snf7(L199D), vps20(L188D) | URA3 (pRS416) snf7(L199D), vps20(L188D) | This study |
| pMB394 | snf7(L199D), vps20(L188D) | LEU2 (pRS415) snf7(L199D), vps20(L188D) | This study |
| pPN3 | VPS20 | LEU2 (pRS415) VPS20 | This study |
| pVPS4(I18D) | vps4(I18D) | URA3 (pRS416) vps4(I18D) | This study |
| pVPS4(L64D) | vps4(L64D) | URA3 (pRS416) vps4(L64D) | This study |
| pVPS4(I18D, E233Q) | vps4(I18D, E233Q) | URA3 (pRS416) vps4(I18D, E233Q) | This study |
| pVPS4(L64D, E233Q) | vps4(L64D, E233Q) | URA3 (pRS416) vps4(L64D, E233Q) | This study |
| pGO45 | GFP-CPS | URA3(pRS426) GFP-CPS1 | Odorizzi et al. (1998) |
| pMC48 | MIT-GFP | URA3(pRS416) P(CPS1)-vps4(MIT)-GFP | This study |
| pMC50 | MIT(I18D)-GFP | URA3 (pRS416) P(CPS1)-vps4(MIT(I18D))-GFP | This study |
| pAH31 | GST-DID2 | (pGEX-KG) GST-DID2 | This study |
| pAH32 | GST-CT(DID2) | (pGEX-KG) GST-DID2(113-204) | This study |
| pMB411 | GST-vps20(C) | (pGEX-KG) GST-vps20(101-221) | This study |
| pMB412 | GST-snf7(C, L199D) | (pGEX-KG) GST-snf7(101-240)(L199D) | This study |
| pMB413 | GST-snf7(C) | (pGEX-KG) GST-snf7(101-240) | This study |
| pMB414 | GST-vps20(C, L188D) | (pGEX-KG) GST-snf7(101-221)(L188D) | This study |
| pAS85 | snf7(L199D), vps20(L188D), MIT-GFP | LEU2 (pRS415) snf7(L199D), vps20(L188D), P(CPS1)-MIT-GFP | This study |
a All strains are Saccharomyces cerevisiae are except the one marked E. coli.
DNA Manipulations
Plasmids used in this study are listed in Table 1. All plasmids were constructed using standard cloning techniques. Plasmids obtained by PCR-based cloning techniques were confirmed by DNA sequencing. Point mutations were introduced using Stratagene QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA). The pRS4XX shuttle vectors used in this study have been described previously (Christianson et al., 1992). pEGFP-C1 was from Clontech Laboratories (Palo Alto, CA). MIT-green fluorescent protein (GFP) was constructed by fusing EGFP to the Eco47III site of VPS4. GFP-VPS4ΔMIT-type constructs were obtained by fusing a fragment containing the PRC1 promoter and GFP of pGO36 into the Eco47III site of a VPS4-containing plasmid.
Procedures
Fluorescence microscopy was performed on a deconvolution microscope (DeltaVision, Applied Precision, Issaquah, WA). The distribution of MIT-GFP was quantified from deconvoluted Z-stack projections of cells using an open-source GIMP 2.6.6 program (www.gimp.org). To achieve a more uniform level of protein expression in cells MIT-GFP was expressed from a CPS1 promoter (pMC48 and pMC50). The percent signal on endosomes was calculated by dividing endosomal fluorescence intensity by total cellular fluorescence intensity. Using an unpaired t test, p values were calculated using Instat software. Subcellular fractionation experiments were performed as described previously (Dimaano et al., 2008), except for subcellular fractionation experiments localizing Vps4, where spheroplasted cells were lysed by osmotic stress in Pop buffer (100 mM KCl, 50 mM KAc, 20 mM PIPES, pH 6.8, 5 mM MgAc2, 100 mM sorbitol) containing 0.1 mM AEBSF and Complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN). When localizing ESCRT-III components, spheroplasts were lysed by douncing with 15–20 strokes in a glass homogenizer in PBS containing Complete protease inhibitor cocktail. Immunoprecipitation experiments were performed as described previously (Babst et al., 2002). For in vitro binding experiments glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified by affinity purification using standard methods on glutathione-Sepharose 4 Fast Flow resin (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The purification of Vps4E233Q, Ist1, and Vta1 were previously described (Babst et al., 1998; Azmi et al., 2006; Dimaano et al., 2008). To test the interaction between proteins in vitro, 40 μg of purified GST fusion protein was prebound to ∼15-μl bead volume of glutathione-Sepharose 4 Fast Flow resin. Equimolar amounts of purified test proteins were then added to the resin in GST pulldown buffer (100 mM KAc, 5 mM MgAc2, 20 mM HEPES, pH 7.4) to a final volume of 150 μl and incubated for 10 min at room temperature with gentle mixing; 1 mM ATP or ADP was included as indicated. An unbound sample was taken, and the bound protein was washed extensively in GST pulldown buffer. The bound protein was eluted by boiling for 5 min in SDS-PAGE sample buffer (2% SDS, 0.1 M Tris, pH 6.8, 10% glycerol, 0.01% bromophenol blue, 5% ß-mercaptoethanol), and 10 μl of each sample was separated by SDS-PAGE and stained by Coomassie Brilliant Blue. The liquid overlay assay was performed as previously described (Darsow et al., 2000).
RESULTS
The recruitment of Vps4 subunits from the cytoplasm to ESCRT-III and the consequent oligomerization of Vps4 into the active dodecameric state are not well understood. The study of this process is complicated by the number of Vps4 interactions that have been reported, some of which overlap and compete with each other (Figure 1A). Furthermore, many Vps4 interaction studies have been performed in vitro in the presence of only one or a few of the ESCRT factors, which raises questions of the relevance of these observations for the in vivo situation. Therefore we dissected the Vps4 interactions in vivo by expressing different truncated or mutated forms of Vps4 in a variety of ESCRT deletion strains (see Table 2). These data resulted in a model for the recruitment and assembly of Vps4 on MVBs (Figure 1B).
Figure 1.
Interactions between Vps4 and its substrate and regulators. (A) Vps4 interaction network based on previous studies (Scott et al., 2005b; Azmi et al., 2008; Kieffer et al., 2008; Bajorek et al., 2009a; Xiao et al., 2009). (B) Model for the recruitment and assembly of Vps4. The numbers indicating New Interactions or Lost Interactions refer to the numbers in A. (C) Alignments of the putative MIM1 and MIM2 motifs of yeast and mammalian ESCRT-III subunits (yeast Ist1 does not contain an obvious MIM2 consensus sequence). Mutations used in this study are marked in red.
Table 2.
