Faithful meiotic chromosome segregation requires pairing, synapsis and recombination of homologous chromosomes. In mammals, chromosomal non-disjunction increases with age. A mutation in Caenorhabditis elegans him-19 mimics these age-dependent chromosome segregation defects and might therefore further our understanding of this phenomenon.
Abstract
From a screen for meiotic Caenorhabditis elegans mutants based on high incidence of males, we identified a novel gene, him-19, with multiple functions in prophase of meiosis I. Mutant him-19(jf6) animals show a reduction in pairing of homologous chromosomes and subsequent bivalent formation. Consistently, synaptonemal complex formation is spatially restricted and possibly involves nonhomologous chromosomes. Also, foci of the recombination protein RAD-51 occur delayed or cease altogether. Ultimately, mutation of him-19 leads to chromosome missegregation and reduced offspring viability. The observed defects suggest that HIM-19 is important for both homology recognition and formation of meiotic DNA double-strand breaks. It therefore seems to be engaged in an early meiotic event, resembling in this respect the regulator kinase CHK-2. Most astonishingly, him-19(jf6) hermaphrodites display worsening of phenotypes with increasing age, whereas defects are more severe in female than in male meiosis. This finding is consistent with depletion of a him-19-dependent factor during the production of oocytes. Further characterization of him-19 could contribute to our understanding of age-dependent meiotic defects in humans.
INTRODUCTION
Meiosis is a specialized cell division used by diploid organisms to generate haploid gametes. In a series of coordinated events during prophase I, chromosomes find their correct (homologous) partners, form the synaptonemal complex (SC) between them to stabilize homologous pairing, and undergo meiotic recombination (Petronczki et al., 2003). In most organisms, homologous chromosomes are brought together by a DNA double-strand break (DSB)-dependent mechanism (Peoples-Holst and Burgess, 2005; Zickler, 2006). Caenorhabditis elegans, in contrast, does not rely on DSBs to achieve homologous chromosome pairing and SC assembly (Dernburg et al., 1998). Chromosomal pairing centers (PCs), located near one end of each chromosome, promote both homology recognition and synapsis (MacQueen et al., 2005; Phillips et al., 2005, 2009; Phillips and Dernburg, 2006). The SC consists of lateral elements containing HIM-3, HTP-1, HTP-2, and HTP-3 (Couteau et al., 2004; Couteau and Zetka, 2005; Goodyer et al., 2008; Martinez-Perez et al., 2008) and a central element consisting of SYP-1, -2, -3, and -4 (Colaiacovo et al., 2003; Smolikov et al., 2007a,b, 2009). It stabilizes pairing of homologous chromosomes after they have found the correct partner. However, the high tendency of the SC to self-assemble irrespective of homology requires tight regulation of this process to avoid locking with the wrong partner (Loidl, 1994; Zickler and Kleckner, 1999; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Penkner et al., 2007; Smolikov et al., 2007b). Pairing centers and the associated ZIM proteins (such as ZIM-1, -2, and -3 and HIM-8) have a proofreading ability to promote synapsis only when the matching chromosome is found (MacQueen et al., 2005; Phillips et al., 2005; Phillips and Dernburg, 2006). The SC stabilizes juxtapositioning of chromosomes undergoing crossing over and ensures the maturation of crossovers into chiasmata. The chiasmata, in turn, provide the physical connections that allow homologous chromosomes to segregate faithfully (Petronczki et al., 2003).
Defects in meiosis can result in aneuploidy, a frequent cause of reproductive failure and congenital birth defects. In humans, strong correlations between maternal age and occurrence of aneuploidies, notably trisomies are found. How maternal age increases aneuploidy is not fully elucidated, but it seems likely that there are multiple causes of human age-related nondisjunction at multiple stages during oocyte development. Weakened cohesion, a failure in checkpoint control, altered crossover patterns, abnormal cell cycle control, and a defective spindle assembly checkpoint have been suggested to contribute to aging effects (Hunt and Hassold, 2008; Hassold and Hunt, 2009). Despite the short life span of the metazoa Drosophila and C. elegans, deterioration of the fidelity of meiotic segregation with female age has been observed and possibly models the age-dependent meiotic defects in human females (Rose and Baillie, 1979; Andux and Ellis, 2008; Subramanian and Bickel, 2008).
Here, we describe him-19(jf6), a mutation that shows age-dependent aggravation of meiotic defects in female C. elegans. In him-19, homologue recognition, recombination and SC formation is more affected in females than in males, and the mutant phenotype is more severe in older worms. Interestingly, an SC polymerization defect displayed by the mutant is partially rescued by γ-radiation, which extends the recent discovery of a cryptic DSB-dependent SC polymerization mechanism in C. elegans (Smolikov et al., 2008).
MATERIALS AND METHODS
Worm Strains and Culture Conditions
All C. elegans strains were cultured under standard conditions at 20°C (Brenner, 1974). N2 (Bristol) was used as a wild-type background; CB4856 (Hawaiian) was used for single-nucleotide polymorphism (SNP) mapping, and TY2441 yls34 (Pxol-1::GFP+rol-6 (pRF4)) was used as the screening strain. The following genotypes were used in this work: LGI: prom-1 (ok1140), him-19(jf6), him-19(tm3538); LGIV: spo-11(ok79), spo-11(me44), nT1[unc-?(n754) let-? qIs50](IV;V), fem-3(e1996); LGV: syp-2(ok307), chk-2(gk212), chk-2(me64); and Df: hDf10 dpy-5(e61), unc-29(e403)/hT2[dpy-18(h662)] I; +/hT2[bli-4(e937)] III.
Characterization of the him-19(jf6) Allele
The him-19(jf6) allele was recovered from an ethane methyl sulfonate (EMS) mutagenesis screen performed with the strain TY2441. After EMS mutagenesis, 5000 F1 individuals were single plated, and F2 hermaphrodites were scored for the occurrence of green eggs (expressing Pxol-1::GFP) (Kelly et al., 2000). Meiotic mutants were recovered as heterozygous animals. By using SNP mapping, Y 95B8 was located on LGI left arm. Candidate open reading frames (ORFs) were sequenced from cDNAs. Y95B8A.11 in the homozygous jf6 did not produce the same polymerase chain reaction (PCR) product size as the wild type. Sequencing of the genomic sequence revealed a point mutation in the intron. Injection of the rescue construct could increase the viability of the homozygous mutant in the first day of its reproduction span by ∼30%. him-19(jf6) and him-19(tm3538) can be maintained in a homozygous stock. The two alleles of him-19 do not complement.
Cosuppression
Expression of ORF Y95B8A.11 was inhibited by transgene-mediated cosuppression (Dernburg et al., 2000). A PCR product comprising the gene locus of him-19 was coinjected with the rol-6(su1006) marker. Offspring expressing the roller phenotype were selected, and a line showing a meiotic defect was established.
cDNA Preparation and Analysis
Total RNA was isolated from adult hermaphrodites using TRIzol Reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. mRNA purification was performed with Dynabeads Oligo (dT)25 (Invitrogen), following the manufacturer's instructions.
cDNA libraries were made with RETROscript kit (Ambion, Austin, TX). To test the expression of Y95B8A.11 in wild type and him-19(jf6) mutants, primer pairs MJ1976 5′-ATA CGC AGA ACG CCG TGA AC-3′ and MJ1979 5′-GAT TCG ATC CCT CGA CCA TTC-3′ were used.
Cytological Preparation of Gonads
Hermaphrodites were cut open to release the gonads in 5 μl of 1× phosphate-buffered saline (PBS) on a microscope slide and fixed by addition of an equal volume of 7.4% formaldehyde. The drop was immediately covered with a coverslip, and gentle pressure was applied. The coverslip was removed after freezing preparations in liquid nitrogen and slides were then transferred to methanol. For staining of chromatin and chromosomes, slides were mounted in Vectashield anti-fading medium (Vector Laboratories, Burlingame, CA) containing 2 μg/ml 4,6-diamidino-2-phenylindole (DAPI).
