Inhibitory phosphorylation of Cdh1 by CDK and Polo kinase has been proposed to inactivate APC-Cdh1. Through an exact gene replacement approach, we find CDK, but not Polo, phosphorylation of Cdh1 to be a critical regulatory mechanism. APC-Cdh1 inhibits multiple aspects of spindle morphogenesis, and its activity is modulated by endogenous ACM1.
Abstract
Anaphase promoting complex (APC)-Cdh1 targets multiple mitotic proteins for degradation upon exit from mitosis into G1; inhibitory phosphorylation of Cdh1 by cyclin-dependent kinase (CDK) and Polo kinase has been proposed to prevent the premature degradation of substrates in the ensuing cell cycle. Here, we demonstrate essentiality of CDK phosphorylation of Cdh1 in Saccharomyces cerevisiae by exact endogenous gene replacement of CDH1 with CDK-unphosphorylatable CDH1-m11; in contrast, neither Cdh1 polo kinase sites nor polo interaction motifs are required. CDH1-m11 cells arrest in the first cycle with replicated DNA and sustained polarized growth; most cells have monopolar spindles. Blocking proteolysis of the Cin8 kinesin in CDH1-m11 cells does not promote spindle pole body (SPB) separation. In contrast, expression of undegradable mitotic cyclin results in both SPB separation and the restoration of isotropic growth. A minority of CDH1-m11 cells arrest with short bipolar spindles that fail to progress to anaphase; this can be accounted for by a failure to accumulate Cdc20 and consequent failure to cleave cohesin. Bipolar spindle assembly in CDH1-m11 cells is strikingly sensitive to gene dosage of the stoichiometric Cdh1 inhibitor ACM1. Thus, different spindle-regulatory pathways have distinct sensitivities to Cdh1, and ACM1 may buffer essential CDK phosphorylation of Cdh1.
INTRODUCTION
Ubiquitin-mediated proteolytic degradation is fundamental to proper eukaryotic cell cycle progression. From late mitosis through early G1, the anaphase promoting complex (APC) is essential for cell cycle-relevant proteolytic degradation, and its activity is targeted to appropriate substrates by the evolutionarily conserved coactivators Cdc20 and Cdh1 (Visintin et al., 1997). APC-mediated degradation occurs in two sequential waves, the first of which is coordinated by Cdc20 and the second by Cdh1 (Kramer et al., 2000). In mitosis, Cdc20 promotes cleavage of the anaphase inhibitor Pds1, leading to separation of sister chromatids, as well as an initial decline in B-type cyclin levels (Cohen-Fix et al., 1996; Lim et al., 1998). APC-Cdc20 is active specifically in the presence of high cyclin-dependent kinase (CDK) levels (Kramer et al., 2000; Rudner and Murray, 2000). On mitotic exit, cyclin levels fall, and control of the APC passes from Cdc20 to Cdh1. In contrast to APC-Cdc20, APC-Cdh1 is active only when the effective CDK activity is lower, because it is phosphorylated and inactivated by CDK (Zachariae et al., 1998).
Cdh1 is responsible for the degradation of mitotic B-type cyclins, polo kinase Cdc5, Cdc20, and numerous other proteins, including several involved in spindle stability and assembly (Schwab et al., 1997; Charles et al., 1998; Shirayama et al., 1998; Hildebrandt and Hoyt, 2001; Huang et al., 2001; Woodbury and Morgan, 2007; Benanti et al., 2009). Cdh1 activation helps disassemble the spindle and reduce B-type cyclin levels to a low state, allowing loading of replication origins, thus restoring the system to the initial G1 state. It is likely that most proteins that are degraded under the control of Cdh1 in M/G1 must be synthesized anew in the subsequent cell cycle, which could make APC-Cdh1 inactivation obligatory (Crasta et al., 2006).
Overexpression of CDK-unphosphorylatable Cdh1 blocks construction of a bipolar mitotic spindle (Crasta et al., 2008). Mitotic spindle construction requires duplication of spindle pole bodies (SPBs; functionally equivalent to metazoan centrosomes), followed by disassembly of the bridge connecting them and separation to opposite poles of the nucleus. Subsequently, sister chromatid separation and anaphase spindle elongation separates chromosomes into the progeny. The spindle is then disassembled; each cell inherits a single SPB, which then starts the cycle anew. Degradation of many spindle proteins in late anaphase is dependent on Cdh1. However, in the absence of Cdh1, spindle disassembly is only delayed, not blocked (Visintin et al., 1997). This delay may decrease fidelity of chromosome segregation (Ross and Cohen-Fix, 2003). Notably, the plus-end kinesins Cin8, and possibly Kip1, are Cdh1 targets (Gordon and Roof, 2001; Hildebrandt and Hoyt, 2001; Crasta et al., 2006). cin8 mutants display chromosomal instability and spindle defects, and cin8 kip1 strains are inviable (Hoyt et al., 1992). Cin8 and Kip1 are implicated in SPB separation, as strains with both kip1 and the temperature-sensitive allele cin8-3 retain a half-bridge at the restrictive temperature (Hoyt et al., 1992). Mutations in the minus-end–directed kinesin Kar3, which opposes the forces generated by Cin8 and Kip1, allow for separation of SPBs in a cin8-3 kip1 background (Saunders and Hoyt, 1992). Overexpression of Cin8 is sufficient to separate SPBs in the presence of overexpressed unregulated Cdh1 (Crasta et al., 2006), suggesting that degradation of these proteins may account for the ability of unregulated Cdh1 to block spindle formation. Cdh1 also targets the spindle-stabilizing proteins Ase1 and Fin1 for degradation (Juang et al., 1997; Woodbury and Morgan, 2007).
Cdh1 is regulated at least in part through multisite CDK phosphorylation (Zachariae et al., 1998; Jaspersen et al., 1999). Cdh1 contains 11 putative CDK sites, and CDK-phosphorylated Cdh1 loses the ability to interact with the APC (Kramer et al., 2000). Overexpression of Cdh1-m11 (with all 11 CDK consensus phosphorylation sites mutated to unphosphorylatable alanine residues) is lethal, resulting in a cell cycle arrest with replicated DNA and without mitotic spindles, and with constitutive Cdh1-APC association (Zachariae et al., 1998); overexpression of wild-type Cdh1 with a stronger promoter causes a similar arrest (Visintin et al., 1997). However, it remains unclear whether this is an important regulatory mechanism at endogenous Cdh1 levels. Transformants with a plasmid carrying CDH1-m11 expressed from the CDH1 promoter were reported to be viable, suggesting that CDK phosphorylation of Cdh1 is not essential at endogenous expression levels (Jaquenoud et al., 2002).