Mutations used in this study
| Name | Mutation | Affected interactions (see Figure 1A) |
|---|---|---|
| Vps4I18D | Ile(18) to Asp | 1, 10 |
| Vps4L64D | Leu(64) to Asp | 2, 10 |
| Vps4ΔMIT | aa 2-87 deleted | 1, 2, 10 |
| Vps4S377A | Ser(377) to Ala | 8 |
| Vps4E233Q | Glu(233) to Gln | None (ATP locked) |
| Vps2ΔC | aa 222-end deleted | 2 |
| Vps24ΔC | aa 214-end deleted | 2 |
| Vps20L188D | Leu(188) to Asp | 1 |
| Snf7L199D | Leu(199) to Asp | 1 |
Analysis of the Vps4 MIT Interactions
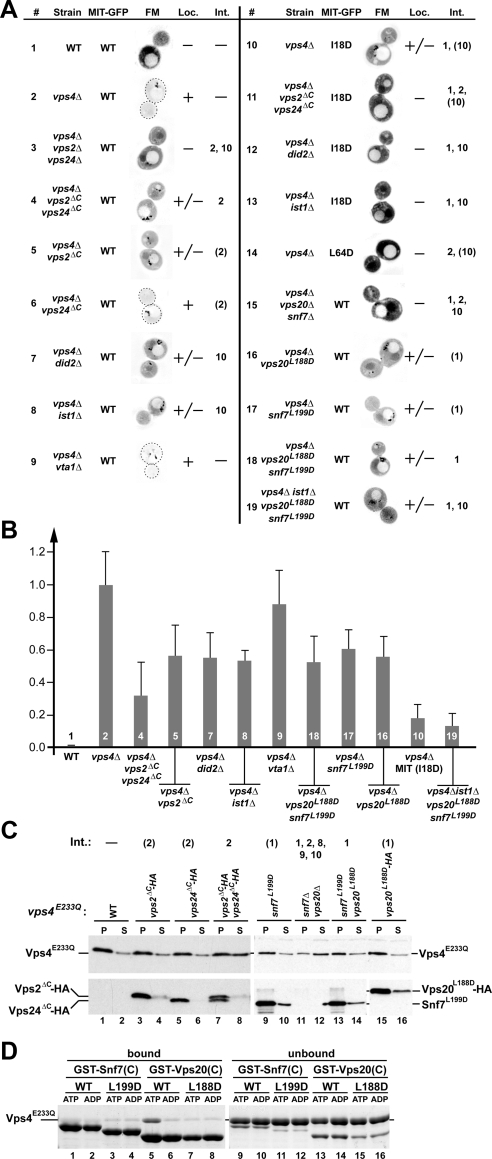
The MIT domain of Vps4 has been shown to interact with six different proteins of the ESCRT machinery: the four subunits of ESCRT-III (Vps2, Vps20, Vps24, Snf7) and two factors that are related to ESCRT-III subunits and have been implicated in the endosomal recruitment of Vps4, Did2, and Ist1 (Figure 1A; Shim et al., 2007; Azmi et al., 2008; Dimaano et al., 2008; Kieffer et al., 2008; Xiao et al., 2008). These six factors bind via two distinct binding motifs, MIM1 and MIM2, to two different surface areas of the MIT domain (Figure 1C). This arrangement allows the MIT domain to simultaneous bind to MIM1 and MIM2. To test which of the MIT interactions are involved in the recruitment of Vps4 to ESCRT-III, we expressed a MIT-GFP fusion construct in yeast strains deleted for VPS4 alone or in combination with other ESCRT mutations. These yeast cells were analyzed by fluorescence microscopy, and the resulting pictures were judged by the ratio of MIT-GFP signal localized either to the cytoplasm or to the aberrant endosomes formed in these mutant strains (class E compartments; Figure 2A). Enlarged examples of the microscopy pictures are shown in Supplemental Figure 1. Furthermore, for a subset of MIT-GFP localization experiments, the ratio of endosomal-to-cytoplasmic localization was quantified (Figure 2B).
Figure 2.
MIM1 and MIM2 interactions contribute to the recruitment of Vps4 to ESCRT-III. (A) Fluorescence microscopy analysis of the Vps4 MIT domain fused to GFP (MIT-GFP). The wild-type, MIM1 mutant (L64D), or MIM2 mutant (I18D) version of MIT-GFP was expressed in different yeast strains (see Table 1), and the extent of endosomal localization was determined (Loc.). For better visualization the fluorescence microscopy pictures were inverted and the intensity was adjusted to the individual brightness range (black is the brightest signal). Vps4 interactions affected by the different mutations are listed (Int., numbers are based on the interactions in Figure 1A). Numbers in parentheses indicate partially disrupted interactions. (B) Quantification of endosome-localized MIT-GFP relative to wild type (0.0) and vps4Δ (1.0). The data shown represent the results of at least 15 individually analyzed cells. Numbers refer to the experiment number in A. (C) Subcellular fractionation of different yeast strains expressing vps4E233Q into soluble, cytoplasmic fraction (S) and pelletable, membrane-associated fraction (P). Fractions were analyzed by Western blot using antibodies specific for Vps4 (top panels), Snf7 (bottom panel, lanes 9–14) and the HA-tag (bottom panels, lanes 1–8 and 15–16). Vps4 interactions affected by the different mutations are listed (Int., numbers are based on the interactions in Figure 1A). Numbers in parentheses indicate partially disrupted interactions. (D) In vitro Vps4 interaction studies using wild-type and MIM2 mutant forms of GST-Vps20(C) (fusion of GST with C-terminal half of Vps20) or GST-Snf7(C) immobilized on GSH-Sepharose. Vps4E233Q was added in the presence of ATP or ADP to immobilized proteins; bound and unbound fractions were analyzed by SDS-PAGE and Coomassie staining.
Previous studies have shown that vps4Δ cells accumulate ESCRT-III and its associated proteins Did2 and Ist1 on endosomal membranes (Babst et al., 2002; Nickerson et al., 2006; Dimaano et al., 2008). Thus the majority of MIT-GFP localized to class E compartments in cells deleted for VPS4 and to cytoplasm in wild-type cells (Figure 2, A and B, 1 and 2). Additional deletions of DID2 or IST1 resulted in a partial redistribution of MIT-GFP to the cytoplasm (Figure 2, A and B, 7 [p < 0.0001] and 8 [p < 0.0001]). This result is consistent with previously published models in which Did2 and Ist1 function together in the recruitment of Vps4 (Dimaano et al., 2008; Rue et al., 2008). In contrast, deletion of VTA1 did not interfere with recruitment of the MIT domain (Figure 2, A and B, 9) supporting a later role for this Vps4-interacting protein.
The ESCRT-III subunits Vps2 and Vps24 both contain C-terminal MIM1 interaction sites (Obita et al., 2007; Stuchell-Brereton et al., 2007). To study the effect of loss of MIM1, 3′ truncations of VPS2 and VPS24 were constructed that lacked the codons for the last 11 amino acids (vps2ΔC, vps24ΔC, Figure 1C, Table 2). These mutations did not affect stability or MVB recruitment of Vps2 and Vps24 (Figure 2C, lanes 3–6). A partial redistribution of MIT-GFP to the cytoplasm was observed in vps4Δ strains that lacked the Vps2 MIM1 motif (Figure 2, A and B, 5; p < 0.0001). The loss of Vps24 MIM1 had no effect on MIT-GFP localization (Figure 2A, 6). However, when combined with vps2ΔC the deletion of Vps24 MIM1 showed a small but significant increase in MIT-GFP relocalization (Figure 2, A and B, 4; p < 0.0048). These data indicated that, although both MIM1 sites are functional, the Vps2 MIM1 site is more important for Vps4 recruitment than the Vps24 MIM1 motif.