Immunostaining
For immunostaining, gonads prepared in 1× PBS on a microscope slide were fixed in a series of methanol, methanol:acetone (1:1), and acetone for 5 min each at −20°C. Then, they were immediately transferred to 1× PBS, 0.1% Tween 20 (PBS-T) without drying. Preparations were transferred twice to fresh PBS-T for 5 min and blocked with 3% bovine serum albumin (BSA) in 1× PBS for 30 min at 37°C in a humid chamber. Primary antibody was incubated overnight at 4°C in a moist chamber. Antibodies were diluted in 1× PBS and 3% BSA. After three washes in 1× PBS-T, secondary antibodies were applied. After 60-min incubation, slides were washed and mounted in Vectashield supplemented with DAPI (see above).
Fluorescence In Situ Hybridization (FISH)
Pooled cosmid R119, and PCR-amplified 5S rDNA, were used as probes for the left end of chromosome I and the right arm of chromosome V, respectively (Pasierbek et al., 2001). Cosmid DNA was directly labeled with cyanine (Cy)3 by using the BioNick Labeling System (Invitrogen) according to the manufacturer's instructions. 5S rDNA was labeled by PCR with digoxigenin-11-dUTP during PCR amplification. Slides were dehydrated by incubation in increasing ethanol concentrations and air-dried overnight. Labeled probe DNA and cytological preparations were denatured separately before the probe was added to the slides for hybridization. The protocol is described in detail in Pasierbek et al. (2001). Hybridized digoxigenin-labeled probes were detected by secondary antibody, anti-digoxigenin fluorescein isothiocyanate (1:500). Cy3 (red)-labeled probes were detected directly. Slides were mounted in Vectashield containing 2 μg/ml DAPI.
Microscopy and Evaluation
Preparations were examined with an Axioskop epifluorescence microscope (Carl Zeiss, Jena, Germany). Images were recorded with a charge-coupled device camera (Photometrics, Tucson, AZ). Evaluation of cytological phenotypes was performed in animals kept at 20°C 48 h post-larval stage 4 (L4) selection. For multicolor immunostaining and FISH pictures, monochrome images were captured separately for each emission wavelength. Three-dimensional (3D) stacks of images were taken (MetaVue software; Molecular Devices, Sunnyvale, CA), deconvolved (AutoDeblur software; AutoQuant Imaging, Troy, NY), and projected (Helicon Focus software; http://helicon.com.ua/heliconfocus/). FISH signals were scored as one signal when the distance between two spots was smaller than the size of the signal, whereas dots with distances larger than the size of the signal were counted as two signals. SYP-1 stretches were measured on deconvolved recorded images using the measuring tool of Photoshop (Adobe Systems, Mountain View, CA).
Irradiation Experiments
Strains 48 h post-L4 were treated with a dose of 5 krad of γ-rays from a 60Co source. Gonads for RAD-51 immunostaining were prepared 2 h after irradiation. Gonads for SYP-1 immunostaining were released 6 h after irradiation.
RESULTS
Identification of him-19 as a Novel Gene Required for Bivalent Formation
We performed a clonal genetic screen for mutants with homology recognition defects by using Pxol-1::GFP as a reporter for X chromosomal nondisjunction (Kelly et al., 2000). xol-1 is expressed only in male embryos. A higher incidence of male (Him) progeny is a consequence of X chromosome missegregation and can thus reflect various problems in meiotic divisions. Defects in homologue recognition, synapsis, recombination, chiasma stability or combinations thereof can lead to reduced chiasma formation and hence chromosomal nondisjunction. Mutants with green fluorescent protein (GFP)-positive embryos were selected and subsequently underwent cytological screening. him-19(jf6) was one of the recessive alleles found, showing meiotic defects segregating an elevated number of males, and microscopic analysis confirmed meiotic irregularities.
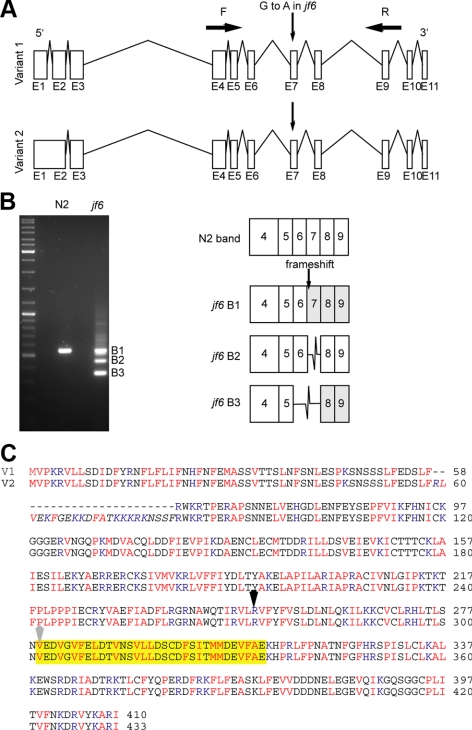
SNP mapping (Wicks et al., 2001) localized the mutation to the left end of chromosome I, and subsequent sequence analysis revealed that the jf6 allele contains a G-to-A transition at coordinate 898498 of a novel gene, encoded by the putative open reading frame Y95B8A.11. In contrast to the www.wormbase.org annotated predictions, our RT-PCR of the wild-type Y95B8A.11 revealed that the predicted ORF has two natural splice variants: one version encodes a putative protein of 413 amino acids (containing 11 exons), and the second variant derived from inclusion of intron 1 can encode 433 amino acids (containing 10 exons) (Figure 1, A and C). The two predicted proteins encoded by these two alternatively spliced mRNAs were termed HIM-19a and HIM-19b. The him-19(jf6) mutation is located at the last nucleotide of intron 6 and therefore destroys the splice acceptor site of the downstream exon 7. RT-PCR revealed several splice variants in him-19(jf6) worms. Sequencing of these PCR products indicated the use of a cryptic splice acceptor at position +2 in intron 7, skipping of exon 7, or joint skipping of exons 6 and 7. Use of the cryptic splice acceptor and skipping of exons 6 and 7 results in frame shifts of the predicted protein, leading to premature stops at 272 and 293 aa, respectively. Skipping of exon 7 removes predicted amino acids 279–311 of variant A (302–334 for variant B) but maintains the open reading frame (Figure 1, B and C).
Figure 1.
Splicing defects induced by the him-19(jf6) mutation. (A) Diagrams of the two naturally spliced variants of Y95B8A.11. Version 1 includes 11 exons. Version 2 has 10 exons. The G-to-A mutation in him-19(jf6) occurs before the exon 7. Primer pairs MJ1976 and MJ1979 were used to amplify the region from jf6 cDNA and to detect the splice variants the mutant. (B) Gel picture of the PCR products amplified with primers F and R (from A). The wild-type product matches the sequence of exon 4–9 consecutively. The three bands in the mutant lane correspond to the various splice versions affected by the disruption of the splice acceptor site. Although band 1 has the same size of the wild-type band, one nucleotide is missing in the exon 7. The next “AG” splice acceptor site is used for exon 7. The product of this band 1 will run into a premature stop codon. Band 2 and band 3 are missing the exon 7 and exon 6 and 7, respectively. (C) Amino acid alignment of the variants 1 and 2. The 23 amino acids unique in version 2 are italicized. Basic amino acids are highlighted in blue: H, K, and R. Hydrophobics in red: V, I, L, M, F, A, and P. The yellow box corresponds to exon 7. B2 in B is missing the 33 amino acids in the highlighted box and continues in frame. The black arrow indicates the start of exon 6 where B3 in Figure 1B and the knockout mutant's disruptions start. The gray arrow indicates where the B1 in Figure 1B starts its frame shift.