Other mechanisms may control Cdh1 activity. Polo kinase (Cdc5) phosphorylation has been proposed to be essential for Cdh1 inactivation (Crasta et al., 2008). Cdh1 is also exported from the nucleus at the time of its inactivation, under control of the Msn5 transporter, which probably sequesters it from access to many of its targets and from the APC itself (Jaquenoud et al., 2002). This transport is dependent on CDK phosphorylation of Cdh1. Finally, the stoichiometric inhibitor Acm1 accumulates early in the cell cycle, forming a stable complex with Cdh1 and preventing APC interaction (Martinez et al., 2006). Interestingly, Acm1 may itself be a Cdh1 target (Enquist-Newman et al., 2008), although there are conflicting reports on this subject (Hall et al., 2008; Ostapenko et al., 2008).
The multitude of Cdh1-regulatory mechanisms, as well as its diverse substrates, make Cdh1 an important regulatory hub. Therefore, it is important to determine the most physiologically significant inputs and outputs for Cdh1 regulation; however, much previous work relies heavily on overexpression of Cdh1, its targets, or both: overexpression can obscure physiological relevance even of authentic regulatory mechanisms. Here, we use exact gene replacement to clarify the critical regulatory mechanism(s) controlling Cdh1, to rigorously determine the phenotype of unregulated Cdh1 at endogenous levels, and to dissect the roles of the multiple Cdh1 targets in control of spindle and cell morphogenesis.
MATERIALS AND METHODS
Strains, Plasmids, and Yeast Methods
All strains used in this study are derivatives of Saccharomyces cerevisiae W303 and are listed in Supplemental Table 1. Plasmids used are listed in Supplemental Table 2. Standard methods were used for strain and plasmid construction throughout. The CDH1-m11 and control recombination alleles in Figure 1B were constructed by integration of BglII-digested FC695 into 2151-1C (cdh1::HIS3), selection on 5-fluoroorotic acid (FOA) for loss of URA3 followed by screening for retention of TRP1, and then crossing to 1960–2B (sic1::HIS3). Tetrad analysis revealed His+ Trp+ strains to be inviable (sic1::HIS3 cdh1-m11::TRP1), confirming CDH1 function to be disrupted in the cdh1-m11::TRP1 allele. Segregant 3023–2 (cdh1-m11::TRP1) was transformed with BglII-digested FC697 to create 3023-2-2 (cdh1-m11::TRP1-URA3-cdh1-m11::LEU2, the control recombination allele in Figure 1B). FOA selection on this strain resulted in Leu+ Trp−, Leu− Trp+, and Leu+ Trp+, but not Leu− Trp− recombinants, as expected from restriction to homologous recombination. The Leu+ Trp− recombinant 3023-2-697 was transformed with BglII cut FC695 to create 3023-2-1 (cdh1-m11::LEU2-URA3- cdh1-m11::TRP1, the experimental recombinational allele in Figure 1B). FOA-resistant popouts from this strain were exclusively Leu+ Trp− or Leu− Trp+, confirming integration at the cdh1-m11::TRP1 locus and suggesting lethality of intact CDH1-m11 (see text). CDH1-m11 exact gene replacements ultimately recovered using either cdc23-1 or GAL-ACM1 to prevent Cdh1-m11-induced lethality (see text) were confirmed by polymerase chain reaction (PCR) product length and DNA sequencing, and they demonstrate no recombination with a marked CDH1 locus in tetrad analysis.
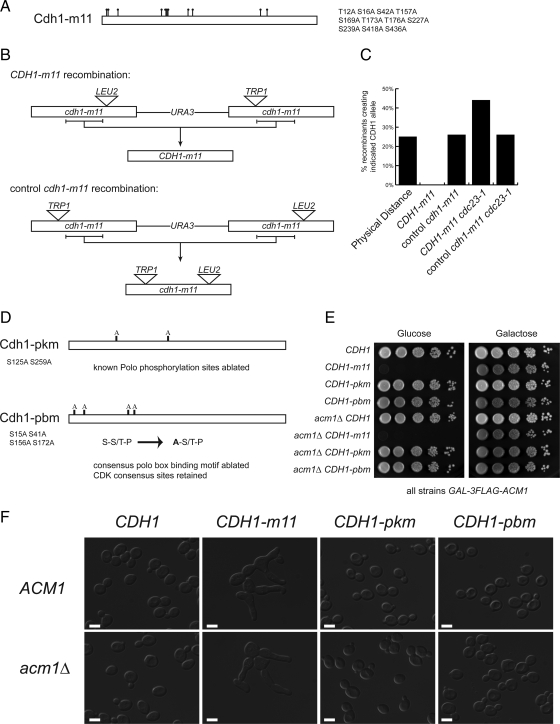
Figure 1.
Cdh1 inhibition requires CDK but not Cdc5 phosphorylation. (A) Schematic of the CDK-unphosphorylatable Cdh1-m11 protein. Dots indicate mutated putative CDK sites. (B) Recombination-based strategy used to obtain CDH1-m11 as an exact gene replacement (top) and control recombination (bottom). Horizontal bracket indicates region of recombination that recreates either CDH1-m11 or the doubly interrupted control cdh1-m11; recombination outside of the bracketed region results in singly interrupted cdh1-m11 alleles in both recombination schemes. (C) Percentage of CDH1-m11 or control, disrupted cdh1-m11 alleles, recovered as determined by selectable markers. Intact CDH1-m11 was not recovered in CDC23 strains. At least 100 recombinants of each genotype were analyzed. (D) Schematic of CDH1-pkm, which has the known Cdc5 phosphorylation sites ablated (top) and CDH1-pbm, which has the Polo box binding motifs mutated so as to eliminate Polo binding but retain the contained CDK sites. (E) Tenfold serial dilutions performed on strains containing galactose-inducible Acm1 and the indicated CDH1 exact gene replacements. (F) DIC images of strains from E after 8 h in glucose. Note the hyperpolarized growth present only in CDH1-m11 strains. Bars, 5 μm.
Time Courses
CDH1-m11 GALL-HA-ACM1 time courses were performed by arrest in YPG + 10 nM α-factor for 135 min at 30°C, followed either by glucose addition or resuspension in YPD + 10 nM α-factor for 30 min. Both procedures were found to result in complete clearance of exogenous Acm1 by Western blot and had identical SPB phenotypes. For time-lapse microscopy, strains were washed three times in SC media, placed onto SC + glucose agar pads, and imaged as described in Bean et al. (2006). For bulk culture time courses, cells were removed from α-factor by three washes in cold YEP and released into YPD at 30°C. For fluorescent microscopy in these time courses, cells were fixed at room temperature for 15 min using a paraformaldehyde buffer, washed twice with sorbitol-phosphate buffer, and otherwise handled as described in Drapkin et al. (2009).
