We propose a general model for the role of the Hsp90 ATPase cycle in proteostasis in which Aha1 regulates the dwell time of Hsp90 with client by integrating chaperone function and client folding energetics by modulating ATPase sensitive N-terminal dimer structural transitions.
Abstract
The activator of Hsp90 ATPase 1, Aha1, has been shown to participate in the Hsp90 chaperone cycle by stimulating the low intrinsic ATPase activity of Hsp90. To elucidate the structural basis for ATPase stimulation of human Hsp90 by human Aha1, we have developed novel mass spectrometry approaches that demonstrate that the N- and C-terminal domains of Aha1 cooperatively bind across the dimer interface of Hsp90 to modulate the ATP hydrolysis cycle and client activity in vivo. Mutations in both the N- and C-terminal domains of Aha1 impair its ability to bind Hsp90 and stimulate its ATPase activity in vitro and impair in vivo the ability of the Hsp90 system to modulate the folding and trafficking of wild-type and variant (ΔF508) cystic fibrosis transmembrane conductance regulator (CFTR) responsible for the inherited disease cystic fibrosis (CF). We now propose a general model for the role of Aha1 in the Hsp90 ATPase cycle in proteostasis whereby Aha1 regulates the dwell time of Hsp90 with client. We suggest that Aha1 activity integrates chaperone function with client folding energetics by modulating ATPase sensitive N-terminal dimer structural transitions, thereby protecting transient folding intermediates in vivo that could contribute to protein misfolding systems disorders such as CF when destabilized.
INTRODUCTION
Hsp90 is an abundant and essential protein in eukaryotic cells that is a key element of the protein homeostasis or proteostasis program that is involved in the synthesis and maintenance of the human proteome (Balch et al., 2008; Hutt et al., 2009; Powers et al., 2009). Hsp90 is involved in regulating multiple biological processes ranging from steroid hormone maturation to cell signaling (Pearl et al., 2008; Wandinger et al., 2008; Neckers et al., 2009), cancer (Taldone et al., 2008), and diseases of proteostasis (Powers et al., 2009). The proteostasis environment is thought to strongly control the stability of the wild-type or mutant protein fold for function, or to target the protein for degradation (Balch et al., 2008; Powers et al., 2009). Loss- or gain-of-toxic function of Hsp90 clients leads to the disruption of cell, tissue, and/or organismal physiology and therefore can be considered “systems disorder” diseases.
Hsp90 is thought to function in the context of a complex ATPase cycle involving numerous cochaperones (Pearl and Prodromou, 2006; Pearl et al., 2008). How its ATPase activity is regulated or the consequence of the ATPase cycle on client protein folding remains a major question in the field. Eukaryotic Hsp90 comprises three domains: an N-terminal domain that harbors the adenine nucleotide-binding pocket followed by an unstructured linker region that joins the middle domain and a C-terminal domain that maintains Hsp90 in a dimeric state (Pearl and Prodromou, 2006; Wandinger et al., 2008; Neckers et al., 2009). In the nucleotide-free or ADP-bound forms, Hsp90 is in the open state; in the presence of nonhydrolysable ATP analogs such as 5′-adenylyl-β,γ-imidodiphosphate (AMP-PNP), Hsp90 is restricted to the closed state. Incubation of human Hsp90 in vitro in the absence of nucleotide results in its continuous cycling between open and closed states; thus, Hsp90 has a flexible fold. On client interaction in an as yet unknown manner, dimerization through its N-terminal domains in response to the binding of ATP leads to the closed configuration. Hydrolysis of ATP in the Hsp90 closed state results in large conformational changes that are believed to direct folding and dissociation of a bound substrate (Cunningham et al., 2008; Krukenberg et al., 2008; Hessling et al., 2009; Mickler et al., 2009).
Hsp90–client interactions require numerous cochaperones. Hsp90 cochaperones include p23, Hop, several peptidyl-prolyl-isomerases, and activator of Hsp90 ATPase 1 (Aha1; Pearl and Prodromou, 2006). Aha1 is the most recently identified Hsp90 cochaperone and has been shown to substantially accelerate the ATPase activity of eukaryotic Hsp90 in vitro (Panaretou et al., 2002). Yeast studies have demonstrated that overexpression of an Aha1 prologue, yeast Hch1p, can suppress the temperature-sensitive defect of Hsp82pE381K (the yeast prologue of Hsp90), at elevated temperatures (Panaretou et al., 2002; Lotz et al., 2003; Meyer et al., 2003; Harst et al., 2005). Curiously, the yeast Hsp82pE381K mutant does not have an intrinsic or Aha1-stimulated ATPase defect at any temperature tested. Moreover, there are conflicting reports regarding the ability of yeast Hch1p or the homologous N-terminal domain of yeast Aha1 to stimulate wild-type yeast Hsp90 ATPase activity (Panaretou et al., 2002; Lotz et al., 2003; Meyer et al., 2003; Harst et al., 2005). Owing to the discrepancy between the in vitro and in vivo properties of Hch1 in yeast, the mechanism by which mammalian or yeast Aha1 exerts its biological effect remains controversial (Neckers et al., 2009). However, recent studies using single-molecule approaches suggest that the different conformations coexist in the absence of nucleotide and that Aha1 can strongly influence the pathway even in the absence of ATP (Hessling et al., 2009; Mickler et al., 2009).
The three-dimensional structures of the N-terminal domain of the yeast Aha1 and the C-terminal domain of the human Aha1 have been solved by x-ray crystallography (Meyer et al., 2004) and NMR (PDB ID 1X53), respectively. Furthermore, the structure of the N-terminal domain of Aha1 was solved in a complex with the yeast Hsp90 middle-domain (Meyer et al., 2004). Although stabilization of an Arg residue (Arg380) in Hsp90 that is important for catalysis by Aha1 may potentially contribute to the rate enhancement (Meyer et al., 2004), there is no information on the role of the C-terminal domain of Aha1 in interaction with Hsp90 or for promoting ATPase activity of Hsp90. Indeed, data from isothermal titration calorimetry suggest that the Hsp90–Aha1 interaction requires both full-length Hsp90 and full-length Aha1 to achieve tight binding, raising the possibility that the C-terminal domain of Aha1 plays an important functional role in the adenine nucleotide-dependent Hsp90 conformational cycle.
We have demonstrated a critical role for wild-type (WT) human Aha1 in the misfolding disease cystic fibrosis (CF) (Wang et al., 2006). CF is one of the most common inherited diseases in humans and is caused by loss of function of the cystic fibrosis transmembrane regulator (CFTR) protein that is critical, for regulation of the balance of chloride ions in secretory organs such as the intestine, pancreas and lungs (Riordan, 2005, 2008). CFTR is a multimembrane spanning protein with two six-membrane spanning domains and two cytosolic ATP-binding domains referred to as NBD1 and NBD2 (Riordan, 2005, 2008). CFTR folding requires human Hsp90 (Loo et al., 1998; Fuller and Cuthbert, 2000; Youker et al., 2004; Skach, 2006; Wang et al., 2006; Sun et al., 2008), suggesting a role for Hsp90 in transient protection/stabilization of a folding intermediate(s) during co- and posttranslational translocation into the endoplasmic reticulum (ER) membrane. The most common disease-associated allele of CFTR is a deletion of Phe 508 (ΔF508) in the NBD1 cytosolic domain. Like other energetically destabilized folds in response to mutation, ΔF508 is uncoupled from the proteostasis network (Balch et al., 2008; Powers et al., 2009). The absence of Phe 508 impairs folding, prevents export from its site of synthesis in the ER, and targets CFTR efficiently to the Hs(c)p40 and Hs(c)70 ER-associated degradation (ERAD) pathway (Turnbull et al., 2007). The ER stability and export of both the WT and ΔF508 CFTR are sensitive to levels of WT Aha1, although the mechanism responsible for these events remains to be elucidated (Wang et al., 2006). Thus, characterizing the molecular and structural basis for Aha1 function in regulation of the Hsp90 ATPase activity will be a potential key to understanding the role of Hsp90 in folding of CFTR and progression of CF disease.
Here, we report that Aha1 is composed of two independently functional domains required for client folding in vivo. To elucidate the structural basis for interactions between the full-length mammalian Hsp90 and Aha1 in solution, we have applied covalent cross-linking approaches (Maiolica et al., 2007) and novel mapping techniques in conjunction with mass spectrometry using multidimensional protein identification technology (MudPIT) (Washburn et al., 2001) to generate a molecular footprint of the residues forming the dynamic Aha1–Hsp90 interaction interface in the presence or absence of nucleotide. Our studies suggest that Aha1 accelerates the ATPase hydrolysis by bridging the Hsp90 dimer structure. To address the biological importance of the trans-interaction of Aha1 with Hsp90 in client folding in vivo, we demonstrate that mutations in both N and C domains of Aha1 influence its ability to promote the folding of CFTR and trafficking to the cell surface. We show that a mutation in the C terminus effects the ATPase-stimulating activity and is required for Aha1 function in folding CFTR in vivo. We propose that through trans-dimerization, Aha1 kinetically regulates an ATP hydrolysis-dependent coupling between client folding intermediates and the protective chaperone environment provided by Hsp90 to promote stabilization and trafficking of WT and ΔF508 CFTR. Alteration of Aha1 ATPase function in vivo protects ΔF508 from degradation, suggesting an important role for reduced ATP hydrolysis activity in stabilization of the mutant fold to promote correction of cystic fibrosis.