The mutant phenotype could be phenocopied by transgene-mediated gene silencing (cosuppression) and rescued by transformation with a cosmid carrying the him-19 sequence Y95B8A.11. Moreover, a deletion of Y95B8A.11, him-19(tm3538) failed to complement the jf6 allele as univalents were observed in him-19(tm3538)/him-19(jf6) heterozygotes and the deletion allele displays the same cytological phenotype as the jf6 allele (Supplemental Figure 1). Similarly, him-19(tm3538) when brought over the deficiency also showed univalent formation at diakinesis and a strongly reduced offspring. Together, this confirms that him-19 maps to Y95B8A.11.
The Search for him-19 Orthologues
Classic homology search tools failed to identify him-19 homologues outside C. elegans and Caenorhabditis briggsae, leaving the function of the encoded protein enigmatic. A recently published metastructure comparison, however, revealed similarities between HIM-19 and the helicase Dhh1(Konrat, 2009; Supplemental Figure 2). Dhh1 is a DEAD-box RNA helicase in the DDX6 family (Cheng et al., 2005). DOZI (development of zygote inhibited) in Plasmodium, another member of the DDX6 family, is found in a complex with mRNA species in cytoplasmic bodies of female, blood-stage gametocytes (Mair et al., 2006). Whether HIM-19 has RNA-helicase activity remains to be determined.
Defects in him-19 Aggravate with Age and Affect Female Meiosis More Severely
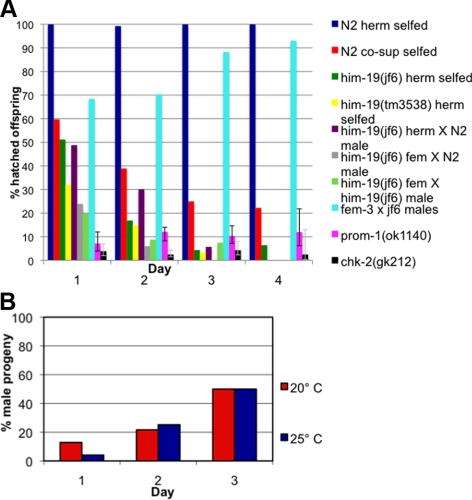
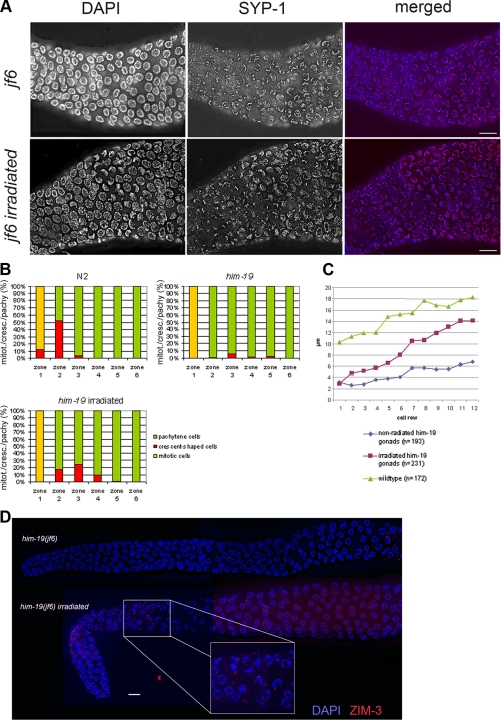
him-19(jf6) homozygous mutants can be maintained as a stable, homozygous strain. Hence, all studies presented here were performed on homozygous mutant offspring derived from homozygous mutant parents. Embryos hatch at a rate of 18.34% (n = 2094 eggs scored from 11 animals) with a high incidence of males occurring in the surviving progeny (19.01%, which is much higher than the 0.2% natural occurrence found in wild type; Hodgkin et al., 1979). Interestingly, the hatch rate decreases sharply with increasing age of the mother hermaphrodite. Although 51.20% (n = 334) of eggs laid on the first day of the reproduction period successfully hatched; only 16.87% (n = 1067) of eggs hatched from the second day brood; the percentage of hatched eggs dropped further to 4.34% (n = 553) on the third day; and only 6.43% (n = 140) of eggs from the fourth day survived (Figure 2A). As viability decreases, the number of males increases among the hatched animals, consistent with an overall increase in chromosome nondisjunction. On day 1, 12.87% of the progeny produced were males (n = 171); on day 2, 22.16% of the progeny produced were males (n = 176); and 50% of the progeny produced were males on day 3 (n = 24; Figure 2B). The deteriorating phenotype observed in the jf6 allele could also be observed in wild-type worms subjected to transgene mediated cosuppression of ORF Y95B8A.11 (Figure 2A). To test whether a similar age-dependent worsening of phenotypes is observed in other meiotic mutants, we scored viability of chk-2 and prom-1 mutants (MacQueen and Villeneuve, 2001; Jantsch et al., 2007). Unlike him-19(jf6), viability of prom-1(ok1140) offspring did not drop with maternal age (Figure 2A). Also, the chk-2(gk212) viability remained steadily low throughout the reproductive span of the mother (Figure 2A). In fact, the 95% confidence intervals might even suggest a slight increase in viability of the offspring with increasing maternal age, at least for prom-1.
Figure 2.
Aging phenotype of the him-19(jf6) mutant. (A) Graph illustrates the percentages of hatch rate (y-axis) of the assorted strains counted over the reproductive time span (x-axis). Fewer eggs hatched in the later broods in the mutant and in wild type worms injected with DNA encoding Y95B8A.11 leading to transgene-mediated gene silencing (cosuppression). Crosses of wild-type females with him-19(jf6) males are less affected than crosses performed with him-19(jf6) females or self crosses. Viability of the offspring of prom-1(ok1120) or chk-2(gk212) remained steadily low over the time frame investigated. Gray lines in bars for prom-1 and chk-2 indicate the 95% confidence intervals. Detailed numbers of brood size and hatch rates for each time point are shown in Supplemental Table 1. (B) Graph describes the percentage of male progeny (y-axis) in various broods (x-axis). At 20°C, the him-19(jf6) hermaphrodites had 12.87% of male progeny on day 1 (n = 171), 21.67% on day 2 (n = 180), and 50.0% on day 3 (n = 24). At 25°C, the him-19(jf6) hermaphrodites had 4.11% of male progeny on day 1 (n = 243), 25.22% on day 2 (n = 115), and 50.0% on day 3 (n = 6).
The comparably mild phenotype observed in young him-19(jf6) cannot be attributed to a maternal effect as all experiments were performed on homozygous offspring from homozygous hermaphrodites. The age-dependent reduction of hatched eggs scored from the him-19 mutant was repeatable over generations. This suggests a factor is replenished in each generation helping eggs laid at earlier times to do better.