Temperature sensitive scc1-73 and corresponding controls were synchronized as described above but shifted to 37°C after 30 min of release. MET3pr-CLB2-kd time courses were carried out similarly except that pregrowth, arrest and 60 min of release were carried out in 0.2g/l methionine (10× standard concentration). MET3-CLB2-kd was then induced by washing three times into methionine-free medium.
Immunoblots
Western blots were performed using standard methods. Antibody concentrations used were as follows: anti-Pgk1, 1:10,000 (Invitrogen, Carlsbad, CA); anti-hemagglutinin (HA) 12CA5, 1:1000 (Roche Diagnostics, Indianapolis, IN); rabbit polyclonal anti-Clb2, 1:10,000; Myc 9E10, 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA); Clb5 yN-17 1:200 (Santa Cruz Biotechnology); Cdc5 yC-19, 1:4000 (Santa Cruz Biotechnology); and horseradish peroxidase-conjugated secondary antibodies at 1:4000. Enhanced chemiluminescence signal was measured with DarkBox (Fujifilm, Greenwood, SC) with a charge-coupled device camera and quantified using MultiGauge software (Fujifilm). Measured values were normalized to Pgk1 loading controls, and identical reference samples were loaded on separate gels to allow cross-gel normalization and comparison. Fold changes were determined by the ratio of the Pgk1-normalized values.
Microscopy
Fluorescence and differential interference contrast (DIC) images were acquired using an Axioplan 2 microscope (Carl Zeiss, Thornwood, NY) with a 63× 1.4 numerical aperture Plan-Apochromat objective. Camera and microscope were interfaced with the OpenLab software (Improvision, Coventry, United Kingdom), which was also used for spindle separation length measurement. For SPB imaging, seven optical sections were taken at 0.3-μm spacing. Quantification of SPB intensity was performed using automated custom software in MATLAB (The MathWorks, Natick, MA).
RESULTS
Inhibitory CDK Phosphorylation of Cdh1 Is Essential
CDH1-m11, which lacks all CDK phosphorylation sites (Figure 1A), is lethal when overexpressed but has been reported to allow viability when carried on a plasmid under control of its endogenous promoter, suggesting the former result to be an artifact of overexpression (Zachariae et al., 1998; Jaquenoud et al., 2002). To determine rigorously whether CDK-mediated Cdh1 phosphorylation was required for viability at endogenous expression levels in budding yeast, we sought to create an exact chromosomal gene replacement of CDH1 with CDH1-m11. We used a recombination-based approach, in which two copies of cdh1-m11, each rendered nonfunctional by insertion of different selectable markers at different positions, were arranged in tandem at the endogenous locus. Recombinants between the two copies can be selected and simply scored for retention of the insertional markers. CDH1-m11 exact gene replacements should lack both markers (Figure 1B); no such recombinants were obtained (Figure 1C). The critical region for this recombination did yield frequent recombinants using an identical cassette with similarly interrupted cdh1-m11 alleles in the opposite order. Recombinants using this control allele will all be nonfunctional due to retention of insertional marker(s). These results suggested that intact CDH1-m11 is severely deleterious as an exact gene replacement.
CDH1-m11 gene replacement could be deleterious due to unregulated APC activation, although previous studies suggested an additional APC-independent mechanism for lethality of overexpressed Cdh1-m11 (Thornton et al., 2006). Cdc23 is an essential subunit of the APC; cdc23-1 is hypomorphic for APC-Cdh1 activity even at the permissive temperature (Schwab et al., 2001). In contrast to failure of recovery of CDH1-m11 recombinants in a CDC23 background, CDH1-m11 cdc23-1 recombinants were readily obtained and confirmed to be exact by mapping and sequencing of PCR products from the recombinants. When we attempted to cross these recombinants to CDC23 strains, doubly heterozygous diploids were not obtainable, suggesting that CDC23 and CDH1-m11 made a lethal combination (even with both heterozygous) and that CDH1-m11 lethality is APC dependent.
We performed a high-copy suppressor screen for CDH1-m11, by transforming a wild-type strain with a genomic library, crossing the pool of transformants to a CDH1-m11 cdc23-1 strain at the permissive temperature, and selecting for viable diploids. High-copy ACM1 was isolated multiple times in independent clones from the genomic library, but no other strong positives were obtained.
Confirming this result, we could readily construct GAL-ACM1 CDH1-m11 strains that were viable on galactose medium (GAL-ACM1 on, Acm1 overexpressed) but inviable on glucose medium (GAL-ACM1 off, only endogenous levels of Acm1 present). A high-copy plasmid suppression screen for viability of such a strain on glucose medium once again only yielded multiple ACM1 clones. These results suggest (but do not prove) that Acm1 may be the only regulator able to restrain activity of CDK-unphosphorylatable Cdh1.
Cdc5 Phosphorylation of Cdh1 Is Not Required for Cell Viability
Cdc5 has been reported to act in concert with CDK phosphorylation to mediate complete Cdh1 inhibition (Crasta et al., 2008). Cdc5 can phosphorylate Cdh1 on serines 125 and 259 (Crasta et al., 2008). It has been proposed that phosphorylation of Cdh1 on these sites is required for complete Cdh1 inactivation, to allow for SPB separation and mitotic spindle assembly. Furthermore, Cdc5-mediated inhibition of Cdh1 was reported to be essential in the absence of ACM1 (Crasta et al., 2008). However, these experiments were all carried out under conditions of overexpression. Therefore, we created an exact gene replacement ablating these two known Cdc5 phosphorylation sites (Figure 1D, CDH1-pkm). We initially introduced this gene replacement into a cdc23-1 background (see above) and confirmed the structure of the CDH1-pkm allele by sequencing of PCR products. We then crossed this allele into a CDC23 GAL-ACM1 background. In contrast to results with CDK-unphosphorylatable CDH1-m11, CDH1-pkm CDC23 strains were not dependent on ACM1 overexpression for viability (Figure 1, E and F), and we observed Mendelian recovery of fully viable CDH1-pkm CDC23 segregants lacking GAL-ACM1 (data not shown).
It has been argued that endogenous Acm1 restrains Cdh1 in the absence of Cdc5 phosphorylation (Crasta et al., 2008). However, CDH1-pkm acm1 strains were viable with no obvious growth or morphological defects (Figure 1, E and F). Efficient degradation of the major mitotic cyclin Clb2 is Cdh1 dependent (Schwab et al., 1997; Visintin et al., 1997), and CDH1-pkm acm1 strains accumulate and destroy Clb2 with normal kinetics (Supplemental Figure 1, A and B). It was reported that the lethality of Cdh1 lacking Cdc5 phosphorylation sites was the result of an inability to separate SPBs. However, we observed wild-type proportions of cells with separated and unseparated SPBs in asynchronous cultures of CDH1-pkm acm1 strains (Supplemental Figure 1, C and D).