MATERIALS AND METHODS
Materials and Instrumentation
Methyl PEG4-NHS ester, 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (Sulfa-NHS), and N-5-azido-2-nitrobenzoyloxysuccinimide (ANB-NOS) were purchased from Thermo Fisher Scientific (Waltham, MA). Trypsin gold mass spectrometry grade enzyme was purchased from Promega (Madison, WI). LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific) was used for analysis in the foot-printing experiments and an Orbitrap mass spectrometer (Thermo Fisher Scientific) for the cross-linking experiments. Surface plasmon resonance (SPR) binding studies we performed on a Biacore 2000 biosensor instrument (GE Healthcare, Piscataway, NJ).
Cloning, Mutagenesis, and Protein Purification
The coding sequence of Aha1 (Embank accession NM_012111) was amplified by polymerase chain reaction (PCR) to create vectors designed to express the N-terminal, C-terminal, and full-length domains by using primer pairs S1/A1, S2/A2, and S1/A2, respectively (S1, 5′-gagagacatatggccaagtggggtgagggagac-3′; S2, 5′-gagagacatatgatcttacctacaatgaatggagag-3′; A1, 5′-gagagaggatcctcactagccctgggtgaactctg-3′; and A2, 5′-gagagaggatcctcactaaaataagcgtgcgccatagcc-3′). PCR products were cloned into pET11d (Invitrogen, Carlsbad, CA) between the NcoI and BamHI sites. The resulting plasmids, termed pET11dHishAha1N, pET11dHisAha1C and pET11dHisAha1FL, were then transformed into Escherichia coli BL21(DE3) cells for recombinant protein expression. The pET14b plasmid coding for the human Hsp90β (to be referred to in the text throughout as simply Hsp90) was kindly provided by Sophie Jackson (University of Cambridge, Cambridge, United Kingdom). Glutathione transferase (GST)-tail, a GST fusion of the 29 amino acid cytoplasmic tail of the vesicular stomatitis virus glycoprotein (VSV-G), was expressed in E. coli by using pGEX2T vector (Aridor et al., 1998, 1999).
The overnight grown culture of recombinant human Aha1 (E. coli BL-21 DE3) was diluted into 1 liter of Luria broth (LB) medium containing carbenicillin (100 μg/ml) and grown for 4 h at 37°C (final OD of ∼0.8). The cells were induced by 0.5 mM isopropyl β-d-thiogalactoside (IPTG) at 30°C for 5 h. After induction, the cells were pelleted and stored at −80°C. Purification of Aha1 was carried out using immobilized metal affinity chromatography (IMAC). Cell pellets were resuspended in 10 mol IMAC buffer A (20 mM NaH2PO4 pH 7.2, 500 mM NaCl, 1 mM MgOAc, and 5 mM β-mercaptoethanol) supplemented with an EDTA-free protease inhibitor tablet (catalog no. 1873580, Complete EDTA-free; Roche Diagnostics, Indianapolis, IN). Cells were then lysed by sonication at 30% power for 3 × 20 s on ice. The resultant lysate was ultracentrifuged at 45,000 rpm for 30 min in a Ti60 rotor (Beckman Coulter, Fullerton, CA), and the cleared cell lysate was fractionated over a 1-mol Ni2+-charged HiTrap Chelating HP column (catalog no. 17-0408-01; GE Healthcare) using an Äkta fast-performance liquid chromatography system with Frac-950 fraction collector (GE Healthcare). Gradient fractionation was carried out with IMAC buffer B (IMAC A with 1 M imidazole). Aha1-containing fractions were pooled, concentrated, and further fractionated by gel filtration chromatography (GFC) using a 100-ml S-100 column (catalog no. 17-0612-01; GE Healthcare) in 25 mM HEPES, pH 7.2, 125 mM KOAc, 1 mM MgOAc, and 1 mM dithiothreitol (DTT).
The overnight grown culture of recombinant human Hsp90β (E. coli BL-21 DE3) was diluted into 1 liter of LB medium containing carbenicillin (100 μg/ml) and grown for 4 h at 37°C (final OD of ∼0.8). The cells were induced by 1 mM IPTG at room temperature for 16 h. After induction, the E. coli cells were pelleted and stored at −80°C. Purification of recombinant human Hsp90 was carried out with the same strategy as described above for Aha1 but with an additional gradient fractionation on a 1-mol Mono Q HR 5/5 column (catalog no. 17-0546-011; GE Healthcare) using Mono Q A buffer (25 mM Tris, pH 7.5, and 1 mM DTT), and Mono Q B buffer (25 mM Tris, pH 7.5, 1 M NaCl, and 1 mM DTT) between the IMAC and GFC steps.
GST and GST-tail were purified using glutathione-Sepharose beads (catalog no. 27-4574-01; GE Healthcare) following the manufacturer's protocol.
Aha1/Hsp90 Methyl-PEG4-NHS Ester Labeling
To solutions of Hsp90 and Aha1 in 25 mM HEPES, pH 7.4, 100 mM NaCl (either separately or after 30 min of preincubation on ice), freshly prepared methyl-PEG4-NHS ester stock solution was added to final 10 mM concentration (final protein concentration, 3.1 μM). After 10 min of incubation at room temperature, the reaction was stopped by addition of excess Tris-HCl. The samples were then acetone precipitated and trypsin-digested for 4 h at 37°C.
Aha1-Hsp90 Zero-Length Cross-Linking
To 10 μM solutions of Aha1 and Hsp90 in 25 mM HEPES, pH 7.4, 100 mM NaCl a freshly prepared mixture of EDC and Sulfa-NHS were added according to manufacturer's specifications. The reactions were cross-linked for 30 min at room temperature followed by the addition of excess quencher Tris-HCl. Proteins were then either run on a 4–12% Bistros precast polyacrylamide gel or acetone-precipitated and trypsin-digested for mass spectrometry (MS) analysis. Samples analyzed by MS were first dissolved in 8 M urea solution prepared in 100% 16O H2O, 95% 18O H2O, or 50% 18O H2O/50% 16O H2O mixture and then trypsin-digested. This ratio of oxygen isotopes in each sample was kept throughout the trypsin digestion.
ANB-NOS Aha1/Hsp90 Cross-Linking
Full-length Aha1 and Hsp90 were dialyzed into 25 mM HEPES, pH 7.4, and 100 mM NaCl for at least 4 h at 4°C. Aha1 was then labeled by addition of 20 mM ANB-NOS stock in dimethyl sulfoxide prepared immediately before use (final concentration, 1 mM). The mixture was incubated for 3 min at room temperature and 2.5 μl of 2 M Tris-HCl was added to all reactions to quench. The labeled Aha1 was then dialyzed for 4 h at 4°C in the dark and added to Hsp90 in an equimolar concentration (final concentration, 4 μM). Reactions were then irradiated from for 1, 2, or 3 min using a 365-nm hand-held lamp. Then, 2 μl of 2 M Tris-HCl was added to each sample to stop the reaction. All samples were run on a 4–12% Bis-Tris-HCl SDS-polyacrylamide gel electrophoresis (PAGE) gels.
Mass Spectrometry and Data Analysis
Each sample (∼100 μg of digested proteins) was analyzed at least three times on a LTQ linear ion-trap MS (Thermo Fisher Scientific) using a three-step MudPIT (Washburn et al., 2001, 2003). Tandem mass spectra (MS/MS) from trypsin-digested cross-linked samples and noncross-linked samples were analyzed using SEQUEST protein identification algorithm and DTA SELECT for the footprinting experiments (see the flow chart in Supplemental Figure S1 for the method analysis of the cross-linking experiments). For the latter, MS/MS from noncross-linked samples were searched against the EBI IPI human database, version 3.23, with its reverse decoy to identify peptides that can be obtain from the individual proteins (Aha1 or Hsp90). The identified Aha1 peptides and Hsp90 peptides were then cross-linked in silico to create a database of artificial cross-linked peptides. The artificial cross-linked peptides database was used for all analyses. Because cross-linked peptides from actually identified noncross-linked peptides reflect more accurately the experimental data set. MS/MS from cross-linked samples were also searched against the EBI-IPI human database. MS/MS matched to the database were subtracted (removed). The remaining MS/MS were then searched against the EBI-IPI human database plus the artificial cross-linked peptides database with their reverse decoys for the discovery of cross-linked peptides.
NMR Analysis
All proteins for the NMR studies were expressed in E. coli BL21 codon plus (Stratagene, La Jolla, CA) cells in minimal M9 media using 15NH4Cl as the sole nitrogen source. The cells were grown to an OD600 of 0.8–1 at 37°C at which point they were induced by the addition of 1 mM IPTG. The cells were then grown for 5–6 h at 25°C and harvested by centrifugation. The proteins were purified using Talon (Clontech, Mountain View, CA) metal affinity resin and subsequently run over a Sephadex 70 gel filtration column equilibrated in NMR buffer [25 mM Na2HPO4, 250 mM NaCl, 2 mM DTT, 2 mM benzamidine, 1 mM EDTA, 1 mM tris(2-carboxyethyl) phosphine hydrochloride, and 0.5 mM phenylmethylsulfonyl fluoride, pH 7.0]. The heteronuclear single quantum correlation spectra were acquired at 35°C on a Enova 600MHz spectrometer (Varian, Palo Alto, CA) equipped with a triple resonance cold probe and processed using NMRpipe.