C. elegans is a sequential hermaphrodite that first produces sperm cells and later oocytes for self-fertilization. Male germ cell development initiates before larval L4 stage, and sperm is stored in the spermathecae concomitantly with the initiation of ovulation (Schedl 1997). Given the age-dependent phenotype of him-19, we wanted to investigate whether the jf6 mutation affects spermatogenesis and oogenesis differently. To test this, him-19(jf6) hermaphrodites were crossed to wild-type males. Viability of the outcrossed progeny dropped similar to that of selfed him-19(jf6) animals indicating that the wild type sperm contributes little to overall viability of the offspring. To exclude the possibility that him-19(jf6) animals were self-fertilized, we repeated the experiment in a feminized background. fem-3(e1996) transforms XX hermaphrodites into fertile females that fail to produce sperm (Hodgkin, 1986). Such him-19(jf6)/him-19(jf6) (LG I); fem-3(e1996)/fem-3(e1996) (LG IV) females were mated to wild-type males, or homozygous him-19(jf6) males. Similar to hermaphrodite worms, viability of the outcrossed progeny dropped throughout the reproductive span of the mother (Figure 2A). Viable progeny of the female mutant (11.37%; 255 eggs scored from 4 animals) was lower than that of the hermaphrodite mutant irrespective of whether the cross was performed with N2 of jf6 males. This suggests that female mutants have a more penetrant phenotype than hermaphrodites. Control fem-3(e1996) females mated to wild-type males produced 100% viable progeny (748 eggs scored from 4 animals; data not shown). him-19(jf6) males mated to fem-3(e1996) showed a moderate reduction in viability of the progeny to 78.95% (1425 eggs scored from 7 animals). Thus, disruption of him-19 not only causes a phenotype that is worsening over time but also apparently affects the female germline more severely than the male germline.
him-19 Is Required for Homology Recognition
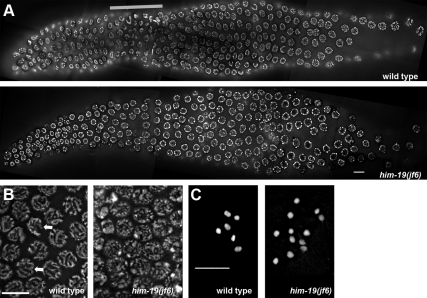
In C. elegans, zygotene (the stage in which synapsis of homologous chromosomes initiates) is characterized by crescent-shaped nuclei where chromatin is located to one side of the nucleus (Hubbard and Greenstein, 2005). This zone is typically referred to as the transition zone (TZ). In him-19(jf6), nuclei with clustered chromatin could only be found rarely (Figure 3A). If present, they were dispersed over the whole gonad. Moreover, him-19(jf6) nuclei of early pachytene lacked parallel chromatin tracks, suggesting unpaired chromosomes (Figure 3B). In late pachytene, however, partial parallel tracks are discernible (Supplemental Figure 5). Wild-type worms have six bivalents at diakinesis (Figure 3C). Consistent with the declining rate of hatched eggs, him-19(jf6) worms had six to nine DAPI-stained structures at diakinesis 17 h post-L4 (average, 6.25; n = 48). At 48 h post-L4, 10–12 (average, 11.33; n = 41) DAPI-stained structures were discernible (Figure 3C). Thus, the number of univalents at diakinesis increases as animals age (Table 1). Hence, all cytological analyses carried out in this study were done on hermaphrodites grown at 20°C, 48 h post-L4 selection. Two-day-old homozygous him-19(jf6) adults lacked a clear TZ. To test, whether entry into meiosis was affected in him-19(jf6), gonads were stained with antibodies against HTP-3, a chromosome axis component (Pasierbek et al., 2001; Goodyer et al., 2008). This marker protein showed normal association with chromosome tracks in cell rows 15–17 in wild-type and him-19(jf6) individuals (Supplemental Figure 3). Thus, the meiotic pathway is initiated normally in the mutant.
Figure 3.
Gonad organization in wild type and him-19(jf6) mutant. (A) DAPI-stained wild-type gonad and him-19(jf6) mutant gonad. A defined TZ zone is missing in the mutant gonad. Thick gray bar delineates position of the TZ in the wild type. (B) Parallel tracks of synapsed chromosomes are observed in the wild type (left, indicated by arrows) pachytene nuclei. him-19(jf6) (right) pachytene nuclei lack the parallel tracks. (C) Six bivalents are seen in wild-type diakinesis (left). Up to 12 univalents can be detected in the him-19(jf6) mutants (right). Bars, 10 μm.
Table 1.
Number of DAPI-stained structures at diakinesis in him-19(jf6), scored 17 and 48 h after L4a
| DAPI structure | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|
| 17 h (n = 48) | 77.08 | 12.50 | 8.33 | 2.08 | 0 | 0 | 0 |
| 48 h (n = 41) | 0 | 0 | 0 | 0 | 9.76 | 26.83 | 63.41 |
a Data are presented as percentages.
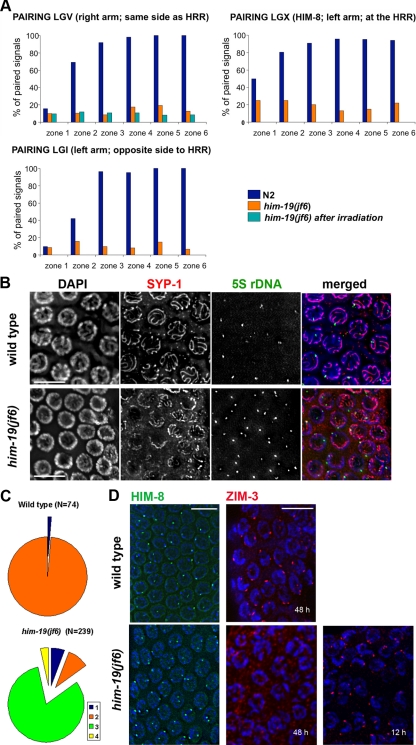
The lack of an organized TZ and the occurrence of univalents at diakinesis are typical features of chromosome pairing mutants. We therefore set out to examine homologous pairing in him-19(jf6) by FISH analysis. Two probes, one probe that detects chromosome I (on the opposite end to the PC) and one probe that detects chromosome V (same side as the PC) were used. For quantification of pairing, images of worm gonads were partitioned into six zones spanning from the mitotic distal tip to the end of the pachytene zone. In 2-day-old wild-type adults homologous pairing begins in zone 2 and synapsis occurs gradually after zone 2, resulting in only one FISH signal per cell in prophase I. In the him-19(jf6), however, more than one FISH signal was mostly observed in meiotic prophase. Autosomal juxtapositioning never increased above the background level in the mitotic region (zone 1), indicating that him-19(jf6) mutants are defective in establishing autosomal homologous pairing (Figure 4A).
Figure 4.
Homologous chromosome pairing is defective in the jf6 mutant. (A) Diagrams representing percentage of nuclei with homologous pairing of chromosome I, V, and X in N2, him-19(jf6), and irradiated him-19(jf6) (FISH performed 6 h after irradiation). Gonads are divided into six equal zones from the distal tip cell to diplotene (x-axes), and the percentage of pairing (y-axes) is determined by FISH probes or α-HIM-8 antibody. Levels of pairing in him-19(jf6) do not rise significantly above those in mitosis. (B) him-19(jf6) shows nonhomologous synapsis. Late pachytene nuclei are first stained with the α-SYP-1 antibody and subsequently tested by FISH with the 5S ribosomal locus probe. In him-19(jf6), a large portion of unpaired FISH signals is associated with SYP-1, indicating nonhomologous synapsis or polymerization on unpaired chromosomes. Bars, 10 μm. (C) Quantitative scoring of the 5S FISH signal in association with SYP-1 stretches in pachytene nuclei. The categories were as follows: 1 (blue), unpaired FISH signals with no association with SYP-1; 2 (orange), paired FISH signals in association with SYP-1; 3 (green), unpaired FISH signals in association with SYP-1; and 4 (yellow), paired FISH with no association with SYP-1. In the him-19(jf6) mutant, the most abundant category is the unpaired FISH signal in association with SYP-1. This either indicates frequent occurrence of nonhomologous synapsis or SYP-1 polymerization along unpaired chromosomes. (D) Immunostaining of HIM-8 (green) and ZIM-3 (red) in wild-type and him-19(jf6) pachytene nuclei. HIM-8 localizes in him-19 gonads in wild-type intensity. However, two signals are often detected in the mutant gonads indicating that the homologous pairing of the X chromosome is impaired. Two foci of ZIM-3, corresponding to the paired chromosome I set and chromosome IV set, are usually detected in wild-type TZ and early pachytene. In the him-19(jf6) background, ZIM-3 foci are detected 12 h post-L4 but not 48 h post-L4. Bars, 10 μm.