These results rule out any significant role in Cdh1 inhibition for Cdc5 phosphorylation of S125 and S259, the only known Cdc5 sites in Cdh1. However, there could be other unidentified Cdc5 sites. Although phosphorylation of S125 and S259 was not detected in a mass spectrometry survey, phosphorylation of numerous other non-CDK sites was observed (Hall et al., 2004). Cdc5-dependent phosphorylation of diverse targets requires polo box binding motifs (PBBs) in the substrate. PBBs have the consensus sequence S-pS/pT-P, with the required phosphorylation frequently created by proline-directed CDK activity (Elia et al., 2003). There are four such sites in Cdh1, which were collectively demonstrated to promote binding of Cdc5 to CDK-phosphorylated Cdh1 (Crasta et al., 2008). Therefore, we constructed a CDH1 allele in which the initial serines in the four PBBs were mutated to alanines (Figure 1D). This manipulation is reported to block Cdc5 binding but not CDK phosphorylation (because the initial S is not part of the CDK consensus S/T-P), thus uncoupling CDK from Cdc5 phosphorylation (Crasta et al., 2008); no phenotypic data from expression of this allele have been reported. Using the same strategy as described above for CDH1-pkm, we constructed an exact gene replacement of CDH1 with CDH1-pbm, and we found that this allele had no discernible cell cycle phenotype and no dependence on ACM1 for viability (Figure 1, E and F).
Overall, our results from endogenous expression levels of Cdh1 do not support a physiological cell cycle role for Cdc5-dependent phosphorylation of Cdh1, at endogenous expression levels, in sharp contrast to the essentiality of CDK-dependent phosphorylation of Cdh1. We cannot formally exclude the possibility that there are other Cdc5 phosphorylation sites and/or other nonconsensus PBBs; however, previous biochemical work argues against this (Crasta et al., 2008).
Cdk Phosphorylation of Cdh1 Is Required for Accumulation of Cdh1 Target Proteins, Switch from Polarized to Isotropic Bud Growth, and Spindle Morphogenesis, but Not for DNA Replication
To determine the function of Cdk-mediated phosphorylation of endogenous levels of Cdh1, we arrested GALL-HA-ACM1 CDH1-m11 cells (GALL is a weakened version of the GAL1 promoter; Mumberg et al., 1994) in G1 by using α-factor in galactose medium. We transferred the cells to glucose medium to turn off the GALL promoter and then released the α-factor block. By immunoblot, HA-Acm1 was greatly reduced in α-factor (Enquist-Newman et al., 2008; Hall et al., 2008; Ostapenko et al., 2008) and undetectable after glucose incubation. Both CDH1-m11 and CDH1 control cells released synchronously and with comparable kinetics from the α-factor block, as indicated by bud emergence and expression of Clb5 (Figure 2, A and C). Clb5 is an early-expressed B-type cyclin that promotes DNA replication whose proteolysis is regulated by Cdc20, not by Cdh1 (Shirayama et al., 1999; Wäsch and Cross, 2002). Consistent with timely Clb5 accumulation, kinetics of DNA replication in CDH1-m11 and CDH1 cells were indistinguishable (Figure 2B). Clb5 levels then declined in CDH1-m11 cells for unknown reasons, stabilizing at approximately a quarter of peak.
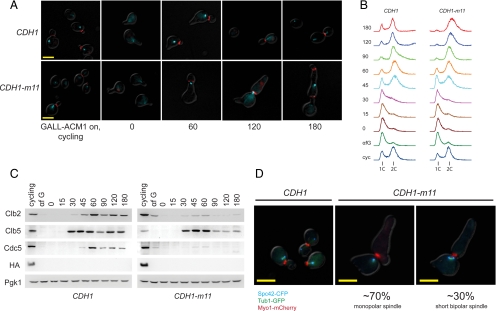
Figure 2.
CDH1-m11 results in a first-cycle arrest with a heterogeneous spindle pole body phenotype. (A) CDH1-m11 or CDH1 cells (both GALL-HA-ACM1) were arrested in G1 with α-factor, depleted of HA-Acm1, and synchronously released. Fluorescence microscopy of Myo-mCherry (red) marking the bud neck and Tub1-CFP (cyan) were taken at the indicated time points after release from α-factor. CDH1-m11 cells multiply bud as indicated by multiple Myo1 rings. Tubulin signal varies in appearance from a point to a short bar, but elongated spindles are not observed. Bar, 5 μm. (B) Bulk DNA flow cytometry of cells as described in A. (C) Immunoblots of cells as described in A detecting the indicated proteins. Pgk1, loading control. (D) Fluorescence microscopy for Spc42-CFP (cyan) marking the SPB, Tub1-GFP (green), and Myo1-mCherry (red); 30% of CDH1-m11 cells form bipolar spindles, as indicated by two separate Spc42 dots connected by intervening tubulin-GFP. Images taken 180 min after release. Bars, 5 μm.
In contrast, accumulation of the mitotic cyclin Clb2, a known Cdh1 target, was significantly reduced in CDH1-m11 cells, with between a 5- and 15-fold reduction in peak Clb2 levels compared with CDH1 controls at 60 min (the range largely reflects the variability in immunoblot background levels and/or small differences in the efficiency of the α-factor block). Clb2 expression drives a switch from polarized to isotropic bud growth (Lew and Reed, 1993), and this is blocked in CDH1-m11 cells (Figure 2A). Accumulation of Cdc5, another known Cdh1 target (Shirayama et al., 1998), was also markedly reduced in CDH1-m11 cells (Figure 2C). Interestingly, the timing of initial accumulation of both Clb2 and Cdc5 was similar in CDH1 and CDH1-m11 cells.
CDH1-m11 cells do not undergo anaphase or cytokinesis. They continue polarized bud growth and rebud as evidenced by accumulation of fluorescent Myo1-mCherry (a bud site marker) at a novel location along the initial hyperpolarized bud (Figure 2A) and/or by a new bud. Spindle morphogenesis seemed defective: using Tub1(β-tubulin)-cyan fluorescent protein (CFP), a range of morphologies from single dots to short bars was detected (Figure 2A).