ATPase Assays
A colorimetric Pi release assay (malachite green) was used with modified assay conditions (Rowlands et al., 2004). In brief, Hsp90 was incubated at the indicated concentration with or without Aha1 at the indicated concentrations at 37°C in 25 mM HEPES, pH 7.2, 125 mM KoAc, 2 mM MgCl2, and 1 mM ATP. Triplicate readings were taken at 0 and 60 min.
Surface Plasmon Resonance
Protein interaction studies were performed with a Biacore 2000. Hsp90 was coupled to a CM5 chip to 300 response units (RUs) and interaction with Aha1 (full length, domains or mutants) at the indicated concentrations was measured in 25 mM HEPES, pH 7.2, and 100 mM NaCl. SPR data for the full-length Aha1 polypeptides (WT and mutants) binding to Hsp90 was fit with a 1:1 Langmuir model using BiaEval software (GE Healthcare) according to the manufacturer's direction. The SPR data for the more weak binding N- and C-terminal domains of Aha1 were fit using a steady state model using BiaEval.
CFTR Trafficking Assays
Overexpression studies were performed as described previously (Wang et al., 2004, 2006).
RESULTS
Aha1 Is Composed of Two Distinct Domains
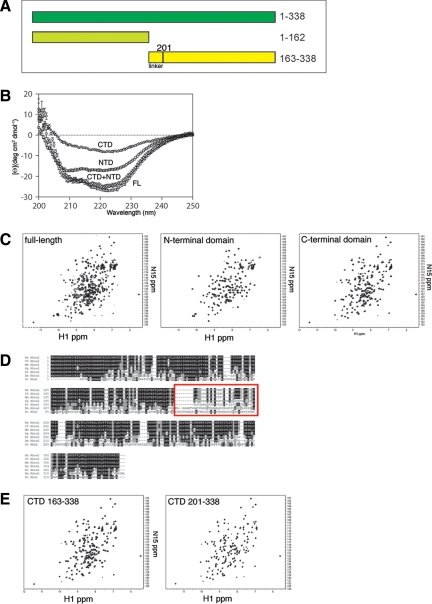
Aha1 is composed of two domains connected by a degenerate an ∼30-amino acid linker (approximately residues 163-200; Figure 1A). Previous studies have reported that the N-terminal domain of Aha1 binds to the middle domain of Hsp90 and stimulates its otherwise very low intrinsic ATPase activity (Panaretou et al., 2002; Meyer et al., 2004). Other studies have concluded that the C-terminal domain of Aha1 is required for ATPase stimulation (Lotz et al., 2003) and confers higher binding affinity for Hsp90 (Meyer et al., 2004). Consequently, the precise role of the C-terminal domain of Aha1 in regulating the adenine nucleotide-dependent conformational cycle of Hsp90 remains controversial.
Figure 1.
Domain structure of Aha1. (A) Schematic of Aha1 domains. Full-length (FL) human Aha1 (amino acids 1-338 shown in dark green), Aha1 N-terminal domain (NTD; amino acids 1-162 shown in light green), and Aha1 C-terminal domain (CTD; amino acids 163-338 shown in yellow). The position of the linker region (residues 163-200) is illustrated. (B) CD spectra of FL (circles), NTD (square), CTD (triangles), and NTD and CTD spectra added (diamonds). (C) NMR spectra of FL (left), NTD (middle), and CTD (right) domains of Aha1. (D) Sequence alignment of Aha1 from different organisms from yeast (bottom row) to human (top row). Prefixes denote the first letters of genus and species name. Red box indicates the position where the putative linker region that lacks evolutionary conservation. See Supplemental Figure S1 for an enlarged view of alignment. (E) NMR spectrum of shortened C-terminal Aha1 domain lacking the linker region (amino acids 201-338; right) next to the NMR spectrum of original C-terminal domain (left).
To study the biochemical properties of the N- and C-terminal domains of Aha1, we created plasmid constructs of residues 1-162 (N-terminal domain) and 163-338 (C-terminal domain) of Aha1 for expression and purification from E. coli (see Materials and Methods; Figure 1A). Circular dichroism suggested folded structures (Figure 1B). When the spectra of the N- and C-terminal domains observed alone were added together, they were equal to the spectrum of the full-length Aha1 protein (Figure 1B). Thus, the two domains retained the same overall secondary structure as when in the context of the full-length protein. To further confirm this conjecture, we performed NMR analysis on full-length Aha1 as well as the N- and C-terminal domains. The NMR spectrum for the full-length Aha1 protein was well dispersed and the N- and C termini gave equally well dispersed spectra further supporting that the two domains are folded properly on their own (Figure 1C). However, the NMR spectrum for the C-terminal domain suggested the presence of an unstructured portion of the polypeptide. Given that full-length Aha1 shows a region of extensive degeneracy (a possible flexible linker) separating the N- and C-terminal domains (Figure 1D; see Supplemental Figure S1 for enlarged image) that corresponded to the residues 163-200 of our C-terminal construct, we generated a new C-terminal construct (residues 201-338) that had this linker peptide removed. The NMR signal of the 201-338 construct was identical to that for the 163-338 construct except for the loss of the poorly dispersed signal contained in the 163-338 data set (Figure 1E). Together, our results suggest that Aha1 is composed of two independently folded domains joined by a flexible linker.
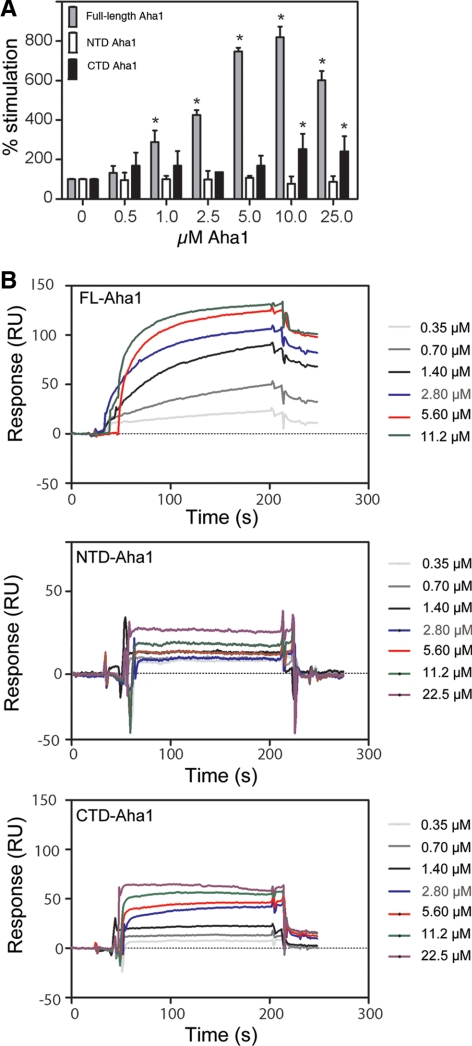
Only Full-Length Aha1 Robustly Stimulates the Hsp90 ATPase Activity
We tested our full-length, N- and C-terminal constructs for their ability to stimulate the ATPase activity of the β isoform of Hsp90 (to be referred to hereafter as simply Hsp90), the house-keeping isoform common to all mammalian cell types. In all experiments reported below, the C-terminal construct of Aha1 contains the linker region. Consistent with previous reports, we found that the full-length protein gave robust ATPase stimulation (Figure 2A). Although we found that the N-terminal domain failed stimulate Hsp90, the C-terminal domain weakly but reproducibly stimulated ATPase activity (Figure 2A). This was surprising because, although there are conflicting reports regarding the ability of the N-terminal domain to stimulate the Hsp90 ATPase activity (Panaretou et al., 2002; Lotz et al., 2003; Meyer et al., 2004), there has never been any activity attributed to the C terminus of Aha1. We also analyzed the ability of Aha1 to interact with Hsp90 using SPR. Full-length Aha1 showed a strong interaction that was fit to a 1:1 Langmuir model and was determined to be 0.5 μM (see Supplemental Table S1 for Kon and Koff values). Because the off rates of the N- and C-terminal domains to Hsp90 were too fast to be measured using this model, binding constants were calculated using a steady-state model applicable for weak interactions (Figure 2B). Using this approach, the N-terminal domain had an affinity for Hsp90 of 2.5 μM, whereas the C-terminal domain containing the linker region showed a slightly reduced affinity of 3.8 μM. This reduction in the binding of the N- and C-terminal domains of Aha1 compared with the intact full-length protein is consistent with the inability of either the N- or C-terminal domains to stimulate Hsp90 ATPase activity in a manner comparable with the full-length protein.
Figure 2.
Effect of increasing Aha1 on Hsp90 binding and ATPase. (A) Hsp90 ATPase assay with increasing amount of full-length (FL), N-terminal domain (NTD), and C-terminal domain (CTD) of Aha1. We incubated 4 μM final Hsp90 with the indicated amounts of Aha1 or domains (final micromolar) for 90 min as described in Materials and Methods. Generation of Pi was detected and measured using a malachite green assay (see Materials and Methods). ATPase activity is expressed as a percentage of Hsp90 alone. Asterisks denote statistically significant increases over Hsp90 alone (two-tailed t test, p < 0.05). Consistent with previous reports (McLaughlin et al., 2004; Onuoha et al., 2008), the intrinsic ATPase activity of Hsp90 was 0.02 min−1 ± 0.005. (B) SPR analysis of Hsp90 interaction with full-length Aha1 (FL-Aha1), and the N-terminal domain (NTD) and C-terminal domain (CTD) of Aha1 (residues 1-162 and 163-338, respectively). Hsp90 was coupled to a CM5 chip as described in Materials and Methods. Raw data are reported as arbitrary RUs (y-axis) as measured at the indicated time (x-axis) in seconds. Binding constants were determined as described in Materials and Methods and Results. See Supplemental Table S1 for Kon and Koff values.