Deletion of both VPS2 and VPS24 abolished MIT-GFP recruitment (Figure 2A, 3), which can be explained by the fact that this strain not only lacks the ESCRT-III MIM1 sites but in addition does not properly localize the other two MIM1-containing proteins Did2 and Ist1 (Nickerson et al., 2006; Dimaano et al., 2008). Similarly, a mutation in the MIT domain shown in mammalian Vps4 to block the MIT–MIM1 interaction (MITL64D; Figure 1C; Stuchell-Brereton et al., 2007) did not localize to class E compartments of vps4Δ cells, supporting the idea that loss of all MIM1 sites both in ESCRT-III as well as in the recruitment factors Did2 and Ist1 abolishes recruitment of MIT-GFP (Figure 2A, 14).
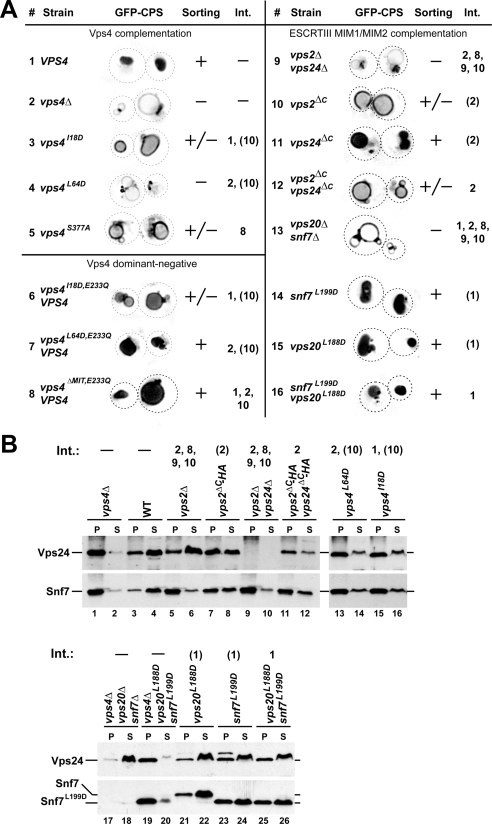
Figure 3.
Phenotypic analysis of mutations affecting Vps4 interactions. (A) Fluorescence microscopy analysis of yeast strains expressing GFP-CPS. The efficiency of GFP-CPS sorting into the lumen of the vacuole is indicated (+, +/−, −). Vps4 interactions affected by the different mutations are listed. (B) Subcellular fractionation of yeast strains into soluble (S) and membrane-associated pellet fractions (P). The samples were analyzed by Western blot using antibodies specific for Vps24 and Snf7. Vps4 interactions affected by the different mutations are listed. (A and B) Int., numbers are based on the interactions in Figure 1A, and numbers in parentheses indicate partially disrupted interactions.
To test the effect of loss of Vps2 and Vps24 MIM1 motifs on the recruitment of full-length Vps4, we fractionated cell extracts by centrifugation, separating the soluble, cytoplasmic pool of Vps4 (S) from the endosomal fraction found in the pellet (P). As previously observed, in wild-type cells the ATP-locked mutant Vps4E233Q accumulated in the pellet fraction, indicating efficient recruitment of Vps4E233Q to ESCRT-III (Figure 2C, lanes 1 and 2; Babst et al., 1998). Similarly, mutant strains that lacked the MIM1 motif of either Vps2 or Vps24 efficiently recruited Vps4E233Q to membranes (Figure 2C, lanes 3–6). The lack of both MIM1 sites caused a partial redistribution of Vps4E233Q to the soluble fraction, whereas Vps2ΔC or Vps24ΔC remained in the membrane-bound pellet fraction (Figure 2C, lanes 7 and 8). These results supported the notion that the ESCRT-III MIM1 motifs are functionally redundant. However, the recruitment defects observed by cell fractionation were less severe than the MIT-GFP localization phenotypes, which can be explained by the presence of Vps4–Vps4 and Vps4–Vta1 interactions that stabilize ESCRT-III–associated Vps4 oligomers (see below).
We tested the importance of the MIM2 interaction motifs in Vps4 recruitment by mutating a conserved leucine residue of the Vps20 and Snf7 MIM2 sites (L188D in Vps20 and L199D in Snf7) that has been previously shown to be important for the MIM2–MIT interaction of the mammalian ESCRT system (Figure 1C; Kieffer et al., 2008). The effect of these mutations on the interaction with yeast Vps4 was first analyzed in vitro using immobilized GST-fusion proteins containing the C-terminal half of either Snf7 or Vps20. Sepharose beads presenting either wild-type or mutant versions of the fusion proteins were incubated with recombinant Vps4E233Q in the presence of either ATP or ADP. The resulting bound and unbound fractions were then analyzed by SDS-PAGE (Figure 2D). Wild-type, but not L188D Vps20 was able to bind to Vps4 in presence of ATP (lane 5), demonstrating that the mutation indeed inhibited the Vps20 MIM2–MIT interaction. In contrast, neither wild-type nor mutant Snf7 bound to Vps4 under the conditions tested. These results are consistent with previously published data from both yeast and mammalian systems that suggested a preferred interaction of Vps4 with Vps20 rather than Snf7 (Azmi et al., 2008; Kieffer et al., 2008).
When expressed in yeast, the mutant proteins Vps20L188D and Snf7L199D were stable and efficiently recruited to endosomal membranes, indicating that the mutations did not interfere with normal protein folding (Figure 2C, lanes 9–10 and 15–16). However, when analyzed by SDS-PAGE the L199D mutation in Snf7 resulted in an apparent size shift, possibly because of the additional negative charge (Figure 3B, lanes 19–22). Mutation of each of the MIM2 motifs resulted in partial loss of MIT-GFP recruitment to ESCRT-III (Figure 2, A and B, 16 [p < 0.0001] and 17 [p < 0.0001]). Similarly, the combination of both mutations impaired endosomal localization of MIT-GFP (Figure 2, A and B, 18; p < 0.0001). Together, these data indicated that both ESCRT-III MIM2 motifs are important for MIT recruitment even though the in vitro data suggested only a minor role of Snf7 in the interaction of ESCRT-III with Vps4 (Figure 2D).
To test the effect of Snf7 and Vps20 MIM2 mutations on the recruitment of full-length Vps4, we performed subcellular fractionation experiments. MIM2 mutations in either Snf7 or Vps20 or the combination of both did not significantly redistribute Vps4 into the cytoplasmic fraction (Figure 2C, lanes 9–10 and 13–16). This result suggested, as observed with MIT-GFP, that MIM1 interactions play a more important role in the recruitment of Vps4 than the MIM2 motifs. Deletion of both SNF7 and VPS20 resulted in a relocalization of Vps4E233Q to the cytoplasm, which is likely caused by the loss of endosomal ESCRT-III and its associated proteins in this strain (Figure 2C, lanes 11 and 12).