To more accurately examine spindle morphogenesis, we used SPC42-CFP and TUB1-GFP to label the SPB and microtubules. In these double-labeled cells, an intact bipolar spindle will appear as two distinct blue Spc42-CFP signals connected by a bridge of green Tub1-GFP (Spc42-CFP and Tub1-GFP fluorescent signals were sufficiently spectrally separated to make this determination). Such spindles were almost uniformly observed in CDH1 controls; at 60 min after release, >80% of cells had clearly separated SPBs (data not shown; Figure 3B). These cells cycle and become asynchronous, displaying either unseparated SPBs with presumptive astral microtubules or separated SPBs connected by a tubulin bridge (Figure 2D). In contrast, 70% of CDH1-m11 cells had a single focus of Spc42-CFP signal (Figures 2D and 6B). Thirty percent of CDH1-m11 cells contain short bipolar spindles that did not progress through anaphase. Discrimination between one and two foci is typically unambiguous; a representative field of these terminally arrested CDH1-m11 cells is provided in Supplemental Figure 2. We expect, from previous work, that a single Spc42 signal in a cell represents duplicated but unseparated SPBs (Fitch et al., 1992; Crasta et al., 2008). Consistent with this idea, Spc42-CFP signal intensity in these single SPBs is approximately double that in the α-factor–blocked cells, which are expected to contain unduplicated SPBs. Consistent results were obtained with SPC42-CFP alone, as well as with SPC29-YFP and untagged SPC42, suggesting that the tags did not significantly affect the results.
Figure 3.
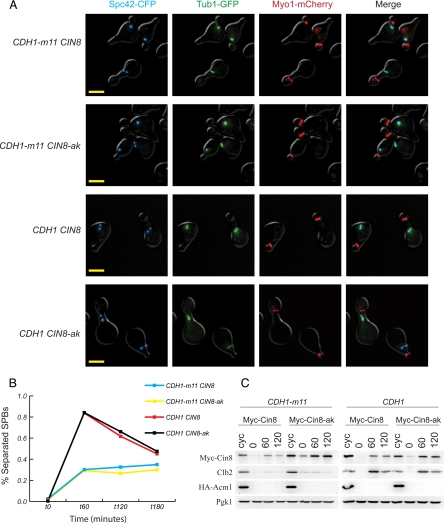
Cdh1-resistant CIN8 does not promote bipolar spindle assembly in CDH1-m11 cells. (A) Fluorescence microscopy of cells with MYC-CIN8 or Cdh1-resistant MYC-CIN8-ak (coding for Myc-Cin8-alaKEN, with KEN box residues mutated to alanine), 60 min after release from α-factor block. Bars, 5 μm. (B) Quantification of SPB separation of cells from A at indicated time points from α-factor release. (C) Immunoblots of released cells. Clb2 and exogenous HA-Acm1 are degraded normally. Cin8-ak is resistant to Cdh1-m11–mediated proteolysis.
Figure 6.
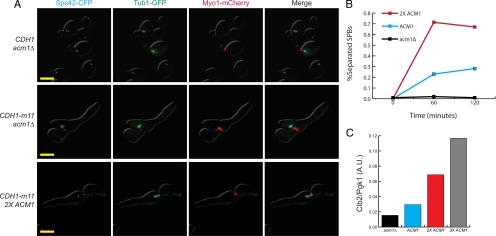
ACM1 gene dosage modulates the CDH1-m11 strain spindle pole body phenotype. (A) Fluorescence microscopy for Spc42-CFP (cyan), Tub1-GFP (green), and Myo1-mCherry (red). In CDH1-m11 acm1 cells, tubulin can be seen emanating from SPBs, but separated SPBs are not observed. 2XACM1 CDH1-m11 cells can separate spindle pole bodies and form bipolar spindles. Strains were treated as described in Figure 2 and kept alive with GALL-ACM1 expression, which was shutoff in α-factor. Images were taken 180 min after release. Bar, 5 μm. (B) Percentage of synchronized acm1, wild type (1X ACM1), and 2X ACM1 cells, all with CDH1-m11, displaying separated spindle pole bodies at indicated time points. (C) Clb2 levels for indicated genotypes, all with endogenous CDH1-m11, at 60 min after release from α-factor, standardized to Pgk1 loading control.
We also noted grossly abnormal nuclear morphology, as monitored with histone H2B-mCherry, in CDH1-m11 cells, whether or not they contained a bipolar spindle (Supplemental Figure 3). Time-lapse microscopy shows H2B-mCherry signal “meandering” along the hyperpolarized bud and the mother cell body. Microscopic observations of fixed cells with labeled SPBs and tubulin suggested that this aberrant nuclear migration may be dependent on astral microtubules, because extended mCherry signal frequently coincided with long microtubules that were not terminated with an SPB. We do not know the reason for this phenotype, which has not been described previously to our knowledge.
Restoring Levels of the Cdh1 Target Kinesin Cin8 Does Not Restore Spindle Pole Body Separation in CDH1-m11 Cells
Because the majority of CDH1-m11 cells arrest with a monopolar spindle, we sought to test whether failure to accumulate a specific spindle-relevant APC-Cdh1 substrate was responsible for the failure to construct bipolar spindles. Previous work (Crasta et al., 2006) suggested that failure to produce a bipolar spindle without restraining Cdh1 activity was specifically due to degradation of the plus-end kinesins Cin8 and Kip1, because a short bipolar spindle could be obtained by overexpression of undegradable Cin8 in the absence of Cdc28 activity, which is required for inhibition of APC-Cdh1. We sought to test this idea more directly, with endogenous levels of expression of both Cdh1-m11 and undegradable Cin8. We used the CIN8-alaKEN allele (Hildebrandt and Hoyt, 2001), in which the KEN box required for Cdh1-mediated Cin8 degradation was mutated to AAA. Myc-tagged alleles of either CIN8 or CIN8-alaKEN were placed at the endogenous locus (with an untagged CIN8 allele downstream) in CDH1-m11 GAL-ACM1 strains. The Myc tag on Cin8 was shown previously to be fully compatible with Cin8 function (Hildebrandt and Hoyt, 2001). By Western blot, Myc-Cin8 levels were reduced approximately fourfold in CDH1-m11 cells compared with CDH1 controls at 60 min; Myc-Cin8-alaKEN was detected in comparable levels in both backgrounds, confirming that the KEN mutation prevents Cdh1-dependent proteolysis of Cin8 (Figure 3C; Hildebrandt and Hoyt, 2001). The CIN8-KED mutation in the KEN box (Hildebrandt and Hoyt, 2001) yielded similar results (data not shown). Thus, reduction of Cin8 in CDH1-m11 cells is specifically due to Cdh1-Cin8-KEN box interaction.