Molecular Footprinting Reveals a Site of Aha1 Interaction with N- and Middle Domains of Hsp90
To develop an understanding of the interaction between Aha1 and Hsp90, we developed a molecular footprinting strategy involving chemical derivatization followed by mass spectrometry analysis (see Materials and Methods). Here, the rapid covalent modification of a reactive group of a derivatizing agent with the side chain containing terminal amine (such as Lys or Arg) found in Aha1 or Hsp90, or the Aha1–Hsp90 complex, was followed by quenching of the reaction, enzymatic digestion of the proteins, and identification of the sites of modification using mass spectrometry and MudPIT (Washburn et al., 2001; Washburn et al., 2003). Theoretically, residues that are protected from derivatization by the interaction of Aha1 with Hsp90 would be evident compared with the sites of derivatization when the Aha1 monomer or the Hsp90 dimer are incubated alone with the derivatizing agent.
After evaluation of a number amine reactive probes, we made use of the short methyl PEG4-NHS ester as the reactive group. This gave adequate mass differences, as well as comparable sequence coverage compared with the unlabeled proteins (our unpublished data). We reasoned that this adduct would not change significantly the surface properties of the proteins as compared with more hydrophobic reagents such as NHS-(CH2)x-biotin, for example, which we found to decrease the MS sequence coverage (our unpublished data). With the methyl-PEG4-NHS-ester we achieved routine labeling of ∼75% of all Lys and Arg in Hsp90 and Aha1 that were evenly distributed across the surface of these proteins.
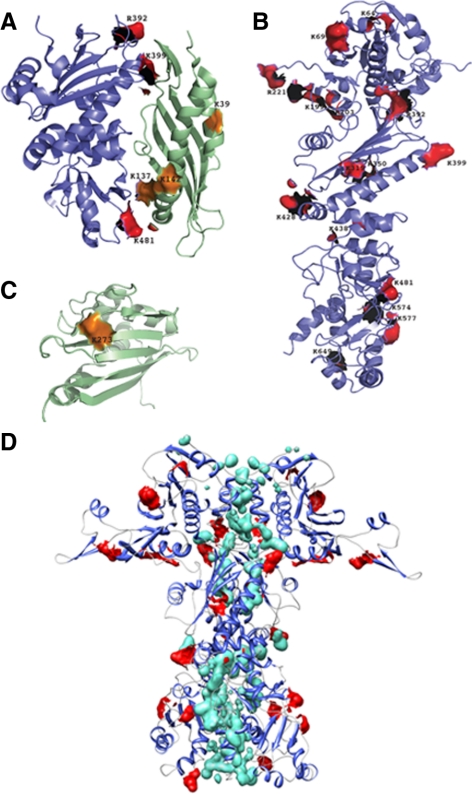
The results of the footprinting experiments in the absence of nucleotide to capture all of the intermediate cycling states are presented in Table 1 and are illustrated in Figure 3, A–D. We observed complete protection from covalent modification of the residues positioned within the footprint of the interaction between the middle domain of Hsp90 and the N-terminal domain of Aha1 observed in the cocrystal structure (Meyer et al., 2003, 2004; Figure 3A). Thus, the footprinting technique recapitulates the previously known interaction site and validates our methodology carried out in solution under physiological conditions. Our analysis, as expected, also revealed a number of protected residues (Figure 3B) located within the dimerization interface between the N-, middle, and C-terminal domains of Hsp90 (Figure 3D). Strikingly, several additional Lys and Arg residues outside the dimerization interface were located in both the N-terminal and middle domains of Hsp90 were protected from modification in the presence Aha1 (Figure 3, B and D). One Lys residue in the C-terminal domain of Aha1 (K273) was protected by Hsp90 (Figure 3C). The presence of a second and new binding site on Hsp90 for the C terminus of Aha1 in addition to the known N-terminal domain interaction of Aha1 with Hsp90 provides an explanation for the observation that only the full-length Aha1 protein can maximally bind and stimulate the Hsp90 ATPase activity (Figure 2).
Table 1.
Foot-printed peptides
| Protein | No. of modified residue andmodification mass difference | Peptide sequencea | No. of copiesobserved | Chargestate | Xcorrb |
|---|---|---|---|---|---|
| 64/219.26 | YESLTDPSK*LDSGKELK | 4 | 3 | 4.9 | |
| 69/219.26 | YESLTDPSKLDSGK*ELK | 4 | 2 | 4.7 | |
| 69/219.26 | LDSGK*ELKIDIIPNPQER | 3 | 2 | 3.5 | |
| 199/219.26 | VK*EVVKK | 1 | 2 | 2.6 | |
| 203/219.26 | VKEVVK*K | 1 | 2 | 2.6 | |
| 221/219.26 | KHSQFIGYPITLYLEKER* | 1 | 2 | 2.6 | |
| 223/219.26 | EK*EISDDEAEEEKGEK | 1 | 3 | 3.9 | |
| Hsp90 | 234/219.26 | EKEISDDEAEEEK*GEK | 1 | 3 | 3.9 |
| 319/219.26 | SLTNDWEDHLAVK*HFSVEGQLEFR | 2 | 3 | 6.7 | |
| 350/219.26 | K*NNIKLYVR | 4 | 2 | 3.4 | |
| 392/219.26 | GVVDSEDLPLNISR* | 3 | 2 | 3.8 | |
| 399/219.26 | EMLQQSK*ILK | 1 | 2 | 2.7 | |
| 428/219.26 | K*FYEAFSKNLK | 2 | 2 | 3.0 | |
| 438/219.26 | FYEAFSKNLK*LGIHEDSTNR | 4 | 3 | 4.9 | |
| 481/219.26 | MKETQK*SIYYITGESK | 2 | 3 | 3.6 | |
| 574/219.26 | K*VEKVTISNR | 1 | 2 | 2.7 | |
| 577/219.26 | VEK*VTISNR | 5 | 2 | 3.3 | |
| 649/219.26 | NDK*AVKDLVVLLFETALLSSGFSLEDPQTHSNR | 1 | 3 | 4.2 | |
| 3/219.26 | MAK*WGEGDPR | 1 | 1 | 2.9 | |
| 39/219.26 | DASNWSTDK*LK | 3 | 3 | 2.5 | |
| Aha1 | 137/219.26 | DEPDTNLVALMK*EEGVKLLR | 2 | 2 | 3.6 |
| 142/219.26 | DEPDTNLVALMK*EEGVK*LLR | 2 | 2 | 3.6 | |
| 182/219.26 | TEFTQGMILPTMNGESVDPVGQPALK* | 3 | 3 | 5.7 | |
| 273/219.26 | HIVMK*WR | 1 | 1 | 2.8 |
a An asterisk denotes modified residue.
b SEQUEST cross-correlation coefficient.
Figure 3.
Molecular footprinting of the interaction between Hsp90 and Aha1. (A) Footprinting was performed as described in Materials and Methods. Protected residues on Hsp90 (shown in red) and Aha1 (shown in orange) are depicted on the crystal structure of the middle domain of Hsp90 (blue) and the N-terminal domain of Aha1 (green) (Protein Data Base [PDB]: 1USV). (B) Hsp90 residues protected from modification in the Aha1–Hsp90 complex are depicted in red on the three-dimensional structure of the Hsp90 (homology model based on the yeast crystal structure [PDB: 2CG9]). (C) Aha1 residues protected from modification in the Aha1–Hsp90 complex are depicted in orange on the three-dimensional structure of the C-terminal domain of the human Aha1 (PDB: 1X53). (D) Homology of the Hsp90 dimer based on the yeast crystal structure (PDB: 2CG9) illustrating the position of protected residues (red) relative to residues forming the dimer interface (green).
Cross-Linking Reveals an Interaction of the Aha1 C Terminus with Hsp90
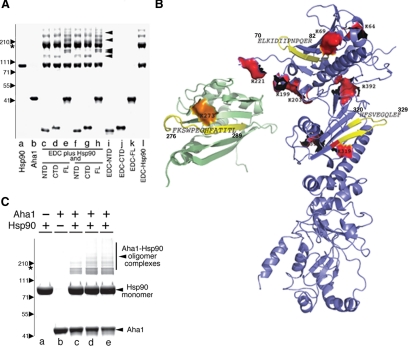
To capture the site(s) of protein–protein interactions between Aha1 and Hsp90, we used a stringent, zero-length cross-linker, EDC. This cross-linker forms a peptide bond between side chains of amino-containing amino acids (such as Lys or Arg) and an acidic residue (Glu or an Asp), residing in immediate proximity in the protein complex. Under physiological salt conditions, we detected higher order complexes (∼5% of total Aha1/Hsp90 added) that reflect interaction of Aha1 full-length or each of the C- and N-terminal domains with either Hsp90 monomer and dimers (Figure 4A, lanes c–h, black arrowheads). These were not detected in control incubations containing either purified Hsp90 (Figure 4A, lane l), full-length Aha1 (Figure 4A, lane k), or the individual C- or N-terminal domains in the presence of EDC (Figure 4A, lanes i and j).