The function of the MIM2 interaction was further analyzed by mutating the MIM2-binding site in the MIT domain of Vps4. A previous study indentified the valine at position 13 of human VPS4A as a crucial amino acid for the interaction between MIM2 and the MIT domain (Kieffer et al., 2008). However, the corresponding mutation in yeast Vps4 (V14D) resulted in an unstable protein, suggesting that this mutation might interfere with protein folding (Supplemental Figure 2). Therefore we changed isoleucine at position 18 to aspartate, a mutation that based on published NMR analysis is predicted to interfere with the MIM2 interaction (Kieffer et al., 2008). The I18D mutation impaired Vps4 function without affecting in vivo protein stability (Figure 3, 3, and Table 3; Supplemental Figure 2). The mutation strongly inhibited the recruitment of MIT-GFP to class E compartments of vps4Δ cells (Figure 2, A and B, 10; p < 0.0001), which is a more dramatic MIT recruitment phenotype than observed in the double MIM2 mutant strain (vps4Δ vps20L188D snf7L199D, Figure 2A, 18). A likely explanation for this result is that the I18D MIT mutation not only affected the interaction with ESCRT-III but also the binding to the recruitment factor Ist1 (interactions 1 and 10, Figure 1A). Consistent with this idea we observed an increased recruitment defect when the vps20L188D snf7L199D mutations were combined with a deletion of IST1 (Figure 2, A and B, 19; p < 0.0001). However, deletion of IST1 caused a complete loss of MITI18D-GFP recruitment to the endosome, which is a more severe phenotype than observed in the vps20L188D snf7L199D ist1Δ strain (Figure 2A, 13 and 19). This result suggested either the existence of an additional unknown MIM2 motif or the possibility that the I18D mutation might affect the MIM1-interaction site of the MIT domain. Combining the I18D MIT mutation with strains lacking Did2 or the MIM1-binding sites in ESCRT-III resulted in loss of MIT-GFP recruitment (Figure 2A, 11 and 12), which suggested that MIM1 and MIM2 interactions act synergistically in the recruitment of Vps4.
Table 3.
CPY-invertase secretion
| Strain | Secretion (%) | Affected Vps4 interactions (Figure 1A) |
|---|---|---|
| Complementation | ||
| Wild type | 0 ± 0 | None |
| vps4Δ | 100 ± 8 | All |
| vps4I18D | 59 ± 8 | 1, 10 |
| vps4L64D | 73 ± 4 | 2, 10 |
| vps4S377A | 12 ± 0 | 8 |
| vps2ΔC | 30 ± 10 | 2 |
| vps24ΔC | 27 ± 6 | 2 |
| vps2ΔC, vps24ΔC | 48 ± 0 | 2 |
| snf7L199D | 3 ± 1 | 1 |
| vps20L188D | 3 ± 2 | 1 |
| snf7L199D, vps20L188D | −1 ± 1 | 1 |
| Dominant-negative | ||
| VPS4 | 0 ± 0 | None |
| vps4E233Q, VPS4 | 100 ± 3 | None |
| vps4I18D, E233Q, VPS4 | 68 ± 5 | 1, 10 |
| vps4L64D, E233Q, VPS4 | 10 ± 0 | 2, 10 |
| vps4ΔMIT, E233Q, VPS4 | −7 ± 0 | 1, 2, 10 |
The in vivo analysis of MIT domain localization to ESCRT-III has demonstrated a high level of redundancy in the Vps4 recruitment system. Not one of the tested MIT interactions has been found to be essential for recruitment. Only mutations that interfere with at least two of the known MIT interactions are able to disrupt binding of the MIT domain to ESCRT-III. Furthermore, the MIM1 and MIM2 interactions act cooperatively in the recruitment of the MIT domain, suggesting that both interactions occur simultaneously on ESCRT-III. In contrast to the microscopy analysis, subcellular fractionations have shown that in vps4Δ MIT-GFP localizes to the soluble, cytoplasmic fraction (data not shown). This result suggested that binding of the MIT domain to ESCRT-III is too weak to maintain the MIT–ESCRT-III interaction during the fractionation procedure, an observation that fits well with the micromolar affinities found in vitro for MIT–MIM interactions (Obita et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008).
Functional Redundancy among the ESCRT-III MIM Motifs
The MIT-GFP localization studies revealed redundancy in the function of the different MIT interaction motifs in Vps4 recruitment. To test if the ESCRT-III MIM sites are also functionally redundant in regard to MVB cargo sorting, we analyzed potential trafficking phenotypes in strains that lack either one or both of the ESCRT-III MIM1 or MIM2 motifs. Newly synthesized carboxypeptidase S (CPS) is a type I transmembrane protein that traffics via the MVB pathway to the vacuolar lumen where it functions as a vacuolar hydrolase (Odorizzi et al., 1998). Fluorescence microscopy of wild-type cells expressing GFP-tagged CPS (GFP-CPS) showed mainly staining of the vacuolar lumen (Figure 3A, 1). In contrast, ESCRT mutants lacked luminal staining of the vacuole but instead accumulated GFP-CPS in aberrant endosomes, the class E compartments, and the limiting membrane of the vacuole (Figure 3A, 2). Cells that expressed the MIM1 mutant form of VPS2, vps2ΔC, exhibited a partial GFP-CPS sorting phenotype, whereas the loss of the VPS24 MIM1 motif showed no obvious MVB trafficking defect (Figure 3A, 10 and 11). Loss of both MIM1 motifs resulted in a sorting defect similar than that of the vps2ΔC mutant strain, consistent with the MIT-GFP localization studies that indicated the Vps2 MIM1 motif plays a more important role in Vps4 recruitment (Figure 3A, 12).
The Vps4 L64D mutation caused a severe GFP-CPS sorting defect indicating that the loss of all MIM1 interactions blocked Vps4 function (Figure 3A, 4). In contrast, the MIM2-interaction mutant Vps4I18D only partially inhibited the activity of Vps4 (Figure 3A, 3), which is consistent with the observed partial loss of MITI18D-GFP recruitment to endosomes (Figure 2, A and B, 10). However, single and double mutants of the ESCRT-III MIM2 motifs resulted in normal GFP-CPS trafficking, further supporting the notion that mutants with weak MIT-localization defects show correspondingly weak (or no) MVB-trafficking phenotypes (Figure 3A, 14–16).
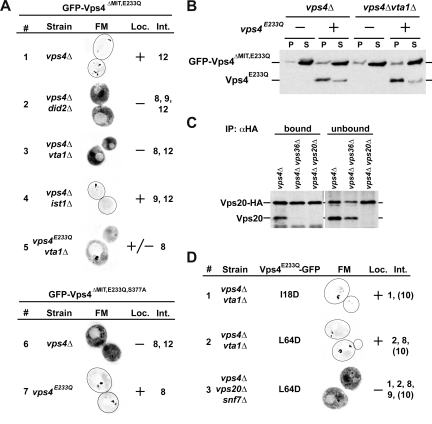
Figure 4.