In addition, pairing of the X chromosome was evaluated using an antibody against the X chromosomal pairing center protein HIM-8 (Phillips et al., 2005). Unpaired HIM-8 signals were frequent in him-19 pachytene nuclei (Figure 4, A and D). Thus, similar to autosomes, also the X chromosome is deficient in homologous pairing in the jf6 background, assuming normal binding of HIM-8 to the X-chromosome (Figure 4A). The fact that HIM-8 staining was positive in the jf6 mutant indicated that the mutant has no problem with the recruitment of the HIM-8 protein to chromosomes (Figure 4D).
Surprisingly, unlike HIM-8, ZIM-3 was not detected on chromosomes in him-19 worms 48 h post-L4 stage (Figure 4D). ZIM-3 protein binds to the PCs of chromosomes I and IV in wild-type animals (Phillips and Dernburg, 2006). Reverse transcription (RT)-PCR showed that the zim-3 message is present in him-19(jf6) (data not shown). Also, ZIM-3 protein was found localized to chromosomes in younger individuals, 12 h post-L4 stage (Figure 4D). This suggests that recruitment of ZIM-3 but not its production is impaired in him-19(jf6).
Synapsis Is Defective in the him-19(jf6) Mutant
HIM-3, a lateral element protein of the SC, and SYP-1, a central element protein of the SC, are indicative of meiotic chromosome axes development and SC formation, respectively (MacQueen et al., 2002; Couteau et al., 2004). These two meiosis-specific proteins were expressed and started to associate with chromosomes in the jf6 mutant similar to wild type (Table 2).
Table 2.
In him-19(jf6), HIM-3 and SYP-1 associate with the chromosomes in a wild-type temporal mannera
| Wild type | him-19(jf6) | |
|---|---|---|
| Initiation of TZ | 18 ± 2.4 (n = 4) | None (n = 6) |
| HIM-3 associates withchromosomes | 17 ± 3.3 (n = 4) | 18.5 ± 6.3 (n = 6) |
| SYP-1 associates withchromosomes | 19.5 ± 3.7 (n = 7) | 23 ± 2 (n = 3) |
a To determine entry into meiosis, gonads were stained for HTP-3, which also associates with chromatin in cell rows 16 and 17 in both the mutant and wild type.
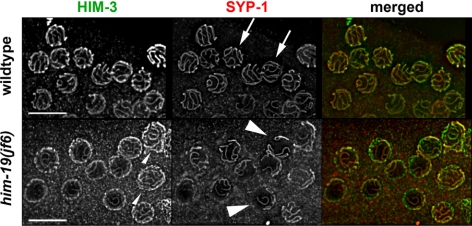
In wild-type pachytene, when all chromosomes are properly paired and synapsed, six HIM-3 tracks and six SYP-1 tracks can be observed (see arrows in Figure 5). Moreover, DAPI staining revealed parallel chromatin tracks. In him-19(jf6) HIM-3 is polymerized along the chromosomes with wild-type kinetics (Figure 5). However, stainings seem less prominent and more HIM-3 tracks are discernible per cell most likely reflecting unpaired homologues (small arrowhead in Figure 5). Consistent with the reduced number of parallel tracks observed by DAPI staining, SYP-1 polymerized in a restricted manner (arrowheads in Figures 5 and 7A). Only short stretches of SYP-1 were seen upon entry into meiosis. From midpachytene onward, SYP-1 stretches polymerized more extensively. Finally, in late pachytene, SYP-1 polymerization reached its maximum (Figure 7). The SYP-1 loading pattern in him-19(jf6) is similar to the one observed in mutants of the meiotic regulator chk-2 (Martinez-Perez and Villeneuve, 2005). Because FISH demonstrated that most homologous regions are unpaired (see above), the extensive synapsis might involve nonhomologous partners. Consistently, partial SYP-1 tracks can frequently be found between DAPI-positive structures, suggesting fragmentary polymerization between paired chromosomes (see arrow in Supplemental Figure 5A). SYP-1 might also polymerize along unpaired chromosomes as described for cra-1 mutants (Smolikov et al., 2008). Regardless, immuno-FISH experiments and stainings with HIM-8 revealed that the partial SYP-1 stretches are formed along/between nonhomologous chromosomes (Figure 4, B and C, and Supplemental Figure 5B).
Figure 5.
Lateral HIM-3, and central SYP-1, components of the SC, are loaded in him-19(jf6). Spread pachytene nuclei were stained with an antibody against HIM-3 an axial element component of the SC. Loading of HIM3 occurs at the same time in wild-type and him-19(jf6) individuals. However, more HIM-3 tracks can be observed in the mutant (small arrowhead). Similarly, the central element component SYP-1 is loaded along the entire chromosome axes in wild-type nuclei (indicated by arrows). In him-19 (jf6), the extent of SYP-1 polymerization is reduced as demonstrated by the relatively short SYP-1 stretches (see arrowheads). Also, compare Supplemental Figure 5 for a more detailed view of SYP-1 loading. Bars, 10 μm.
Figure 7.
Artificially induced DSBs restore chromosome clustering and enhance SYP-1 polymerization. (A) High magnification of the meiotic entry area of the him-19(jf6) gonad and him-19(jf6) 6 h post-γ-radiated and stained with α-SYP-1 antibody. SYP-1 loading is restricted in him-19 mutant. Mutant gonads post irradiated show more complete SYP-1 polymerization than nonirradiated gonads. Images are projections of 3D stacks including the whole depth of nuclei shown. Bars, 10 μm. (B) Bar chart showing the presence of nuclei with chromosome clustering. Nuclei with polarized chromosomes (the crescent-shaped nuclei found at the transition zone) peak at zone 2 in wild-type gonads. him-19 mutants have no defined TZ and only a few nuclei in the clustered chromosome configuration. Six hours after irradiation him-19 mutants show more nuclei with polarized chromosomes. Numbers of nuclei scored are as follows: wild type: zone 1, 165; zone 2, 185; zone 3, 131; zone 4, 136; zone 5, 133; and zone 6, 79. him-19(jf6): zone 1, 194; zone 2, 239; zone 3, 198; zone 4, 189; zone 5, 165; and zone 6, 144. him-19(jf6) after irradiation: zone 1, 139; zone 2, 138; zone 3, 190; zone 4, 231; zone 5, 193; and zone 6, 190. (C) Lengths of SYP-1 stretches were measured per cell row upon initiation of SYP-1 loading. Measurements were taken from projected 3D stacks containing the depth of the nuclei. The him-19(jf6) postirradiation gonads have more extensive SYP-1 polymerization than the nonirradiated mutants of the same age. (D) Gamma irradiation restores ZIM-3 loading in him-19(jf6) in parallel to the restoration of clustered chromatin. Worms irradiated 48 h post-L4 were stained for ZIM-3 2 h after irradiation.