Restoration of Cin8 protein levels by the alaKEN mutation had essentially no effect on the terminal spindle phenotype of Cdh1-m11 cells (Figure 3, A and B). Therefore, restoration of Cin8 at physiological levels is not sufficient to allow bipolar spindle formation in Cdh1-m11 cells, strongly suggesting the existence of other Cdh1 targets that are required for bipolar spindle formation. Previous results suggesting that restoring Cin8 might be sufficient for bipolar spindle formation (Crasta et al., 2006) could be explained by the idea that overexpressed Cin8 could exert a strong pulling or polymerizing force between the two SPBs, or could be due to a lack of equivalence between the hypomorphic CDC28 allele and complete failure of Cdh1 phosphorylation.
Mitotic Cyclins Are Central Regulatory Targets of Cdh1
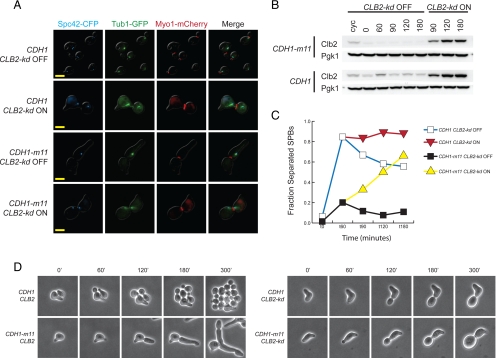
The Cdh1-m11 arrest is associated with degradation of cell cycle regulators (mitotic cyclins, Cdc5) as well as spindle components (see above). Mitotic cyclins modulate numerous cell cycle processes. Some cell cycle defects in CDH1-m11 cells could be due specifically and solely to mitotic cyclin proteolysis. To test this, we placed Clb2-kd, an undegradable version of Clb2 lacking both KEN and destruction boxes and therefore immune to APC-mediated proteolysis (Wäsch and Cross, 2002) under the control of the MET3 promoter, and turned on expression by methionine deprivation in synchronized CDH1-m11 cells, after they were released from α-factor and allowed to bud. Clb2-kd had striking though variable effects on SPB and tubulin morphology. The majority of CDH1-m11 cells in which Clb2-kd was expressed had separated their SPBs as indicated by at least two SPC42-CFP foci, instead of the single focus predominantly observed in controls without Clb2-kd (Figure 4, A and C). This effect was detectable when Clb2-kd levels were similar to those attained with Clb2-kd expressed from the endogenous locus (this level was attained transiently at 30 min after induction; fully induced Clb2-kd levels from the MET3 promoter plateau at ∼ 3-fold the level of Clb2-kd under its endogenous promoter).
Figure 4.
Restoration of mitotic cyclin Clb2 promotes spindle pole body separation and restores isotropic growth in CDH1-m11 cells. (A) MET3pr-Clb2-kd cells, with either CDH1 or CDH1-m11, were synchronized in α-factor, released, and Clb2-kd induced 60 min after release; images were obtained 180 min after α-factor release. Bars, 5 μm. (B) Clb2 immunoblot for cells in A. Clb2 antibody detects both endogenous Clb2 and Clb2-kd. Pgk1 serves as a loading control. (C) Quantification of cells with separated SPBs from A. (D) Single-cell time-lapse microscopy of strains of the indicated genotypes (all exact gene replacements), with minutes after release from α-factor indicated.
Spc42-CFP foci in CDH1-m11 MET3-CLB2-kd cells were sometimes associated with intervening Tub1-GFP signal, as in a normal metaphase spindle; in other cells, little or no polymerized tubulin could be detected. Various other abnormal structures were observed, including multiple (≥3) Spc42-CFP foci. The average SPC-42 signal in individual Spc42-CFP foci in these cells at 180 min after release was approximately half that of CDH1-m11 cells not expressing Clb2-kd. This suggests that in response to Clb2-kd expression, duplicated SPBs separate, resulting in two foci that each contain a level of SPC42 comparable to a normal unduplicated SPB (Supplemental Figure 4). Therefore, failure of SPB separation in CDH1-m11 cells might be specifically due to Cdh1-mediated degradation of Clb2 and other mitotic cyclins. Nevertheless, reintroduction of Clb2 into CDH1-m11 cells results in severe disruption of normal spindle morphogenesis in most cells, perhaps due to alterations in microtubule dynamics (Higuchi and Uhlmann, 2005). Normal spindle morphogenesis requires not only mitotic cyclin stabilization but also stabilization of other proteins, probably including spindle morphogenesis proteins such as Cin8, Ase1 and Fin1; we have not tested the effects of simultaneous stabilization of multiple APC-Cdh1 substrates in CDH1-m11 cells.
Strikingly, the presence of CLB2-kd in CDH1-m11 cells largely eliminated the CDH1-m11 hyperpolarized bud growth phenotype (Figure 4, A and D). Cdc5 protein does not reappear after Clb2-kd expression, suggesting that APC-Cdh1-m11 remains active in the presence of undegradable Clb2.
As noted above, Clb2-kd under the MET3 promoter was not much overexpressed in this experiment compared with the level of Clb2-kd expressed from the endogenous locus. Consistent with this, comparable effects on cell polarity and spindle morphogenesis were obtained in CDH1-m11 cells bearing an exact endogenous gene replacement of CLB2 with CLB2-kd using single cell time-lapse analysis (Figure 4D). CLB2-kd (exact gene replacement) cells have distinct shmoo morphologies (Figure 4D) and are partially defective in the α-factor block-release protocol, precluding clear quantification of bulk cultures.
Thus, restoration of Clb2 to CDH1-m11 cells eliminates the polar bud growth characteristic of these cells, implying that mitotic cyclins may be the sole Cdh1 targets responsible for this phenotype. Restoring Clb2 also results in SPB separation in CDH1-m11 cells; mitotic cyclin degradation is not responsible for all spindle phenotypes of these cells, though, because spindle structure and perhaps microtubule dynamics are profoundly perturbed due to persistent APC-Cdh1 activity even in the presence of stable mitotic cyclins. Thus, a primary role for Cdk-mediated Cdh1 inhibition is to allow mitotic cyclin accumulation; allowing accumulation of mitotic cyclin Clb2, alone among Cdh1 targets, restores isotrophic bud growth and substantially restores bipolar spindle morphogenesis.
Cdk Phosphorylation of Cdh1 Is Essential for Cdc20 Accumulation and Cohesin Cleavage
The majority phenotype of CDH1-m11 cells includes failure to construct bipolar spindles, and this phenotype was shown above to be due largely to failure to accumulate Clb2. However, 30% of CDH1-m11 cells do construct short bipolar spindles that nevertheless fail to undergo anaphase. If these bipolar spindles were aberrant in structure or kinetochore attachment, this could trigger the spindle assembly checkpoint to prevent anaphase. However, deletion of the critical checkpoint component MAD2 had no effect on spindle assembly or function in CDH1-m11 cells (Supplemental Figure 5A).