Figure 4.
Analysis of binding of Aha1 cross-linked to Hsp90. (A) A 4–12% gradient SDS-PAGE of purified Hsp90 (lane a) and Aha1 (lane b). Full-length (lanes e, h, and k), N-terminal domain (lanes c, f, and i), or C-terminal domain (lanes d, g, and j) were incubated in the presence of Hsp90 (c–h) or alone in the presence of zero-length (EDC) cross-linker (lanes i–l) in the presence of 1× (c–e) or 2× (f–h) cross-linker as described in Materials and Methods. No cross-linking was observed in any incubation that only contained full-length, or N- and C-terminal domains of Aha1. Incubation of Hsp90 with EDC revealed higher order oligomers reflecting its known C-terminal and N-terminal interaction motifs. Black arrowheads show the Aha1–Hsp90 complexes captured by the EDC cross-linker. The position of the Hsp90 is indicated by the asterisk. Migration of indicated molecular weight markers are shown to the left of the panel. (B) Hsp90 peptides cross-linked to Aha1 are colored in yellow (also see Table 2). Aha1 peptide cross-linked to Hsp90 is shown in yellow on the structure of the C-terminal domain of the protein (PBD: 1X53). Protected residues identified by footprinting in the Hsp90 N-terminal and middle domain (see Figure 3) are shown in red; Aha1 C-terminal residues protected from modification in Aha1–Hsp90 complexes (see Figure 3) are shown in orange. (C) Full-length Aha1 was modified with ANB-NOS, incubated with unlabeled Hsp90, and complexes were photocross-linked for 1 min (lane c), 2 min (lane d), and 3 min (lane e) and separated using a 4–12% gradient gel as described in Materials and Methods. Higher order oligomers are not observed upon incubation of unmodified Hsp90 (lane a) alone or cross-linker modified Aha1 (lane b) alone. The potential migration position of an Hsp90 dimer based on molecular weight markers (left) is shown by the asterisk (see A). Higher molecular weight bands >220 kDa represent complexes of Aha1 with two or more monomers of Hsp90.
To further characterize the interaction of Aha1 and Hsp90, we mapped the chemically cross-linked sites by using mass spectrometry. In brief, MS/MS were generated from trypsin-digested non–cross-linked and cross-linked samples from incubations containing full-length Aha1 or N- and C-terminal fragments incubated with Hsp90 (see Materials and Methods). To identify cross-linked residues, we used the schema outlined in Supplemental Fig. S2. MS/MS from non–cross-linked samples were searched against the EBI IPI human database, version 3.23, with its reverse decoy to identify peptides that can be obtained from the individual proteins (e.g., Aha1 or Hsp90). The identified Aha1 peptides and Hsp90 peptides were then cross-linked in silico to create a database of artificial cross-linked peptides. We only used the experimentally identified peptides to create the artificial cross-linked peptides database because the cross-linked peptides from experimentally identified non–cross-linked peptides are probably a more accurate reflection of peptides relevant to the cross-linking experiment. MS/MS from cross-linked samples were also searched against the EBI-IPI human database. MS/MS matched to the database were subtracted from the data set. The remaining MS/MS were then searched against the EBI-IPI human database plus the artificial cross-linked peptides database with their reverse decoys for the discovery of cross-linked peptides. These experiments were performed in the absence of presence of ATP nucleotide to access all possible intermediate folding states, or in the presence of ADP or AMP-PNP, to favor populating the Hsp90 dimer to the open or closed states, respectively.
In the absence of nucleotide using either low-ionic strength buffer (25 mM HEPES, pH 7.4) to favor the interactions between Hsp90 and Aha1 (Richter et al., 2008) or in a physiologically relevant ionic strength (50 mM HEPES, pH 7.4, and 100 mM NaCl) to minimize nonspecific charge–charge interactions that might occur in the low ionic strength buffer, we found that only one binding site in Aha1 is the preferred site for cross-linking given prominence of this peptide in both MS/MS analyses (Table 2 and Figure 4B). This site involves the C terminus of Aha1 (residues 277-289; 275FKSWPEGHFATTIL289) with the N terminus of Hsp90 (residues 70-82; 70ELKIDIIPNPQER82). Identical results were obtained in the presence of ATP (Table 2), demonstrating that consistent with previous results (Richter et al., 2008), the interaction of Aha1 with Hsp90 occurs with similar efficiencies in the presence or absence of nucleotide. In the absence of nucleotide, we also detected a second cross-linking event between residues 277-289 (275FKSWPEGHFATTIL289) and a peptide (residues 320-329; 320HFSVESGQLEF329) found in the middle domain of Hsp90, suggesting additional conformational flexibility. In contrast, when cross-linking was performed at physiological salt in the presence of ADP to stabilize the open state, we observed no cross-linking (Table 2), suggesting that interaction of Hsp90 with Aha1 is more restricted in an ADP-conformation favored incubation condition. These results provide an important control for specificity of interactions observed in the presence of absence of ATP.
Table 2.
Cross-linked peptides
| Experiment | Aha1-Hsp90 cross-linked peptides | No. of copies observed | Charge state | Xcorra score |
|---|---|---|---|---|
| No nucleotide | FKSWPEGHFATITL_ELKIDIIPNNPQER | 16 | 2 | 4.1 |
| SWPEGHFATITL_HFSVEGQLEFR | 4 | 2 | 4.1 | |
| ADP | NDb | |||
| ATP | FKSWPEGHFATITL_ELKID | 11 | 2 | 2.8 |
| AMP-PNP | SWPEGHFATITL_ELKID | 8 | 2 | 2.6 |
a SEQUEST cross correlation coefficient.
b None detected.
We performed an additional cross-linking experiment to analyze whether we gain or lose interactions between full-length Aha1 and Hsp90 by incubation in the presence of the nonhydrolysable analogue AMP-PNP. When AMP-PNP is added at the beginning of the incubation of Aha1 and Hsp90, we observe poor and variable recovery of cross-linked species, suggesting that rapid recruitment of the analogue leading to the closed state may preclude interaction of Aha1 and Hsp90. However, when AMP-PNP is added shortly after reincubation of Aha1 and Hsp90 (5–10 min), we recover the identical C-terminal Aha1 and N-terminal Hsp90 cross-linked peptide observed in the presence or absence of ATP (Table 2). Under these conditions we did not detect interactions between the N terminus of Aha1 and the peptide in the middle domain of Hsp90 found in the nucleotide-free condition. These results suggest that Aha1 must interact with Hsp90 in a specific manner as it actively cycles from the open to closed states with Aha1 becoming trapped in a partially or fully closed state in the presence of AMP-PNP. In summary, the limited number of cross-linked species detected had strong overlap with the regions protected by the interaction of Aha1 and Hsp90 using molecular foot printing, supporting the conclusion that Aha1 and Hsp90 have a unique two-domain interaction.
Aha1 Bridges the Hsp90 Dimmer to Facilitate ATP Hydrolysis
The presence of two discrete binding sites for full-length Aha1 on Hsp90 poses several models. One possibility is that Aha1 binds along the surface of one Hsp90 monomer (‘cis'-binding); alternatively, Aha1 may bind across the Hsp90 dimmer interface (‘trans'-binding). To address this question; we performed an additional cross-linking experiment using bifunctional cross-linkers. In brief, we labeled Aha1 with ANB-NOS. This cross-linker has an aminoreactive moiety that can be used to modify surface Lys residues and a UV-activated group that can react nonspecifically with side chains in proximity. Using this reagent, we mildly derivatized Aha1 under substoichiometric labeling conditions to retain its surface properties. After quenching of the reaction and removal of the unreacted ANB-NOS, Aha1 was incubated with Hsp90, and complexes were UV irradiated to cross-link Aha1 to Hsp90 with the photo-reactive moiety of ANB-NOS followed by SDS-PAGE. Because Hsp90 has not been labeled, and as expected, we only recover Hsp90 monomers (Figure 4C). If one molecule of Aha1 binds to a Hsp90 monomer (∼90 kDa), we would expect to observe cis-cross–linked species larger than the Hsp90 monomer (∼130 kDa) but always smaller than the migration of the Hsp90 dimer (∼180 kDa). These species were not observed. If, in contrast, Aha1 binds across the Hsp90 dimer interface in trans, we would expect to see trans-cross–linked species larger (>220 kDa) than that observed for the migration of the Hsp90 dimer (Figure 4C, asterisk). Because Hsp90 is not modified with the cross-linking agent, the only way that species larger than the Hsp90 dimer can be generated are through Aha1 interactions. Indeed, we were able to identify a number of high-molecular-weight bands (>220 kDa) that represent Aha1-cross-linked Hsp90 species migrating more slowly than the Hsp90 dimer (Figure 4C). It remains possible that these slower migrating cross-linked bands reflect an unusual extended state in response to denaturation by the sample buffer before SDS-PAGE. We consider this unlikely given the very low stoichiometry of the cross-linking agent used to covalently modify Aha1; that Aha1 is well folded and compact under these conditions; and that Hsp90 is a native, unmodified protein.
Our combined results demonstrate that the two distinct interaction sites between Aha1 and Hsp90 detected by protection (Figure 3) and cross-linking experiments (Figure 4) reveal that Aha1 functions to bridge the dimer interface of Hsp90 with the N terminus of Aha1 bound to the middle domain of one Hsp90 monomer and the C terminus of Aha1 bound to the other Hsp90 monomer in the dimer pair to stabilize and/or promote ATP hydrolysis.