Initiation of Meiotic Recombination Is Impeded in him-19(jf6)
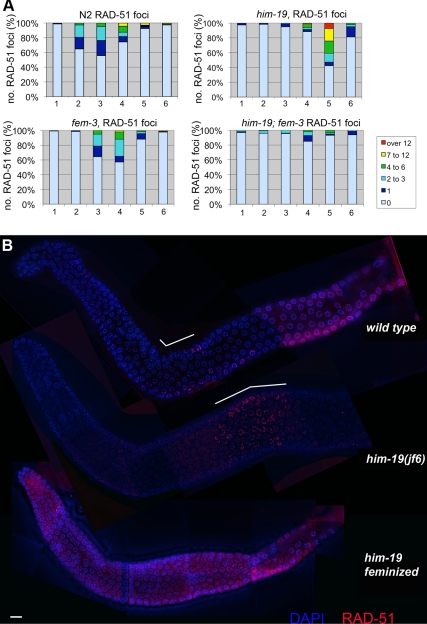
To study the induction of DNA DSBs, we monitored the appearance and disappearance of the strand exchange protein RAD-51 (Alpi et al., 2003; Colaiacovo et al., 2003). Following the time course of RAD-51 foci as shown in Figure 6, RAD-51 foci peak in zone 3 of the wild-type gonad, which corresponds to early pachytene. In 2-d-old him-19(jf6) gonads, RAD-51 foci peak in zone 5, possibly indicating a delay in DSB induction. Most him-19(jf6) cells exhibit over 12 foci. Such high numbers are rarely observed in wild-type gonads and might thus reflect a DSB repair problem in the mutant. Despite the low level of homologous pairing in the mutant, chromosome fragmentation was not observed at diakinesis. This suggests that similar to other pairing mutants, DSBs might be repaired not from the homologous partner but via the sister chromatids (Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Penkner et al., 2007). Alternatively, breaks might get repaired via nonhomologous end-joining. As evident from the reduced fertility described above, the feminized him-19(jf6) mutants have a more penetrant phenotype also with respect to recombination. Placing him-19 in the fem-3 feminized background revealed a strong reduction of RAD-51–positive nuclei. Only few RAD-51–positive stray nuclei were detected (Figure 6). Therefore, in its most penetrant version (in the feminized background), him-19 either suppresses DSBs or delays RAD-51 loading for even longer than in the hermaphrodite. This might also reflect a synthetic effect in the fem-3; him-19(jf6) double mutant preventing RAD-51 loading.
Figure 6.
Progression of recombination is disrupted in the him-19(jf6) mutant. (A) Bar chart of quantitative time course analysis of RAD-51 foci in various mutant backgrounds. Projected 3D stacks containing the depth of nuclei, whole gonads were sectioned in six parts. Zones 1 and 2 include premeiotic nuclei, zone 3 contains mostly nuclei in transition zone, and zones 4–6 are the pachytene nuclei. RAD-51 foci were counted on deconvolved flattened images. Differently colored sections on the bar represent the percentages of nuclei in a given zone with the number of RAD-51 foci indicated in a color code. In the wild type, RAD-51 foci peak around zone3 and never exceed 12 foci per cell. In him-19(jf6), RAD-51 foci peak around zone 5, and frequently more than 12 foci per cell can be observed. In the feminized fem-3; him19(jf6) double mutant, only very few RAD-51 foci can be detected around zone 4. In the fem-3 single mutant, a normal number of RAD-51 foci was observed that peaked around zone 3 and 4, with a slight delay compared with wild-type. Number of nuclei scored for wild type per zone is as follows: zone 1, 165; zone 2, 185; zone 3, 131; zone 4, 136; zone 5, 133; and zone 6, 79. him-19(jf6): zone 1, 194; zone 2, 239; zone 3, 198; zone 4, 189; zone 5, 165; and zone 6, 144. fem-3(e1996): zone 1, 134; zone 2, 149; zone 3, 143; zone 4, 154; zone 5, 118; and zone 6, 95. him-19(jf6); fem-3(e1996): zone 1, 156; zone 2, 122; zone 3, 133; zone 4, 140; zone 5, 117; and zone 6, 81. (B) Immunostaining of RAD-51 (red) in wild type, him-19(jf6) hermaphrodite, and him-19(jf6) female whole gonads, counterstained with DAPI (blue). White bars on top of the wild-type and the him-19(jf6) hermaphrodite gonads indicate the zones rich in RAD-51 foci. Bar, 10 μm.
To determine whether missing RAD-51 foci reflect defective DSB formation or lack of the protein, we artificially induced DSBs by γ radiation. Two-day-old jf6 mutant hermaphrodites were irradiated and immunostained after 1.5 h onward. Abundant RAD-51 foci were seen from the meiotic entry zone onward in irradiated jf6 (Supplemental Figure 4A). Even in fem-3, him-19 mutants, RAD-51 foci were restored (data not shown). This indicates that recruitment of components of the repair machinery, such as RAD-51, is possible and that in the mutant DSBs might fail to get induced or processed with progressing age.
For comparison, we used the chk-2(gk212) mutant which also has a defect in homologous pairing and SC assembly and lacks RAD-51 foci (Rinaldo et al., 1998; MacQueen et al., 2002; Alpi et al., 2003). When chk-2 gonads were γ-radiated, RAD-51 foci were restored as in him-19;fem-3 mutants (Supplemental Figure 4B). This suggests that both chk-2 and him-19 mutants might have problems in inducing or properly processing DSBs.
Artificially Induced DSBs Restore Chromatin Clustering, Reload ZIM-3 to Chromosomes, and Enhance SYP-1 Polymerization in the him-19(jf6) Mutant
Six hours after artificially inducing DSBs by irradiation of him-19(jf6), chromatin clustering was restored in a narrow transition-like zone (Figure 7B). However, although the chromosomes were arranged in the typical crescent, they did not seem to be as condensed as wild-type TZ nuclei.
In the wild type, crescent formation has been related to the presence of an active pairing process (MacQueen and Villeneuve, 2001; Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Penkner et al., 2007). Consistently, the absence of a TZ in mutants such as chk-2, him-3, htp-1, and matefin/sun-1 is accompanied by a lack of presynaptic alignment (MacQueen and Villeneuve, 2001; Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Penkner et al., 2007). Unexpectedly, ZIM-3, the PC protein on chromosomes I and IV, was recruited back to chromosomes upon irradiation in him-19(jf6) (Figure 7D). Despite reclustering of chromosomes and reloading of ZIM-3, FISH experiments showed that homologous pairing was not improved (Figure 4A).
Polymerization of SYP-1 along chromosomes was remarkably improved upon irradiation (Figure 7, A and B). We measured the length of SYP-1 stretches in three-dimensional stacks of nuclei and related it to the cell's position within the gonad. Although below wild-type levels, earlier, more extensive polymerization of SYP-1 was observed upon irradiation of the mutant (Figure 7, A and C). Because no increase in parallel DAPI tracks was discernible upon irradiation, SYP-1 polymerization most likely occurred along unpaired chromosomes. We also analyzed the SYP-1 polymerization rate in a him-19(jf6); spo-11(me44) double mutant devoid of DSBs. In spo-11(ok79) single mutants SYP-1 loading seems normal in pachytene but its departure is disturbed during later stages (Nabeshima et al., 2005). As expected, SYP-1 loading was similar to him-19(jf6) in the him-19(jf6); spo-11(me44) double mutant (data not shown). Also by DAPI staining, the him-19(jf6); spo-11(me44) gonad resembled that of the jf6 mutant. Both a clear transition zone was missing and univalents could be observed at diakinesis. In this respect, him-19 is epistatic to spo-11, indicating that him-19 is acting upstream of spo-11 and is involved in the formation of DSBs.
To see whether rapid SYP-1 polymerization due to induced DSBs is a general feature of pairing mutants, we also irradiated chk-2(gk212) along with the him-19(jf6) mutant. However, 6 h after irradiation, chk-2 gonads did not seem different from the nonirradiated gonads (data not shown). Neither formation of a TZ nor rapid polymerization of SYP-1 was observed. This indicates that him-19 and chk-2 respond differently to artificial DSBs.
DISCUSSION
Here, we have characterized a novel gene encoded by ORF Y95B8A.11 required for proper meiotic prophase I. The gene was named him-19 because the prevalent mutant phenotype is a high incidence of males among the progeny. Remarkably, Him and other phenotypes in this mutant aggravate with increasing maternal age. It is conceivable that the him-19 mutation accelerates the normal ageing effects on meiosis, causing increased chromosomal nondisjunction in aged C. elegans (Rose and Baillie, 1979).