Localization of mutant Vps4 proteins. (A) Localization of MIT-deleted Vps4 (GFP- Vps4ΔMIT,E233Q and GFP-Vps4ΔMIT,E233Q,S377A) in different yeast mutant strains (see Table 1) determined by fluorescence microscopy (FM). (A and D) The extent of observed endosomal localization is indicated (Loc.). Vps4 interactions affected by the different mutations are listed. Int., numbers are based on the interactions in Figure 1A. (B) Endosomal recruitment of MIT-deleted Vps4 in the presence or absence of full-length Vps4 protein determined by subcellular fractionation and Western blot analysis (S, soluble; P, pellet). (C) Immunoprecipitation of Vps20-HA from detergent-solubilized membrane fractions. The resulting bound and unbound samples were analyzed by Western blot using anti-Vps20 antiserum. (D) Fluorescence microscopy (FM) analysis of GFP-tagged full-length Vps4 protein in different mutant strains (Table 1). Numbers in parentheses indicate partially disrupted interactions.
The soluble fusion protein CPY-invertase is synthesized and translocated at the endoplasmic reticulum, transported by the sorting receptor Vps10 from the trans-Golgi to a MVB and finally delivered to the vacuole. ESCRT mutants impair the recycling of Vps10 back to the trans-Golgi, thereby limiting the transport function of this receptor. As a consequence, ESCRT mutants secrete a portion of newly synthesized CPY-invertase, a phenotype that can be detected by a colorimetric assay (Table 3; Paravicini et al., 1992).
The CPY-invertase assay revealed similar functional redundancies of the ESCRT-III MIM1 motifs as observed by the analysis of GFP-CPS trafficking, although there was no clear difference in phenotypic severity between the vps2ΔC and vps24ΔC mutants. Loss of the Vps2 or Vps24 MIM1 motif resulted in a modest secretion phenotype (vps2ΔC and vps24ΔC, ∼30% secretion relative to the secretion of vps4Δ), whereas loss of both motifs caused a more severe phenotype (∼50% secretion relative to vps4Δ). Mutation of the Vps4 MIM1 interaction site (Vps4L64D) further enhanced CPY-invertase secretion (∼70% secretion), consistent with the idea that this mutation interferes with all potential Vps4 MIM1 interactions, including those with Did2 and Ist1.
In contrast to the MIM1 mutants, the ESCRT-III MIM2 mutant strains did not secrete CPY-invertase (Table 3). However, mutating the Vps4 MIM2 interaction site (Vps4I18D) caused detectable secretion (∼60% secretion, Table 3), suggesting that loss of MIM2 interactions both on ESCRT-III and Ist1 together resulted in a synthetic phenotype (interactions 1 and 10, Figure 1A). Together, the phenotypic characterization of the different MIM mutants suggested that MIM1 interactions are more important for Vps4 function than the MIM2 interactions. However, we cannot exclude the possibility that the mutations in the MIM2 motifs retain some functionality.
The immediate consequence of the loss of Vps4 function is the accumulation of ESCRT-III on endosomes (Babst et al., 1997). Therefore, we analyzed the effect of the MIM mutations on the localization of ESCRT-III subunits by fractionating cell extracts into soluble and membrane-associated pellet. In wild-type cells the majority of Vps24 and Snf7 localized to the soluble cytoplasmic pool (S), whereas deletion of VPS4 resulted in the accumulation of both proteins in the pellet fraction (P; Figure 3B, lanes 1–4). Similarly, deleting VPS2 alone or in combination with VPS24 caused a shift of Snf7 pool to the pellet fraction, consistent with the published function of Vps2/Vps24 in Vps4 activity (Figure 3B, lanes 5–6 and 9–10; Babst et al., 2002). In contrast, the majority of Vps24 was found in the soluble fraction in vps2Δ cells, which can be explained by the observation that Vps2 and Vps24 required each other for proper ESCRT-III assembly (Figure 3B, lanes 5 and 6; Babst et al., 2002). Deletion of the VPS2 MIM1 motif resulted in partial accumulation of both Vps24 and Snf7 (vps2ΔC-HA; Figure 3B, lanes 7 and 8), consistent with the partial trafficking phenotype associated with this strain. The additional deletion of the VPS24 MIM1 motif further impaired but did not block the Vps4-dependent disassembly of ESCRT-III (cf. vps2ΔC-HA vps24ΔC-HA with vps4Δ in Figure 3B, lanes 1–2 and 11–12), suggesting that the remaining MIM2 sites were sufficient to maintain some Vps4 activity. Consistent with the observed redundancy among the MIM1/2 interactions we found that mutating either the MIM1 or the MIM2 interaction site in Vps4 only partially inhibited ESCRT-III disassembly (Figure 3B, lanes 13–16).
Loss of the Vps20–Snf7 subcomplex in vps4Δ caused the redistribution of Vps24 to the soluble, cytoplasmic fraction, an expected result based on previous publications (lanes 17 and 18, Figure 3B; Babst et al., 2002). Additional expression of vps20L188D and snf7L199D in this strain restored ESCRT-III accumulation on endosomal membranes, indicating that the mutations in MIM2 did not interfere with the assembly of this protein complex (lanes 19 and 20, Figure 3B). However the mutation of the Snf7 MIM2 motif partially impaired the recycling of ESCRT-III from membranes whereas the corresponding mutation in Vps20 did not interfere with the Vps4-dependent disassembly reaction (lanes 21–26, Figure 3B). These observations suggested that although both MIM2 motifs of ESCRT-III are involved in the binding of the Vps4 MIT domain only Snf7 MIM2 seems to play an important role in the ESCRT-III disassembly reaction.
In summary the fractionation experiments indicated that MIM1 and MIM2 motifs of ESCRT-III act together not only in the recruitment of Vps4 but also in the subsequent ESCRT-III disassembly reaction. Both type of MIT interactions are important for Vps4 activity and loss of MIM1 or MIM2 affect recycling of both ESCRT-III subcomplexes, Vps20/Snf7 and Vps2/Vps24.
Characterization of the Vps4 AAA Domain Interactions
We tested the role of the Vps4 AAA domain interactions on the recruitment of the ATPase to the endosome in vivo by analyzing the localization of an ATP-locked Vps4 protein in which the MIT domain had been exchanged with GFP (GFP-Vps4ΔMIT,E233Q). The only known binding partners of this protein are Vta1, Ist1, and Vps4 itself (interactions 8, 9, 11, and 12 in Figure 1A). In vps4Δ cells GFP-Vps4ΔMIT,E233Q localized mainly to class E compartments (Figure 4A, 1), indicating that the MIT domain is not essential for the localization of Vps4 to endosomes. Loss of the AAA-Ist1 interaction did not affect the recruitment of GFP-Vps4ΔMIT,E233Q to ESCRT-III (vps4Δist1Δ, Figure 4A, 4), which is consistent with previous reports that described a rather weak interaction (Dimaano et al., 2008). In contrast, cells that lacked Vta1 were not able to recruit GFP-Vps4ΔMIT,E233Q to ESCRT-III (Figure 4A, 3). Similarly, lack of Did2 abolished endosomal recruitment of GFP-Vps4ΔMIT,E233Q (Figure 4A, 2). Because Did2 has been shown to be important for endosomal localization of Vta1, the mislocalization of GFP-Vps4ΔMIT,E233Q in vps4Δ did2Δ is likely due to a lack of ESCRT-III–associated Vta1 in this strain (Azmi et al., 2008).