Overexpression of WT Aha1 Prevents WT-CFTR Folding and Export from the ER in a Dose-dependent Manner
In vitro interactions do not necessarily recapitulate interactions observed in the cell given the involvement of additional unknown factors, particularly in the complex cochaperone interactions with Hsp90. Moreover, the unknown stoichiometries involved in biological reactions often provide an additional level of complexity. For example, Hsp90 is present in large excess over Aha1 (>100-fold; see below), has additional cochaperones that assist Hsp90 in folding, and has numerous clients that it must maintain in a physiological functional state (Panaretou et al., 2002)(our unpublished data).
To address the impact of the interaction of Aha1 with Hsp90 as dimer-bridging molecule directing client folding in vivo, we turned to our previous work. This work focused on the role of WT Aha1 and Hsp90 in the stability of the ΔF508 variant of CFTR for trafficking through the exocytic pathway and delivery to the cell surface (Wang et al., 2006). Here, we demonstrated that small interfering RNA (siRNA) silencing of Aha1 expression reduced ERAD of ΔF508 and promoted conversion to band C and delivery to the cell surface (Wang et al., 2006). In contrast, overexpression of Aha1 destabilized both WT and ΔF508-CFTR and promoted ER-associated degradation, suggesting that the Hsp90 ATPase cycle regulated by Aha1 plays a critical role in CFTR folding (Wang et al., 2006). The mechanism responsible for these events remains unknown.
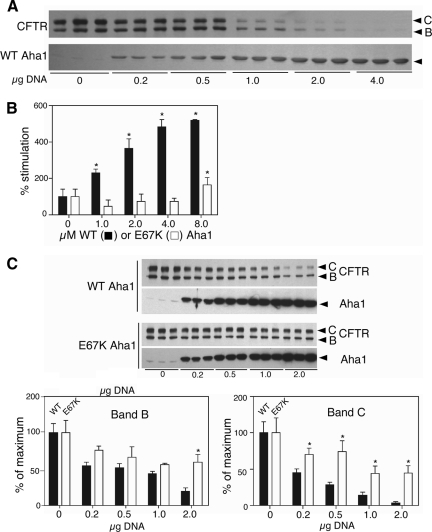
In the current study, we wanted to characterize the effect of Aha1 and N- and C-terminal mutants that disrupt its interaction with Hsp90 on the stability and trafficking of WT-CFTR as a biological measure of domain function. For this purpose, we cotransfected WT-CFTR with increasing amounts of full-length Aha1 by using a vaccinia expression system in human embryonic kidney (HEK)293 cells (see Materials and Methods). Trafficking of CFTR to the cell surface can be readily measured by following the processing of CFTR from the band B, high-mannose glycoform localized to the ER during nascent synthesis by ER-bound ribosomes, to the more slowly migrating band C glycoform that is generated by Golgi-associated enzymes to form complex sugar-containing species before delivery of CFTR to the cell surface (Figure 5A). Similar to results we observed for ΔF508, increasing amounts of WT Aha1 destabilized WT-CFTR in the ER. This is evident by the observed reduced levels of band B. Moreover, increasing WT Aha1 Aha1 interfered with export from the ER to Golgi as indicated by the partial loss of band C even at the lowest levels of expression tested (Figure 5A).
Figure 5.
Effect of Aha1 mutants on CFTR folding and trafficking. (A) Dose-dependent destabilization of CFTR overexpression with the indicated amount of Aha1 plasmid. Western blotting performed with indicated antibody to CFTR (top) or Aha1 (bottom). The three identical samples illustrate the response of cells to overexpression in three separate experiments. (B) Hsp90 ATPase assay using wild-type (black bars) and E67K (white bars) of Aha1 with the indicated amount (μg) of Aha1 as described in Materials and Methods. Asterisks denote statistically significant increase in ATPase activity over Hsp90 alone (two-tailed t test, p < 0.05). (C) Coexpression of increasing amounts of Aha1 (WT or E67K) with WT-CFTR shows that E67K is impaired in its ability to destabilize CFTR. Western blotting performed with indicated antibody to CFTR or Aha1 (top). Quantification of CFTR destabilization by WT (black bars) and E67K (white bars) Aha1 overexpression (bottom). Asterisks denote statistically significant difference between band B or C levels in cells expressing E67K or WT-Aha1 (two-tailed t test, p < 0.05). The three identical samples illustrate the response of cells to overexpression in three separate experiments.
The observation that the steady-state level of band C preceded the loss of band B restricted to ER in response to increasing Aha1 (Figure 5A) is consistent with the interpretation that the Aha1-sensitive Hsp90-dependent step(s) required for the stability of WT-CFTR in the ER is required to generate a folding competent conformation for export (Wang et al., 2004, 2008). Using absolute quantification (single ion reaction monitoring mass spectrometry; Janecki et al., 2007) approaches, we determined that Hsp90 constitutes approximately ∼1–2% of the total protein content in the lysates prepared from the HEK293 cells, whereas Aha1 is a minor pool (<0.01%). Quantitative Western blotting showed that band C CFTR loss could be detected with as little as approximately threefold overexpression (the lowest plasmid concentration tested; Figure 5A). This represents a change in stoichiometry with Hsp90 from ∼1:100 to ∼1:30 but was dose dependent and much stronger with increased expression of Aha1. These results likely reflect competition Aha1 with a large pool of dynamic Aha1–Hsp90–client complexes. These experiments suggest that increased levels of Aha1 relative to its endogenous concentration impair the ability of even WT-CFTR to fold into an export competent state that is protected from ERAD.
The N-Terminal Domain of Aha1 Is Critical for Destabilization of WT CFTR Folding
Because the relationship between the in vivo properties of WT Aha1 and its Hsp90 ATPase activity are controversial and the mechanism(s) facilitating client folding remain largely unknown, we tested whether destabilization of WT-CFTR folding by Aha1 overexpression was dependent on Hsp90 by generating Aha1 mutants. We were particularly interested in whether binding and/or ATPase activity were required for stabilization of WT CFTR folding. Whereas loss of binding would be predicted to result in loss of interaction Hsp90 and hence, loss of ATPase stimulating activity, it is unclear whether binding is sufficient and that the ATPase-stimulating activity conferred by the C-terminal domain is, in addition, required in CFTR folding.
A previous study identified a mutation in the N-terminal domain of yeast Hch1p (D53K) that impaired its ability to stimulate yeast Hsp90 (Meyer et al., 2004). We made the equivalent mutation in human Aha1 (E67K) and tested for stimulation of Hsp90 ATPase activity. No stimulation was observed, similar to that observed previously for yeast D53K (Figure 5B). Using SPR, we found that the E67K mutant had an extremely slow association rate (at least 15-fold slower than WT) and did not reach equilibrium binding under the range of conditions and concentrations tested (1–30 μM; our unpublished data), limiting our ability to accurately assess the kinetics of binding of the E67K mutant to Hsp90. However, it is clear that binding is severely impaired and this result explains the inability of the E67K mutant of Aha1 to stimulate the Hsp90 ATPase activity. We then tested this mutant by examining its effect on WT-CFTR export from the ER and trafficking to the cell surface. Here, we found that overexpression of the E67K mutant, compared with WT Aha1, was less efficient in promoting WT-CFTR degradation of band B at the highest expression level tested (Figure 5C, left). We observed statistically significant protection against the loss band C at the lowest concentration tested (Figure 5C, right). These results demonstrate that interaction of Aha1 through the N-terminal domain with Hsp90 is at least one critical event required for promoting efficient folding and export of WT-CFTR from the ER.
Mutations in the C Terminus of Aha1 Impair Hsp90 ATPase Stimulation without Significantly Altering Binding to Hsp90
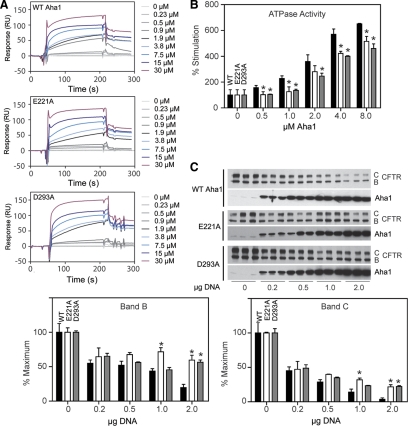
To address the role of the C-terminal ATPase activation domain of Aha1 on WT-CFTR export, we made a series of mutants based on primary sequence alignments and conserved residues between Aha1 proteins from different species (Figure 1D and Supplemental Figure S1). Several of these mutants are found in regions that are likely to form the interaction region of the C-terminal domain of Aha1 with the N terminus of Hsp90 (Figures 3 and 4). Many mutants did not express well in mammalian cells, probably owing to the loss of critical side chains necessary for folding based on the structure of Aha1 C- and N-terminal domains. However, several mutants not only seemed to be expressed to levels within 50% or greater of wild-type Aha1 but were markedly impaired in their ability to destabilize WT-CFTR (Supplemental Figure S3A) and ΔF508-CFTR (Supplemental Figure S3B). Notably, these included C207S, E221A, E267K, D293A, E297A, T298A, and E313A, residues that may contribute to the stability of the Aha1 interaction with Hsp90 (Figures 3 and 4B).