The Pleiotropic Phenotype Suggests a Regulatory Function for him-19
him-19 plays a role in homology recognition, synapsis, and DSB initiation or processing during meiotic prophase I. Presence of the SC components HIM-3 and SYP-1 clearly indicates that meiosis is initiated in the mutant. However, him-19 individuals lack the crescent-shaped organization of chromatin in TZ nuclei, which is characteristic for various pairing mutants. Clustering of chromatin to one side of the nucleus is believed to be an indication of ongoing homology search (MacQueen and Villeneuve, 2001; Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Penkner et al., 2007). Consistently, homologous alignment is impaired in the mutant.
The PC protein HIM-8 is normally recruited to X chromosomes in the him-19 mutant, whereas the autosomal PC protein ZIM-3 is loaded in young adults but fails to load in older individuals. This could contribute to the autosomal alignment and synapsis defect. Still, not all pairing-defective mutants show mislocalization of ZIM proteins (e.g., Penkner et al., 2007). Phillips and Dernburg (2006) reported diffuse but aberrant association of ZIM proteins with chromosomes in chk-2 mutants. The fact that proper pairing of the X remains low despite HIM-8 loading suggests an alignment defect independent of HIM-8 loading.
SYP-1 polymerization is restricted in the him-19 mutant, resembling the situation in the chk-2 mutant. Only short stretches of SYP-1 are observed, consistent with previous reports showing that during early to midpachytene some regulatory mechanisms prevent nonhomologous synapsis, whereas synapsis in late pachytene is indifferent to homology (Rasmussen, 1986; Loidl et al., 1990; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). At later pachytene when the constraint is removed, SYP-1 polymerizes more extensively. This polymerization might either occur with a wrong partner or along unpaired chromosomes (Rasmussen, 1986; Loidl et al., 1990). ZIM proteins are known to be involved in pairing, but they also serve as sites where synapsis initiates (Rasmussen, 1986; Loidl et al., 1990; MacQueen et al., 2005; Phillips et al., 2005). The aberrant SYP-1 staining in the him-19 mutant might be a consequence of the failure of ZIM proteins to localize to autosomes (Figure 4D). However, as exemplified for HIM-8 localization, recruitment of pairing center proteins by itself is not sufficient for proper pairing and SYP-1 loading.
Some pairing defective mutants, such as him-3, form RAD-51 foci with wild-type kinetics (Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). This is not the case for him-19 and chk-2. chk-2 null mutants lack RAD-51 foci (Colaiacovo et al., 2002; Rinaldo et al., 2002; Alpi et al., 2003). In him-19(jf6), the zone following the mitotic zone lacks RAD-51 foci almost entirely. Instead, an increased number of RAD-51 foci is observed at the more proximal gonad end. These foci most likely stem from cells that were in the transition zone in young adults when DSBs could still be formed. An increased number of foci might indicate less efficient DSB repair due to nonhomologous synapsis. However, because chromosome fragmentation is not observed in him-19 mutant diakinesis, breaks are eventually repaired, most likely by using the sister chromatid as a template. In a fem-3 background, him-19(jf6) worms lack RAD-51 foci almost entirely. Because γ-radiation restores RAD-51 loading throughout the mutant gonad, the absence of RAD-51 foci might indicate that DSBs are completely lacking in him-19(jf6); fem-3 individuals.
The him-19;syp-2 and him-19;spo-11 double mutants both display the phenotype of the him-19 mutant as judged by DAPI staining (lack of a clear TZ and formation of univalents at diakinesis) (this work; data not shown). This means that him-19 is epistatic to both syp-2 and spo-11 and that HIM-19 is placed upstream of the SC central region protein and DSBs induction protein. In addition to synapsis and DSB induction, homologous alignment and loading of ZIM-3 are also disrupted in the him-19(jf6) background. This places him-19 even further up in the meiotic chain of events.
C. elegans germline proliferation and meiotic entry is controlled by assorted RNA regulators, posttranscriptional repressors, and translation factors. GLD-1, -2, and -3 and NOS-3 are the key regulators for meiotic entry (Kadyk and Kimble, 1998; Eckmann et al., 2004; Hansen et al., 2004; Kimble and Crittenden, 2007). HIM-19 might play a role in coordinating homologous pairing, synapsis and DSBs induction/repair by controlling expression of other meiotic master players early after meiotic entry. Alternatively, HIM-19 could be a chromatin associated protein providing independent contributions to those processes. Due to its phenotypic similarity to CHK-2, encoding a protein kinase, one could also speculate that it regulates the initiation of specific meiotic events.
DSB Induction Restores ZIM-3 Foci and SYP-1 Polymerization, but Not Homologous Pairing
Studies of C. elegans chromosomal PCs have shown that they are crucial for homologous pairing and synapsis initiation (MacQueen et al., 2005; Phillips et al., 2005, 2009). However, in a background that is deficient of homozygous PCs, a high residual level of SC-polymerization is still observed (MacQueen et al., 2005). spo-11 mutants have neither pairing nor synapsis defects indicating the dispensability of DSBs for these processes (Dernburg et al., 1998). Aged him-19(jf6) mutants do not show autosomal PC protein localization (as shown here with ZIM-3) and display a homologous pairing defect. However, upon induction of artificial DSBs, ZIM-3 is recruited back to pairing centers and more nuclei with crescent-shaped chromatin can be made out. Despite this restoration, homologous pairing is still not improved. Thus, localization of ZIM proteins is crucial but not sufficient for homologue alignment. A similar case was observed in the him-8(me4) mutant: HIM-8 is recruited to the X chromosome's PCs but pairing of the X is defective (Phillips et al., 2005).
The synaptic defect at early to midpachytene in the him-19 mutant is probably due to the fact that not all ZIMs are recruited to pairing centers (MacQueen et al., 2005; Phillips et al., 2005). Reloading of ZIM-3 to pairing centers upon irradiation seemingly promotes SYP-1 polymerization, whereas homologous alignment is not improved by irradiation. This further proves that DSB formation and pairing are independent events in C. elegans and that in this system ongoing repair most likely does not contribute to distinguish “self and nonself” but rather promotes SC polymerization. Our work further extends the observations made with the cra-1 mutant suggesting the existence of a cryptic DSB-dependent SC polymerization mechanism in C. elegans (Smolikov et al., 2008).
A Model for the Oogenesis-directed and Age-dependent Action of him-19
C. elegans is a sequential hermaphrodite that produces sperm in late L4 stage before switching to oocyte production in the adult. Mature sperm is stored in the spermathecae and used to fertilize oocytes (Schedl, 1997). In him-19 mutants, male meiosis is less affected, whereas feminized mutants are more affected than early mutant hermaphrodite meioses. Several models can be envisaged to explain this phenomenon.
First, a factor provided during spermatogenesis might mitigate the mutant phenotype during the first day of adulthood, allowing the rescue of female meiosis. In principle, this could be a homologue expressed during spermatogenesis. Given that the similarity of HIM-19 to Dhh-1 could only be identified using the metastructure concept, one would have to assume that it is rather distantly related proteins that might exhibit similar molecular functions. However, even in the fem-3, him-19 double mutants (which lack a male gonad) we can detect a higher hatch rate in the very first brood, making the rescue of subsequent meioses by a protein expressed during spermatogenesis rather unlikely.
An alternative explanation model relies on the fact that neither the mutant created by deletion nor the jf6 allele created by us represent a complete null phenotype. In both cases, the first 240 amino acids accounting for >60% of the predicted protein are unaffected. The truncated protein might therefore still have residual activity.