To further corroborate the role of Vta1 in GFP-Vps4ΔMIT,E233Q localization, the previously published mutation S377A was introduced into the beta-loop of Vps4, a mutation predicted to interfere with Vta1 binding (Scott et al., 2005b). When expressed in yeast, the Vps4S377A mutant protein was stable and present at normal levels, indicating that the mutation did not interfere with protein folding (Supplemental Figure 2). As expected, the mutation resulted in a partial GFP-CPS and a weak CPY-invertase trafficking phenotype (Figure 3A, 5, and Table 3), similar to the phenotype observed in cells deleted for VTA1 (Azmi et al., 2006; Saksena et al., 2009). When introduced into GFP-Vps4ΔMIT,E233Q, the resulting mutant construct GFP-Vps4ΔMIT,E233Q,S377A did not localize to endosomes in vps4Δ cells (Figure 4A, 6), which is consistent with the loss of Vta1 interaction. Together, the data supported the model that Vta1 localizes to ESCRT-III via Did2 and that the ESCRT-III–associated pool of Vta1 is sufficient to recruit MIT-deleted Vps4.
Figure 5.
Ist1 and Did2 form a stable complex. GST-tagged full-length Did2 (GST-Did2) or C-terminal half of Did2 [GST-CT(Did2)] was immobilized on GSH-Sepharose, and purified Ist1, Vta1, or Vps4E233Q protein was added. The resulting bound and unbound fractions were analyzed by SDS-PAGE and Coomassie staining.
Introducing full-length Vps4E233Q into vps4Δvta1Δ cells partially restored endosomal localization of GFP-Vps4ΔMIT,E233Q (vps4E233Q vta1Δ; Figure 4A, 5). Similarly the presence of Vps4E233Q suppressed the localization defect of Vps4ΔMIT,E233Q,S377A (Figure 4A, 7). These results indicated that Vps4-Vps4 interactions were sufficient to recruit the ATPase to ESCRT-III.
In contrast to the microscopy studies, subcellular fractionation experiments GFP-Vps4ΔMIT,E233Q localized almost exclusively to the soluble, cytoplasmic fraction in presence or absence of either Vps4E233Q or Vta1 (Figure 4B). This discrepancy is likely caused by the dissociation of GFP-Vps4ΔMIT,E233Q during the fractionation procedure, suggesting that the lack of the MIT domain weakened the interaction not only between Vps4 and ESCRT-III but also the interactions within the Vps4 oligomer.
In summary, the localization studies using MIT-deleted Vps4 suggested that Vps4 could be recruited to ESCRT-III either via binding to Vta1 or via interactions with other ESCRT-III–associated Vps4 subunits. However, loss of the MIT domain resulted in reduced stability of the ESCRT-III–associated Vps4 oligomer.
Vps4 Oligomer and ESCRT-III Make Multiple Contacts
Recently, a model for the ESCRT-III structure has been proposed that suggested a single linear polymer consisting of the ordered assembly of one subunit of Vps20, followed by ∼10 subunits of Snf7, followed by about three subunits of Vps2, and finished by about two Vps24 proteins (Teis et al., 2008; Saksena et al., 2009). It is difficult to model how this linear arrangement of ESCRT-III subunits would allow the simultaneous interaction of both MIM motifs with the Vps4 MIT domains, which based on our observations are critical for the Vps4-dependent ESCRT-III disassembly. Therefore, the predicted ESCRT-III composition was tested using immunoprecipitation experiments to determine if ESCRT-III indeed contains a single Vps20 subunit. For this purpose we expressed HA-tagged VPS20 (VPS20-HA) and untagged VPS20 in the same strain, immunoprecipitated Vps20-HA from detergent-solubilized membranes and analyzed the resulting samples by Western blot using anti-Vps20 antiserum. The result demonstrated that Vps20 coimmunoprecipitated with Vps20-HA from vps4Δ cells, a mutant strain that accumulates ESCRT-III on the endosomal membrane (Figure 4C). In contrast, vps4Δ cells that lacked either the ESCRT-II subunit Vps36 or Vps20 itself did not show coimmunoprecipitation of Vps20 with Vps20-HA. These results indicated that ESCRT-III contains at least two subunits of Vps20, suggesting a more complex structure for ESCRT-III than previously proposed.
A more complex ESCRT-III structure could allow for several MIT domains of a Vps4 oligomer to contact ESCRT-III simultaneously via both MIM1 and MIM2 interactions, a model that is supported by our localization studies of full-length Vps4. The microscopy data indicated that Vps4E233Q-GFP localized to endosomes efficiently even in presence of only the MIM1 or MIM2 interaction (I18D or L64D mutant in vps4Δ vta1Δ, Figure 4D). This observation is in contrast to the studies of MIT-GFP shown in Figure 2A, which demonstrated that the loss of MIM1 or MIM2 severely inhibited the localization of the MIT domain to endosomes. Only deletion of the Snf7-Vps20 subcomplex, which results in a complete loss of ESCRT-III on endosomes, blocked the localization of Vps4E233Q-GFP to endosomes (vps4Δ vps20Δ snf7Δ, Figure 4D).
Together, the data indicated that the binding of full-length Vps4 to ESCRT-III is less sensitive to loss of MIT interactions than observed with a single MIT domain. This difference is likely due to the oligomerization of full-length Vps4 that allows the formation of several MIT-ESCRT-III interactions per Vps4 complex. These multiple interactions would act additively, thereby dramatically enhancing overall affinity of Vps4 to its substrate.
Indications for Functionally Distinct Ring Structures in the Vps4 Oligomer
The Vps4E233Q mutant protein is unable to hydrolyze the bound ATP and exhibits a dominant-negative phenotype in vivo (Babst et al., 1998). This dominant-negative effect is likely caused by the assembly of the Vps4E233Q mutant proteins together with wild-type Vps4 into the oligomeric structure, thereby inhibiting the function of the assembled protein complex. Interestingly, both CPY-invertase secretion assays and GFP-CPS sorting analysis indicated that deletion of the MIT domain eliminated the dominant-negative phenotype of Vps4E233Q (vps4ΔMIT,E233Q, Figure 3A, 8, and Table 3) even though microscopy studies demonstrated that Vps4E233Q lacking the MIT domain is recruited to ESCRT-III via binding to full-length Vps4 (Figure 4A, 5 and 7). Similarly, mutations in the MIT domain that interfere with binding to ESCRT-III and Ist1 (L64D and I18D) suppressed the dominant-negative phenotype of Vps4E233Q (Table 3 and Figure 3A, 6 and 7). Because our data suggested that the MIT domain plays an important role in the initial recruitment of Vps4 to ESCRT-III, we speculate that without the MIT domain Vps4 is preferentially incorporated into the second, ESCRT-III-distal ring of the Vps4 oligomer. If correct, this would argue that the function of the Vps4 oligomer is not inhibited by the presence of ATP-locked subunits in the second ring but is blocked by the presence of Vps4E233Q in the ESCRT-III–bound ring.