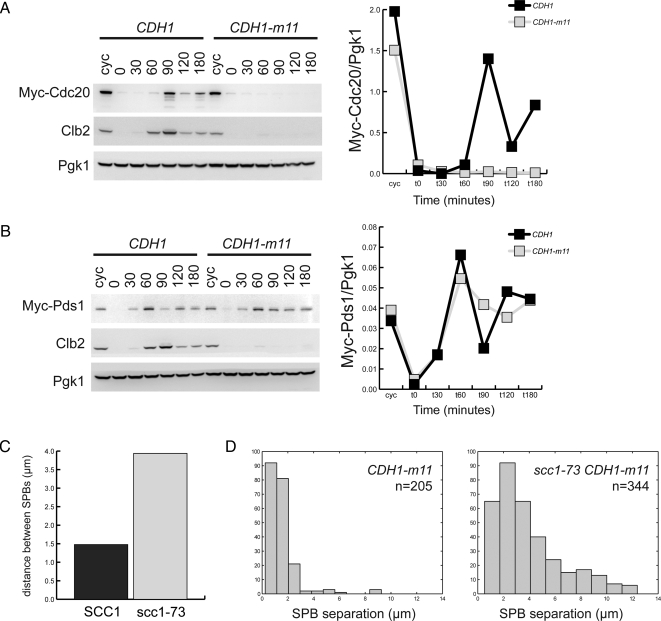
A failure of cohesin cleavage not dependent upon checkpoint activation could also explain failure of anaphase. Cdh1 may target Cdc20 for degradation (Huang et al., 2001). Cdc20 promotes anaphase by degradation of the separase inhibitor Pds1, allowing cleavage of the cohesin complex subunit Scc1; sister chromatids can then separate upon loss of cohesion. Failure to accumulate sufficient Cdc20, if it results in an inability to clear Pds1, could account for persistent short bipolar spindles. We find that CDH1-m11 cells fail to accumulate Cdc20 (Figure 5A).
Figure 5.
CDH1-m11 cells accumulate Pds1 but not Cdc20, and lengthen their spindles upon cohesin inactivation. (A) Left, immunoblots against strains synchronously released from α-factor with endogenously tagged Cdc20 and either CDH1 or CDH1-m11. Right, quantification of normalized Myc-Cdc20 levels from immunoblots. (B) Left, immunoblots against strains synchronously released from α-factor with endogenously tagged Pds1 and either CDH1 or CDH1-m11. Right, quantification of Myc-Pds1 levels standardized to Pgk1 loading control. (C) Average distance between separated SPBs in CDH1-m11 SCC1 and temperature-sensitive CDH1-m11 scc1-73 cells, 2 h after release from α-factor block. (D) Histogram of distance between SPBs from C.
If failure to accumulate Cdc20 accounts for anaphase failure, then Pds1 should remain at high levels in CDH1-m11 cells. Indeed, we find these cells unable to clear Pds1, with levels comparable with those found in cycling cells (Figure 5B). In the control CDH1 culture, a transient drop in Pds1 is rapidly followed by reaccumulation due to entry into the next cell cycle and loss of cell cycle synchrony; in contrast, the CDH1-m11 culture shows accumulation of Pds1 to a high level that drops only slightly, despite the continued uniform cell cycle arrest of these cells. The kinetic differences in Pds1 accumulation between CDH1 and CDH1-m11 cultures are reproducibly observed.
Thus Pds1 accumulation and consequent failure of cohesin cleavage could account for anaphase failure in sporadic CDH1-m11 cells with bipolar spindles. Consistent with this idea, scc1-73, a temperature-sensitive allele of a cohesin complex subunit, promotes increased spacing between SPBs in CDH1-m11 cells with duplicated SPBs at the restrictive temperature, indicating that inability to cleave cohesin contributes to the short bipolar spindle phenotype (Figure 5, C and D; Supplemental Figure 5B).
Cdc20 was reported to promote Pds1 proteolysis much more effectively than mitotic cyclin proteolysis, and Cdh1 was reported to have the opposite specificity (Visintin et al., 1997). Our results are consistent with this idea, because Pds1 persists in the face of unregulated Cdh1-m11.
Acm1 Cooperates with Cdk Phosphorylation in Regulating Spindle Pole Body Separation
The heterogeneous SPB phenotype of CDH1-m11 cells suggested the possibility that the level of APC-Cdh1 activity in these cells is close to a threshold for spindle morphogenesis. We reasoned that endogenous Acm1 might titrate a sufficient level of Cdh1-m11 to keep the system near this threshold. Consistent with this idea, CDH1-m11 acm1 cells completely failed to separate SPBs: <1% of cells, compared with ∼30% in ACM1 cells (Figure 6, A and B).
If Cdh1-m11 is near a threshold for inhibition by Acm1, then increasing ACM1 gene dosage should strongly shift the CDH1-m11 strain arrest phenotype. To test this in a GALL-ACM1 CDH1-m11 background, we performed an ends-in recombination of a genomic segment containing ACM1 at the URA3 locus, thereby allowing for multiple tandem integrations. Transformants were tested for ability to accumulate biomass on glucose (GALL-ACM1 off), and for distinguishable levels of biomass accumulation we assessed ACM1 copy number by quantitative PCR. We found clones with two, three or five copies of ACM1 (including the endogenous locus). Five copies of ACM1 fully rescued viability of CDH1-m11 cells, consistent with the high-copy plasmid suppression results described above. However, two or three copies were essentially insufficient for rescue (∼3 to 4 log drop in colony formation upon shutoff of GAL-ACM1 expression; data not shown).
Despite lack of rescue of overall viability, 2X ACM1 CDH1-m11 GALL-ACM1 cells were almost all able to form a short bipolar spindle upon GALL-ACM1 shutoff (Figure 6, A and B). Strikingly, these cells nevertheless almost quantitatively failed to progress to anaphase. These results suggest that multiple events in spindle morphogenesis and function are inhibited by Cdh1-m11, because failure of short spindle formation could be quantitatively uncoupled from subsequent anaphase failure by increased ACM1 gene dosage. It is likely that different events regulated by Cdh1 have distinct thresholds for inhibition, presumably due to different sensitivity of targets to Cdh1-driven proteolysis.
DISCUSSION
CDK Phosphorylation of Cdh1 Is Essential for Accumulation of Cdh1 Targets and for Spindle Morphogenesis
Multiple mechanisms of Cdh1 regulation have been proposed. CDK-mediated phosphorylation of Cdh1 inhibits Cdh1-APC interaction (Zachariae et al., 1998), and promotes export Cdh1 nuclear export (Jaquenoud et al., 2002). The Acm1 protein is a stoichiometric Cdh1 inhibitor that blocks Cdh1-APC interaction (Martinez et al., 2006). Cdc5 (polo kinase) may phosphorylate and inhibit Cdh1 (Crasta et al., 2008).