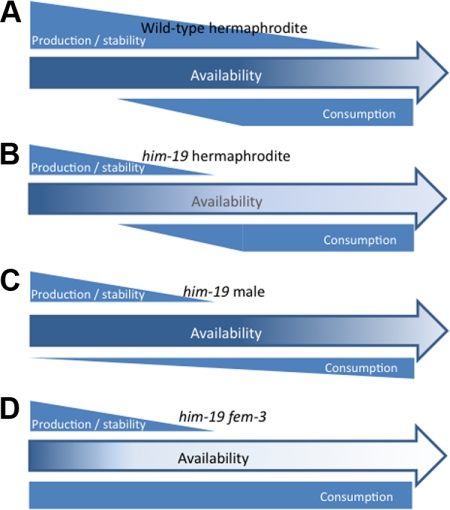
In one scenario, truncation of the protein might decrease its stability (Figure 8). HIM-19 synthesis might start during spermatogenesis where it is only marginally used but mainly stockpiled for use during female meiosis. A reduced stability of the protein might therefore explain the age dependent aggravation of phenotypes in female meiosis. In feminized fem-3 worms initiation of HIM-19 synthesis and its use/degradation would coincide with the initiation of female meiosis. Therefore, only very little HIM-19 would be available at the beginning of meiosis in fem-3 worms, explaining the severe phenotype observed at the initiation of meiosis.
Figure 8.
Model of HIM-19 activity. Production of HIM-19 or a HIM-19-dependent (or HIM-19 stabilized) factor X is initiated in the young gonad. Both the deletion him-19(tm3538) and point mutation him-19(jf6) allele allow production of the aminoterminal 60% of HIM-19, which might still allow residual activity of the protein. The mutated protein might either be less stable or less active leading to reduced activity or production of factor X. Assuming a stabilizing function of HIM-19 on factor X, both truncation or even complete loss of HIM-19 would lead to a more rapid degradation of factor X. (A) In the wild type, HIM-19/Factor X levels are sufficient to support normal meioses (almost) to the end of the reproductive lifespan. (B) In the him-19 hermaphrodite, HIM-19 is less stable, produces less of its dependent factor X, or leads to destabilization of factor X. Consumption starts with the onset of female meioses. The available stock becomes depleted in old animals. (C) Male meiosis is less dependent on HIM-19/factor X; hence, the production to consumption ratio is sufficiently high to ensure mostly normal meioses in him-19 males. (D) In fem-3 worms, HIM-19/factor X consumption by female meioses starts in the young gonad. Depletion of HIM-19/factor X occurs earlier leading to more severely affected meioses.
In another, but somehow more plausible scenario, HIM-19 would act as a cofactor required for the synthesis, modification, or stabilization of a hypothetical factor X, essential for female meiosis (Figure 8). Therefore, in hermaphrodites without or with compromised HIM-19, synthesis or modification of factor X during male meiosis would be less efficient and it would then be depleted during female meiosis. In fem-3 worms, synthesis of factor X would coincide with its depletion during female meiosis, therefore never reaching amounts that allow significant rescue of the phenotype. Alternatively, if HIM-19 was a stabilizing component to factor X, factor X might be degraded at a faster rate than in wild type, therefore showing a stronger effect on later meiosis than on early meiosis. This model would also be consistent with a complete loss of function in the him-19(jf6). In all but the first models, HIM-19 and/or the proposed factor X would only play a minor role during male meiosis, therefore leaving it almost unaffected by the mutation. The auxiliary role of HIM-19 would be in line with its homology to an RNA helicase proposed by its metastructure. Whichever model applies, future experiments will be required to decipher the pathway in which HIM-19 is involved.
Supplementary Material
ACKNOWLEDGMENTS
We thank Japanese National BioResource for C. elegans for providing the him-19 knockout mutant. Many thanks to Anne Villeneuve (Stanford University), Tim Schedl (Washington University in St. Louis), Monique Zetka (McGill University), Abby Dernburg (University of California, Berkeley), and Anton Gartner (University of Dundee) for strains and antibodies. We are grateful to Christian Pflügl, Pawel Pasierbek, Markus Ladurner, Johanna Kober, Daniela Gallerano, and Michael Haberler for technical assistance. We thank Maria Novatchkova for bioinformatics support. L. T. was funded through the Doctoral Program of the Austrian Science Foundation (to M. J.). This work was supported by grants LS05009 Wiener Wissenschafts-, Forschungs-, und Technologiefonds (WWTF), P21338 (Fonds zur Förderung der wissenschaftlichen Forschung [FWF]), and an Elise Richter grant (FWF) (to V. J.), SFB 1706 (to M.F.J.) and P17329 (to J. L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0811) on January 13, 2010.
REFERENCES
- Alpi A., Pasierbek P., Gartner A., Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Andux S., Ellis R. E. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Coller J., Parker R., Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A. M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M. P., Stanfield G. M., Reddy K. C., Reinke V., Kim S. K., Villeneuve A. M. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics. 2002;162:113–128. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Couteau F., Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A. M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dernburg A. F., Zalevsky J., Colaiacovo M. P., Villeneuve A. M. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Eckmann C. R., Crittenden S. L., Suh N., Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J. D., Boulton S. J., Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr. Opin. Pediatr. 2009;21:703–708. doi: 10.1097/MOP.0b013e328332c6ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E.J.A., Greenstein D. T.C.e.R. Community: WormBook, editor. Introduction to the germ line. WormBook. 2005. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Hunt P. A., Hassold T. J. Human female meiosis: what makes a good egg go bad? Trends Genet. 2008;24:86–93. doi: 10.1016/j.tig.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Jantsch V., Tang L., Pasierbek P., Penkner A., Nayak S., Baudrimont A., Schedl T., Gartner A., Loidl J. Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol. Biol. Cell. 2007;18:4911–4920. doi: 10.1091/mbc.E07-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., Villeneuve A. M. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Konrat R. The protein meta-structure: a novel concept for chemical and molecular biology. Cell Mol. Life Sci. 2009;66:3625–3639. doi: 10.1007/s00018-009-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. Cytological aspects of meiotic recombination. Experientia. 1994;50:285–294. doi: 10.1007/BF01924012. [DOI] [PubMed] [Google Scholar]
- Loidl J., Ehrendorfer F., Schweizer D. EM analysis of meiotic chromosome pairing in a pentaploid Achillea hybrid. Heredity. 1990;65:11–20. [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., Villeneuve A. M. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M., Dernburg A. F. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Villeneuve A. M. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., Khan S. M., Dimopoulos G., Janse C. J., Waters A. P. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A. F., Villeneuve A. M. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A. M. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Colaiacovo M. P. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Peoples-Holst T. L., Burgess S. M. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M. F., Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Dernburg A. F. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Meng X., Zhang L., Chretien J. H., Urnov F. D., Dernburg A. F. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 2009;11:934–942. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., Meneely P. M., Dernburg A. F. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. W. Chromosome interlocking during synapsis—a transient disorder. Tokai J. Exp. Clin. Med. 1986;11:437–451. [PubMed] [Google Scholar]
- Rinaldo C., Bazzicalupo P., Ederle S., Hilliard M., La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C., Ederle S., Rocco V., La Volpe A. The Caenorhabditis elegans RAD51 homolog is transcribed into two alternative mRNAs potentially encoding proteins of different sizes. Mol. Gen. Genet. 1998;260:289–294. doi: 10.1007/s004380050897. [DOI] [PubMed] [Google Scholar]
- Rose A. M., Baillie D. L. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics. 1979;92:409–418. doi: 10.1093/genetics/92.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T. Developmental Genetics of the Germ Line. In: Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., editors. C. elegans II. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Hurlburt A., Rogers E., Villeneuve A. M., Colaiacovo M. P. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics. 2007a;176:2027–2033. doi: 10.1534/genetics.107.076968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Schild-Prufert K., Hurlburt A., McDonald K., Engebrecht J., Villeneuve A. M., Colaiacovo M. P. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007b;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., Colaiacovo M. P. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 2008;4:e1000088. doi: 10.1371/journal.pgen.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., Colaiacovo M. P. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 2009;5:e1000669. doi: 10.1371/journal.pgen.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V. V., Bickel S. E. Aging predisposes oocytes to meiotic nondisjunction when the cohesin subunit SMC1 is reduced. PLoS Genet. 2008;4:e1000263. doi: 10.1371/journal.pgen.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.