Binding of Vps4, Vta1, and Ist1 to Did2
Did2 binds to Vps4, Vta1, and Ist1 via its C-terminal MIM1 motif (Figure 1A; Howard et al., 2001; Stuchell-Brereton et al., 2007; Azmi et al., 2008; Bajorek et al., 2009b; Xiao et al., 2009). To compare the strength of these interactions, in vitro GST pulldown experiments were performed. GST fused to Did2 was purified and immobilized on GSH-Sepharose. Recombinant Ist1 or Vta1 was added in a one-to-one ratio to the immobilized GST-Did2 and bound and unbound fractions were analyzed by SDS-PAGE. In these experiments, Ist1 bound with an approximate 1:1 ratio to Did2 (Figure 5, lane 9). In contrast, the binding of Vta1 to GST-Did2 was inefficient, and a majority of the protein was found in the unbound fraction (Figure 5, lane 8). Because Vps4 and GST-Did2 comigrate in SDS-PAGE gels, additional interaction studies were performed using the C-terminal half of Did2 [CT(Did2), amino acids 113-204]. Similar to the results observed with full-length Did2, GST-CT(Did2) bound very efficiently to Ist1, but only small amounts of Vta1 were detected in the bound fraction (Figure 5, lanes 2 and 3). The hydrolysis mutant Vps4E233Q bound poorly to GST-CT(Did2) both in the presence of ATP and ADP (Figure 5, lanes 4 and 6). However, the presence of Ist1 increased the amount of Did2-bound Vps4E233Q (Figure 5, lanes 5 and 7). This increase in Vps4 binding was more pronounced in the presence of ATP compared with ADP, which is consistent with the previously observed nucleotide dependence of the Vps4-Ist1 interaction (Dimaano et al., 2008). These results suggested that in the presence of Ist1 the observed Vps4-Did2 interaction is not direct but mediated by Ist1.
Together, the in vitro binding studies suggested that Ist1 has much higher affinity to Did2 than Vta1 or Vps4. Therefore, we predicted that in the cytoplasm Ist1 is the preferred binding partner of Did2. Furthermore, previous studies have shown that high cytoplasmic levels of Did2 interfere with endosomal localization of Ist1, supporting the notion that Did2 binds to Ist1 in vivo in the cytoplasm (Dimaano et al., 2008). In contrast, high Did2 expression levels did not affect the endosomal localization of Vta1 or Vps4 (data not shown). In the presence of ATP, Vps4 demonstrated a high-affinity for Ist1 (Figure 5; Dimaano et al., 2008), which suggested that in the cytoplasm Did2, Ist1, and Vps4 might form a complex that is then recruited to ESCRT-III. However, attempts to obtain direct evidence for the presence of this complex in the cytoplasm by coimmunoprecipitation experiments or gel filtration analysis were not successful (data not shown), which might be a consequence of the transient nature of the Vps4-Ist1-Did2 complex.
DISCUSSION
The recruitment of Vps4 to ESCRT-III is mediated to a large extent by interactions between the Vps4 MIT domain and the four subunits of ESCRT-III, as well as Ist1 and Did2. All six proteins interact with the MIT domain via MIM1 or MIM2 motifs, which bind to two different surface areas of the MIT domain (Figure 1, A and C). These interactions are rather weak, with Kd values in the micromolar range (Stuchell-Brereton et al., 2007; Kieffer et al., 2008), suggesting that several interactions act synergistically to achieve Vps4 recruitment and assembly.
The binding of Ist1 to Did2 is required for the localization of Ist1 to ESCRT-III (Dimaano et al., 2008; Rue et al., 2008). Overexpression of DID2 results in accumulation of Ist1 together with Did2 in the cytoplasm, indicating that the Did2-Ist1 interaction is not limited to ESCRT-III but can occur in solution (Dimaano et al., 2008). This observation is consistent with our in vitro data demonstrating the formation of a stable interaction between Ist1 and full-length Did2, which suggested that this interaction occurs in the closed conformation of cytoplasmic Did2 (in contrast to the open conformation of ESCRT-III–associated Did2; Figure 5). Therefore, we propose that Ist1 and Did2 interact in the cytoplasm and that this complex binds to Vps4, which facilitates its recruitment to ESCRT-III. In this complex the C-terminal MIM1 and MIM2 motifs of Ist1 bind to the Vps4 MIT domain, whereas the Ist1 ELYC domain interacts with both Did2 and the AAA domain of Vps4 (interactions 7, 9, and 10, Figure 1A). This trimeric complex is recruited to ESCRT-III via the interaction between Did2 and the Vps2–Vps24 subcomplex (step A in Figure 1B). In this model the direct interaction between Vps4 and Did2 implicated in previous studies, does not play a role in Vps4 recruitment (interaction 3, Figure 1A).
One of the two MIT domains of Vta1 has been shown to bind to the MIM1 motif of Did2 (Azmi et al., 2008). This motif is accessible in the open, or ESCRT-III–associated conformation of Did2. Therefore, we predict that Vta1 is recruited to the ESCRT-III–associated Did2-Ist1-Vps4 complex (step B in Figure 1B). This recruitment might initialize the assembly of Vps4 from the Ist1-Did2–bound state into the active oligomeric form. Because Ist1 apparently competes with Vta1 and ESCRT-III for Vps4 binding (Figure 1A), we propose that Vps4 assembly requires rearrangement of the existing Vps4 interactions (step C in Figure 1B). The MIT domain switches from the MIM1 and MIM2 interactions with Ist1 to MIM1 and MIM2 interactions with ESCRT-III (MIM1 of Vps2–Vps24 subcomplex and MIM2 of Vps20–Snf7 subcomplex). The Vps4 AAA domain changes its interaction from the Ist1 ELYC domain to the dimerization domain of Vta1 (VHL domain; Azmi et al., 2006). These rearrangements replace the recruiting factor Ist1 with the assembly factor Vta1 and the substrate ESCRT-III.
In contrast to cytokinesis, which requires Ist1 function (Agromayor et al., 2009; Bajorek et al., 2009a), the MVB pathway showed only minor defects in ist1Δ cells (Dimaano et al., 2008). This observation suggested that at the MVB, Vps4 can be directly recruited by ESCRT-III– and Did2-associated Vta1 (step C bypassing A and B in Figure 1B). This redundancy seems to be common in the Vps4 system of the MVB pathway. For example, only the loss of two or more Vps4 MIT interactions severely impaired the recruitment and function of Vps4 (Figures 2 and 3, Table 3). Mainly, the phenotypic severity of the different MIT and ESCRT-III interaction mutants followed the severity of the MIT-GFP recruitment defects observed in these strains. Exceptions were the MIM2 mutations in Vps20 and Snf7 that resulted in reduced endosomal recruitment of MIT-GFP and increased accumulation of ESCRT-III, but no obvious effects on MVB trafficking.