Mutants impaired in their ability to destabilize CFTR were selected for purification from E. coli for further testing in vitro. Only the E221A and D293A mutants were soluble when purified, as many of the mutants either did not express well in E. coli or rapidly aggregated into higher molecular weight species during or after purification. Unlike the E67K mutant, both the E221A and D293A Aha1 mutants were capable of interacting with Hsp90 by SPR (Figure 6A) in a manner similar to that observed for wild-type Aha1 (0.89 and 0.44 μM, respectively; see Supplemental Table S1 for Kon and Koff values). Interestingly, both of these mutants were partially impaired in Hsp90 ATPase stimulation (Figure 6B). To more carefully assess their ability to influence WT-CFTR export, we coexpressed increasing amounts of E221A and D293A with WT-CFTR in HEK293 cells (Figure 6C). At an ∼5- to 10-fold increased expression relative to endogenous Aha1 (∼1:10, Aha1:Hsp90), these mutants were statistically significantly impaired compared with WT Aha1 in their ability to prevent CFTR folding and export.
Figure 6.
Analysis of E221A and D293A mutants in vitro. (A) SPR analysis of the indicated amount of E221A and D293A mutants of Aha1 with Hsp90 as described in Materials and Methods. (B) Hsp90 ATPase assay with the indicated amount (micromolar) of WT (black bars), E221A (white bars), and D293A (gray bars) Aha1. Asterisks denote statistically significant differences between WT and mutants at the same concentration (analysis of variance [ANOVA], p < 0.05). (C) Coexpression of increasing amounts of WT, E221A, and D293A Aha1 with WT-CFTR. Western blotting performed with indicated antibody to CFTR or Aha1 (top). Quantification of CFTR destabilization in response to WT (black bars), E221A (white bars), and D293A (gray bars) Aha1 overexpression (bottom). Asterisks denote statistically significant differences between band B or C levels in cells expressing E221A and WT Aha1, or D293A and WT Aha1 (ANOVA, p < 0.05).
The combined data suggest that the activity of both the N- and C-terminal domains of Aha1 in binding Hsp90 or stimulating the ATPase activity of Hsp90, respectively, are important for Hsp90 regulated folding and export of CFTR. These interactions are probably aggravated by the energetically destabilizing Phe 508 deletion in ΔF508 CFTR leading to cystic fibrosis.
DISCUSSION
An important question for understanding the Hsp90 system in client management in vivo is the role of Aha1 in regulating Hsp90 ATPase activity. We have presented evidence that Aha1 requires both the N- and C-terminal domains for full activation of Hsp90. Moreover, we have demonstrated that the C-terminal domain of Aha1 binds Hsp90 and have localized the binding site on the Hsp90 N-terminal domain using two novel structural approaches, cross-linking and footprinting, in conjunction with mass spectrometry and MudPIT to detect highly dynamic interactions occurring under physiological conditions. Intriguingly, we have now provided evidence that binding of Aha1 probably occurs in trans across the dimer interface, raising the possibility that Aha1 stimulates ATPase activity by modulating the structural interface of the Hsp90 dimer clamp to promote client folding (Krukenberg et al., 2008; Neckers et al., 2009). In support of this model, we have provided evidence that a two domain interaction is critical for the biological function of Aha1 in regulating Hsp90-dependent folding and trafficking of CFTR in vivo, a client whose defective variant fold is responsible for cystic fibrosis (Wang et al., 2006).
Aha1 Bridges the Hsp90 Dimer Interface
The striking overlap of data from two different approaches, molecular footprinting and cross-linking, performed in solution under near native conditions, now implicates the importance of both the N- and C-terminal domains of Aha1 in recognition and regulation of Hsp90 activity. Given that cross-linking detected only a limited number of peptides, our data suggest a precise interaction of Aha1 with Hsp90 during client folding. Moreover, incubation of Aha1-containing a zero-length, photoactivatable cross-linker with untagged Hsp90 led to the appearance of higher order oligomers consistent with the interpretation the Aha1 bridges the Hsp90 dimer interface. Our evidence suggests that this complex has at least 1 Aha1 monomer per Hsp90 dimer, although we cannot rule out the possibility that each dimer may contain two Aha1 monomers.
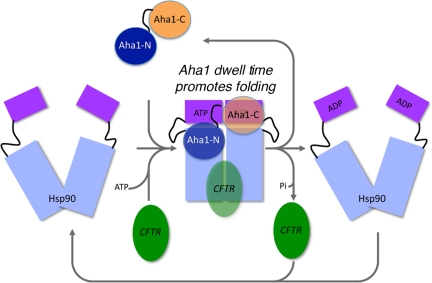
Our results now provide a model for the role of Aha1 C-terminal domain in the stimulation of Hsp90 ATPase activity during client folding events. Biochemical and structural studies currently support the view that the Hsp90 conformational cycle is coupled to the hydrolysis of ATP (Pearl and Prodromou, 2006; Krukenberg et al., 2008; Richter et al., 2008; Hessling et al., 2009; Mickler et al., 2009; Neckers et al., 2009; Vaughan et al., 2009). Given our surprising results that suggest that full-length Aha1 can bridge the dimer interface, we now propose that the C terminus of Aha1 can help prime dimerization of the Hsp90 N-terminal domains, an event likely critical for either stabilizing client binding and/or promoting a folding step(s) thought to be sensitive to ATP-binding and hydrolysis (Richter et al., 2008; Vaughan et al., 2009). In this view, association of Aha1 with the Hsp90 dimer occurs via two separate interactions- the N-terminal domain of Aha1 binding to the middle domain of Hsp90 (Meyer et al., 2004) and the C-terminal domain of the same Aha1 molecule binding in trans to the adjacent Hsp90 N terminus (shown here; Figure 7). This is consistent with the binding of yeast Aha1 to the yeast Hsp90 dimer by Buchner and colleagues using biochemical and structural approaches (Retzlaff et al., 2010). Based on the effects of mutants in both the N-terminal and C-terminal domains of Aha1 on Hsp90 association or ATPase activity, respectively, we suggest that trans-binding of full-length Aha1 stabilizes the catalytic ATP-bound intermediate of Hsp90 and thereby accelerates ATP hydrolysis (Figure 7). Aha1-dependent ATP hydrolysis by Hsp90 would relieve the molecular embrace formed at the N-terminal dimer interface, releasing the client in timely manner for subsequent step(s) in its folding pathway (Shiau et al., 2006; Cunningham et al., 2008).
Figure 7.
Aha1 functions as molecular referee to promote Hsp90 ATPase activity regulating client folding and WT/ΔF508 CFTR trafficking. Aha1 binds in trans to the dynamically cycling Hsp90 dimer. Binding of the N-terminal domain of Aha1 occurs to the middle domain of Hsp90. The C-terminal domain of Aha1 binds to the N terminus of Hsp90 to promote ATP hydrolysis by stabilization of the N-terminal dimer interface. We propose that the differential dwell time (Mickler et al., 2009) of the client-Hsp90 complex in response to the Aha1 ATPase activating activity is a critical event in dictating success or failure of folding of WT and ΔF508 CFTR, and other clients by the Hsp90 chaperone–cochaperone system. The dwell time for required for proper client folding is probably a key determinant of the activity of the proteostasis program.
Why is an ATPase-activating protein such as Aha1 essential for Hsp90 activity? Bacterial HtpG (the homologue of Hsp90) has a high intrinsic ATPase and lacks the unstructured amino acid linker domain between the N-terminal ATPase and the middle domain of this molecule (Shiau et al., 2006; Krukenberg et al., 2008). In contrast, the presence of a flexible hinge region in eukaryotic Hsp90 may make the trans positioning of the N-terminal and the middle domains entropically less favorable resulting in the observed low intrinsic rate of ATP hydrolysis (Meyer et al., 2003). Although the intrinsic basal rate of hydrolysis of yeast and mammalian Hsp90s differ, yeast being more active, this could reflect evolutionary fine-tuning of eukaryote specific folding pathways that require additional modulation by Aha1 to achieve a functional state. Thus, stabilization of N-terminal region dimerization by Aha1 may be used to regulate the intrinsic activation of eukaryotic Hsp90 N-terminal dimerization to expand the diversity of folds chaperoned by the Hsp90 ATPase cycle to direct the proteostasis program. This is an issue that is particularly acute in higher eukaryotes given the extreme cellular diversity of the folding problem. Here, variable timing of ATPase activity may facilitate folding of kinetically and thermodynamically divergence folding intermediates. This conclusion is supported by the increasing complexity of the folding problem during evolution and by the marked increase in cochaperone components required for Hsp90-dependent function comparing prokaryotes with invertebrates and vertebrates (Morimoto and Cuervo, 2009).
Physiological Role of Aha1 as a Molecular Referee in Hsp90-dependent Client Folding In Vivo in Health and Disease
We have shown in vivo that by either reducing the binding of Aha1 to Hsp90 by mutation of the N-terminal domain, or by partial reduction of the ATPase-stimulating activity of Aha1 by mutation of the C-terminal domain, we reduce the inhibition of folding of ΔF508 and WT CFTR after overexpression of WT Aha1 in vivo. These results not only support the model whereby both the N- and C-terminal domains of Aha1 play critical roles in function in vivo but raise the possibility of a specific role for the Aha1 C-terminal domain in promoting ATPase activity independent of Hsp90 binding in vivo through the N-terminal domain. We propose the important concept that the client “dwell time” in the Hsp90 folding complex (Hessling et al., 2009; Mickler et al., 2009) in vivo probably needs to be kinetically coupled through the ATPase timer activity of the C-terminal domain of Aha1 to the specific energetics of client folding intermediates (Wang et al., 2006; Powers et al., 2009; Figure 7). Thus, we propose that Aha1 functions as “molecular referee” to gauge client-Hsp90 chaperone interactions to direct productive folding, an activity that could be similar to the role of Hsp40 and nucleotide exchange factors in regulating client folding by Hsp70 (Vos et al., 2008; Vembar et al., 2009).