Based on our mutational analysis, all four potential ESCRT-III MIT interaction motifs are functional. However, loss of Vps2 MIM1 or Snf7 MIM2 resulted in more severe phenotypes than observed with mutations in the MIT interaction motifs of Vps24 and Vps20, suggesting that Vps2 and Snf7 contain the major Vps4-binding sites. This finding contradicted in vitro data, indicating Vps20 as a major binding partner for Vps4 (Figure 2D, Azmi et al., 2008; Kieffer et al., 2008) and demonstrated the importance for in vivo studies to determine the relevance of interactions observed in vitro.
The high level of redundancy in the Vps4 recruitment system is further illustrated by the fact that Vps4 lacking the MIT domain is still able to localize to endosomes. Our data showed that MIT deleted Vps4 was able to localize to MVBs by binding to Vta1 that has been localized via Did2 to ESCRT-III (Figure 4). However, the deletion of the MIT domain resulted in loss of Vps4 function, suggesting the MIT domain functions not only in Vps4 localization but also in the ESCRT-III disassembly reaction.
Our data indicated that efficient recruitment of Vps4 and subsequent ESCRT-III disassembly required MIM1 and MIM2 MIT interactions to occur simultaneously, an observation that has strong implications in the ESCRT-III structure. A recently published model of ESCRT-III proposed a single linear polymer of ∼16 subunits (Teis et al., 2008; Saksena et al., 2009). However, our results indicated a more complex structure for ESCRT-III (Figure 4C), a structure that should allow for the simultaneous binding of several Vps4 MIT domains to ESCRT-III MIM1 and MIM2 motifs.
Vps4 oligomerizes into a complex composed of 12 subunits which form two six-membered rings in a tail-to-tail arrangement (Yu et al., 2008; Landsberg et al., 2009). Based on this structure, one ring interacts via the N-terminal MIT domains with ESCRT-III, whereas the second ring seems to have no direct contact with the substrate, a functional difference that is reflected in the different subunit arrangements of the two rings (Yu et al., 2008). ATP-locked Vps4E233Q is a dominant-negative mutant protein that assembles with wild-type Vps4 and blocks its activity (Babst et al., 1998). However, deletion or mutation of the MIT domain suppressed the dominant-negative phenotype of Vps4E233Q even though the protein was recruited together with full-length Vps4 to the endosomal membrane (Figures 3A and 4A, Table 3). Because the MIT domain plays an important role in the initial recruitment of Vps4 to ESCRT-III, we speculate that the mutant MIT versions of Vps4 predominantly assemble via Vps4–Vps4 and Vps4–Vta1 interactions in the second, ESCRT-III-distal Vps4 ring (Figure 1B, step E). If correct, this model would imply that the presence of ATP-locked Vps4E233Q in the second ring does not interfere with the function of the overall complex. The second, non-ESCRT-III–associated ring might mainly play a structural or regulatory role and thus is not directly involved in the ATP-dependent dissociation reaction. Similarly, type-II AAA ATPases, such as NSF/Sec18 and p97/Cdc48, have been shown to contain two functionally distinct AAA domain rings (reviewed in Erzberger and Berger, 2006). The N-terminal AAA ring hydrolyzes ATP, providing the energy for the disassembly or unfolding of the bound substrate. The second, C-terminal AAA ring, plays a structural role and does not hydrolyze the bound nucleotide.
Together, our studies resulted in a model of the Vps4 interaction network that underlies the recruitment and oligomerization of the ATPase. Both types of interactions, MIM1 and MIM2, play a role in these activities and work together to accomplish the recycling of ESCRT-III subunits and the formation of MVB vesicles. These built-in redundancies ensure the robustness of the system and highlight the importance of Vps4 in the proper function of endosomal protein trafficking and cytokinesis, two essential cellular processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tamara Darsow, Vladimir Lupashin, and Charles Jones for critical reading of the manuscript. We thank Wes Sundquist and Jack Skalicky for assistance with designing the MIT point mutations. This work has been supported by Grant R01 GM074171 from the National Institutes of Health (NIH) and Grant 0530210N from the American Heart Association. D.J.K. and B.A.D. are supported by Grant R01 GM073024 from the NIH.
Abbreviations used:
- AAA
ATPase associated with various cellular activities
- ESCRT
endosomal-sorting complex required for transport
- MIM1/2
MIT-interacting motif 1/2
- MIT
microtubule-interacting and transport
- MVB
multivesicular body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0572) on January 28, 2010.
REFERENCES
- Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell. 2009a;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009b;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Darsow T., Odorizzi G., Emr S. D. Invertase fusion proteins for analysis of protein trafficking in yeast. Methods Enzymol. 2000;327:95–106. doi: 10.1016/s0076-6879(00)27270-4. [DOI] [PubMed] [Google Scholar]
- Davies B. A., Lee J. R., Oestreich A. J., Katzmann D. J. Membrane protein targeting to the MVB/lysosome. Chem. Rev. 2009;109:1575–1586. doi: 10.1021/cr800473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. Ist1 regulates vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger J. P., Berger J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gonciarz M. D., Whitby F. G., Eckert D. M., Kieffer C., Heroux A., Sundquist W. I., Hill C. P. Biochemical and structural studies of yeast Vps4 oligomerization. J. Mol. Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky B. F., Busch G. R., Emr S. D. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard T. L., Stauffer D. R., Degnin C. R., Hollenberg S. M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Emr S.D. The ESCRT Complexes: Structure and Mechanism of a Membrane-Trafficking Network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Landsberg M. J., Vajjhala P. R., Rothnagel R., Munn A. L., Hankamer B. Three-dimensional structure of AAA ATPase Vps 4, advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. N., Martin J. AAA proteins. Curr. Opin. Struct. Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- McDonald B., Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Paravicini G., Horazdovsky B. F., Emr S. D. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35 null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol. Biol. Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and Function of Multivesicular Bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Emr S. D. ESCRTs and human disease. Biochem. Soc. Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Chung H. Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 2005b;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Lawrence L. W. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Shiflett S. L., Ward D. M., Huynh D., Vaughn M. B., Simmons J. C., Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A., Hanson P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Shim S., Merrill S. A., Hanson P. I. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Muller S., Hanisch F. G., Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S. D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Vajjhala P. R., Nguyen C. H., Landsberg M. J., Kistler C., Gan A. L., King G. F., Hankamer B., Munn A. L. The Vps4 C-terminal helix is a critical determinant for assembly and ATPase activity and has elements conserved in other members of the meiotic clade of AAA ATPases. FEBS J. 2008;275:1427–1449. doi: 10.1111/j.1742-4658.2008.06300.x. [DOI] [PubMed] [Google Scholar]
- VerPlank L., Bouamr F., LaGrassa T. J., Agresta B., Kikonyogo A., Leis J., Carter C. A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc. Natl. Acad. Sci. USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Vaughn M. B., Shiflett S. L., White P. L., Pollock A. L., Hill J., Schnegelberger R., Sundquist W. I., Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Chen X. W., Davies B. A., Saltiel A. R., Katzmann D. J., Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the mVB pathway and cytokinesis. Mol. Biol. Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S. C., et al. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- Yu Z., Gonciarz M. D., Sundquist W. I., Hill C. P., Jensen G. J. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J. Mol. Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.