Both N- and C-terminal mutants were dominant negative with as little as ∼10-fold overexpression relative to the endogenous WT Aha1 pool. This corresponds to ∼1:10 M ratio of Aha1 to the typical endogenous Hsp90 pool, suggesting a dynamic interaction in vivo. Thus, altering the kinetics of interaction of Hsp90 with either WT or ΔF508 CFTR folding intermediate(s) by regulating the kinetics of the Aha1 driven ATPase cycle may be an important feature of the folding reaction. One possibility is that endogenous levels of Aha1 ATPase activating activity in the CF lung kinetically predisposes a human Hs(c)p-40/70 bound ΔF508 CFTR folding intermediate(s) to be redirected to ERAD (Younger et al., 2006; Rosser et al., 2008). This conclusion is consistent with our observation that partial silencing of Aha1 expression can markedly stabilize ΔF508-CFTR folding and promote transport to the cell surface (Wang et al., 2006, 2008). As suggested by our mutational analysis of Aha1 function in vivo, a reduction of endogenous Aha1 levels by siRNA may potentially decrease ATP turnover in Hsp90 and thereby increase the client (ΔF508) dwell time (Mickler et al., 2009; (Figure 7). Increased dwell time may favor the generation of a mature Hs(c)p40/70/90/client complex through kinetic stabilization of the ΔF508-fold, thereby generating a substrate for ER export (Wang et al., 2006; Wiseman et al., 2007). The kinetics of turnover by the Hsp90 system may differ substantially between different clients and be dependent on interactions with additional cochaperone assistants such as p23 that seems to bind to the N-terminal domain of Hsp90 and inhibit its ATPase activity, yet is not competitive with Aha1 (Loo et al., 1998; Wang et al., 2006; Holmes et al., 2008). Thus, Aha1 in conjunction with other cochaperones could provide an exceptionally versatile platform in eukaryotes to facilitate protein folding by linking the unique kinetics and thermodynamics of Hsp90-dependent client folding intermediates with Aha1 regulated ATPase-activating activity (Powers et al., 2009).
Understanding how an evolutionarily conserved molecular referee such as Aha1, in conjunction with other folding modulators, facilitates Hsp90 ATPase activity will be critical for the development of Hsp90 as a target for the treatment of a large variety of diseases of proteostasis, including cystic fibrosis and cancer (Powers et al., 2009).
Supplementary Material
ACKNOWLEDGMENTS
We thank Sophie Jackson for the human Hsp90 clone. We thank Jason Schuman (GE Healthcare) for critical advice and help with interpretation of our SPR results. This is The Scripps Research Institute manuscript 5678. This study was supported by National Institutes of Health grants GM-42336 and DK-51870 (to W.E.B.) and HL-079442 (to J.R.Y.) and Cystic Fibrosis Foundation Postdoctoral Research fellowships to A. K., P.L.P., and B. L.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1017) on January 20, 2010.
REFERENCES
- Aridor M., Bannykh S. I., Rowe T., Balch W. E. Cargo can modulate COPII vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 1999;274:4389–4399. doi: 10.1074/jbc.274.7.4389. [DOI] [PubMed] [Google Scholar]
- Aridor M., Weissman J., Bannykh S., Nuoffer C., Balch W. E. Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Cunningham C. N., Krukenberg K. A., Agard D. A. Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J. Biol. Chem. 2008;283:21170–21178. doi: 10.1074/jbc.M800046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W., Cuthbert A. W. Post-translational disruption of the delta F508 cystic fibrosis transmembrane conductance regulator (CFTR)-molecular chaperone complex with geldanamycin stabilizes delta F508 CFTR in the rabbit reticulocyte lysate. J. Biol. Chem. 2000;275:37462–37468. doi: 10.1074/jbc.M006278200. [DOI] [PubMed] [Google Scholar]
- Harst A., Lin H., Obermann W. M. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem. J. 2005;387:789–796. doi: 10.1042/BJ20041283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessling M., Richter K., Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Holmes J. L., Sharp S. Y., Hobbs S., Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- Hutt D. M., Powers E. T., Balch W. E. The proteostasis boundary in misfolding diseases of membrane traffic. FEBS Lett. 2009;583:2639–2646. doi: 10.1016/j.febslet.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecki D. J., Bemis K. G., Tegeler T. J., Sanghani P. C., Zhai L., Hurley T. D., Bosron W. F., Wang M. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal. Biochem. 2007;369:18–26. doi: 10.1016/j.ab.2007.06.043. [DOI] [PubMed] [Google Scholar]
- Krukenberg K. A., Forster F., Rice L. M., Sali A., Agard D. A. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz G. P., Lin H., Harst A., Obermann W. M. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. H., Ventouras L. A., Lobbezoo B., Jackson S. E. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J. Mol. Biol. 2004;344:813–826. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. Structural and functional analysis of the middle segment of hsp 90, implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Meyer P., Prodromou C., Liao C., Hu B., Mark Roe S., Vaughan C. K., Vlasic I., Panaretou B., Piper P. W., Pearl L. H. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickler M., Hessling M., Ratzke C., Buchner J., Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I., Cuervo A. M. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L., Tsutsumi S., Mollapour M. Visualizing the twists and turns of a molecular chaperone. Nat. Struct. Mol. Biol. 2009;16:235–236. doi: 10.1038/nsmb0309-235. [DOI] [PubMed] [Google Scholar]
- Onuoha S. C., Coulstock E. T., Grossmann J. G., Jackson S. E. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J. Mol. Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Panaretou B., et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C., Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- Powers E., Morimoto R., Dillin A., Kelly J. W., Balch W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Ann. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Retzlaff M., Hagn F., Mitschke L., Hessling M., Gugel F., Kessler H., Buchner J. Assymetric activation of the Hsp90 dimer by its co-chaperone Aha1. Mol. Cell. 2010 doi: 10.1016/j.molcel.2010.01.006. in press. [DOI] [PubMed] [Google Scholar]
- Richter K., Soroka J., Skalniak L., Leskovar A., Hessling M., Reinstein J., Buchner J. Conserved conformational changes in the ATPase cycle of human Hsp90. J. Biol. Chem. 2008;283:17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- Riordan J. R. Assembly of functional CFTR chloride channels. Annu. Rev. Physiol. 2005;67:701–718. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- Riordan J. R. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- Rosser M. F., Grove D. E., Chen L., Cyr D. M. Assembly and misassembly of cystic fibrosis transmembrane conductance regulator: folding defects caused by deletion of F508 occur before and after the calnexin-dependent association of membrane spanning domain (MSD) 1 and MSD2. Mol. Biol. Cell. 2008;19:4570–4579. doi: 10.1091/mbc.E08-04-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands M. G., Newbatt Y. M., Prodromou C., Pearl L. H., Workman P., Aherne W. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal. Biochem. 2004;327:176–183. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Shiau A. K., Harris S. F., Southworth D. R., Agard D. A. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Skach W. R. CFTR: new members join the fold. Cell. 2006;127:673–675. doi: 10.1016/j.cell.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Sun F., et al. Chaperone displacement from mutant cystic fibrosis transmembrane conductance regulator restores its function in human airway epithelia. FASEB J. 2008;22:3255–3263. doi: 10.1096/fj.07-105338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taldone T., Gozman A., Maharaj R., Chiosis G. Targeting Hsp 90, small-molecule inhibitors and their clinical development. Curr. Opin. Pharmacol. 2008;8:370–374. doi: 10.1016/j.coph.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull E. L., Rosser M. F., Cyr D. M. The role of the UPS in cystic fibrosis. BMC Biochem. 2007;8(suppl 1):S11. doi: 10.1186/1471-2091-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan C. K., Piper P. W., Pearl L. H., Prodromou C. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. FEBS J. 2009;276:199–209. doi: 10.1111/j.1742-4658.2008.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Jin Y., Brodsky J. L., Hendershot L. M. The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions. J. Biol. Chem. 2009;284:32462–32471. doi: 10.1074/jbc.M109.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M. J., Hageman J., Carra S., Kampinga H. H. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- Wandinger S. K., Richter K., Buchner J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Wang X., Koulov A. V., Kellner W. A., Riordan J. R., Balch W. E. Chemical and biological folding contribute to temperature-sensitive DeltaF508 CFTR trafficking. Traffic. 2008;9:1878–1893. doi: 10.1111/j.1600-0854.2008.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Matteson J., An Y., Moyer B., Yoo J. S., Bannykh S., Wilson I. A., Riordan J. R., Balch W. E. COPII-dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di-acidic exit code. J. Cell Biol. 2004;167:65–74. doi: 10.1083/jcb.200401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Washburn M. P., Ulaszek R. R., Yates J. R., 3rd. Reproducibility of quantitative proteomic analyses of complex biological mixtures by multidimensional protein identification technology. Anal. Chem. 2003;75:5054–5061. doi: 10.1021/ac034120b. [DOI] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D., Yates J. R., 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wiseman R. L., Powers E. T., Buxbaum J. N., Kelly J. W., Balch W. E. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.