Here, we show that Cdk phosphorylation is a central and essential mechanism at endogenous Cdh1 levels, using exact gene replacement. Acm1 binding contributes buffering capacity. We have been unable to detect a contribution of Cdc5 phosphorylation to Cdh1 regulation.
The essentiality of CDK phosphorylation for Cdh1 regulation could be due to a phosphorylation requirement for blockage of Cdh1-APC interaction, for Msn5 interaction and nuclear export, or both. Our experiments do not distinguish between these possibilities, although nuclear export is unlikely to be strictly required for Cdh1 inhibition, because Msn5 is not essential.
CDH1-m11 as an exact gene replacement yields a tight first-cycle arrest with uniform bud morphology and replicated DNA. In contrast to results with CDH1-m11 overexpressors, the spindle phenotype of CDH1-m11 cells is somewhat heterogeneous; this result may have interesting consequences for the role of Acm1 (see below).
Heterozygous CDH1-m11/CDH1 GAL-ACM1 diploids, although inviable upon shutoff of GAL-ACM1, nevertheless undergo efficient meiosis and sporulation without ACM1 overexpression. This may still reflect a requirement for phosphorylation for Cdh1 inhibition, because the meiotic kinase Ime2 inhibits Cdh1 by phosphorylation of different sites (Holt et al., 2007).
Cdk Phosphorylation of Cdh1 Is Required for Multiple Steps of Spindle Morphogenesis
The majority of CDH1-m11 cells fail to assemble a bipolar spindle. Our results fail to confirm the hypothesis (Crasta et al., 2006) that Cdh1-dependent degradation of the plus-end– directed motor Cin8 is sufficient to explain the requirement to inhibit Cdh1 for bipolar spindle morphogenesis. Cin8 is indeed efficiently degraded in CDH1-m11 cells, but undegradable Cdh1-resistant Cin8 (Hildebrandt and Hoyt, 2001) did not restore bipolar spindle formation to CDH1-m11 cells. The most likely reason for the discrepancy between our results and the previous report is that our experiments were carried out at endogenous expression levels. It is possible that stabilization of other spindle proteins degraded by Cdh1, such as Ase1 or perhaps Kip1, might aid spindle formation in CDH1-m11 cells (Crasta et al., 2006), singly or in combination. Ase1 proteolysis may prevent proper targeting of Cin8 to the mitotic spindle (Khmelinskii et al., 2009) even if Cin8 is stabilized. However, CIN8 kip1 ase1 cells are viable (Schuyler et al., 2003). Therefore, Cin8 is sufficient for SPB separation and spindle elongation in the absence of other known microtubule-associated APC-Cdh1 substrates, so if Cdh1-mediated proteolysis of these proteins was sufficient to prevent spindle morphogenesis, then restoration of only Cin8 should have restored bipolar spindle formation, which was not observed in our experiments.
Mitotic cyclins are required for spindle morphogenesis (Fitch et al., 1992). Introducing undegradable Clb2 into CDH1-m11 cells results in apparent SPB separation. Thus, Clb2 is a significant Cdh1-m11 target accounting for the block to spindle morphogenesis. Spindle morphology was defective in most of these cells, implying the existence of other Cdh1 targets (probably including Cin8 and Ase1) that cooperate with mitotic cyclins in spindle morphogenesis.
CDH1-m11 cells that do make a bipolar spindle nevertheless fail to undergo anaphase. Our results suggest that this is probably due to severe depletion of Cdc20 levels by Cdh1-APC from (Shirayama et al., 1998; Huang et al., 2001), with consequent failure of Pds1 proteolysis, leading to failure of cohesin cleavage. Other factors contributing to anaphase failure in bipolar spindle bearing CDH1-m11 cells could include proteolysis of other motor proteins or spindle components due to Cdh1-m11 activity; activation of the spindle checkpoint does not seem to be responsible.
Acm1 Is a Physiological Cdh1 Buffer
Curiously, endogenous levels of ACM1 allow bipolar spindle formation in a minority of CDH1-m11 cells, because deletion of ACM1 eliminates these spindles; in contrast, doubling ACM1 copy number results in bipolar spindle formation in nearly all CDH1-m11 cells. Because varying Acm1 levels results in corresponding changes in Clb2 levels in CDH1-m11 cells (Figure 6C), it is possible that some or all of the SPB separation response to Acm1 levels is mediated through Clb2 levels; alternatively, other Cdh1 targets such as spindle regulatory proteins Cin8, Ase1, and Fin1 may contribute. Reciprocally, the ability of reintroduction of Clb2 into CDH1-m11 cells to restore SPB separation may be due to direct spindle regulation by Clb2, or may be indirect, due to Clb2 regulation of Acm1 levels. The complexity of the network controlling Cdh1, Acm1, and spindle morphogenesis precludes simple answers to such questions.
Acm1 seems to be present at a level just insufficient to inactivate completely CDK-unphosphorylatable Cdh1, at least with respect to bipolar spindle formation, when both proteins are expressed at endogenous levels. It is interesting to consider possible dynamic consequences of this effect. Acm1 levels are tightly cell cycle regulated by changes in transcription and protein stability (Spellman et al., 1998; Martinez et al., 2006). Acm1 levels higher than those in wild-type cells might sporadically allow premature bipolar spindle formation, before full inactivation of Cdh1 by complete CDK phosphorylation (because partial phosphorylation may result in partial Cdh1 activity (Zachariae et al., 1998). Lower levels than wild-type, in contrast, could put a demand on the system for much more efficient and quantitative Cdh1 phosphorylation than would otherwise be required. However, these considerations cannot imply an essential role for regulation of Acm1 levels, because both strong overexpression and deletion of ACM1 are tolerated with little or no overt phenotype.
Substrate Specificity of Cdh1
The APC coactivators Cdc20 and Cdh1 target a distinct but overlapping set of proteins for proteasomal destruction; this substrate specificity probably contributes to the orderly progression through anaphase and exit from mitosis. Because Cdh1 activity is effectively inhibited until late anaphase (Zachariae et al., 1998), it has been unclear whether failure of overexpressed Cdh1 to promote degradation of targets such as Pds1 (Zachariae et al., 1998) was the consequence of true substrate specificity or merely efficient pre-anaphase inhibition of Cdh1 activity. The ability of purified APC-Cdh1 to efficiently ubiquitinate Pds1 (Thornton et al., 2006) argued for the latter. Here, we find that APC-Cdh1 is highly effective at clearing Clb2, Cdc5, and Cdc20, but far less capable of clearing the proposed Cdc20 targets Pds1 and Clb5. The apparent discrepancy between in vitro and in vivo activity of Cdh1 toward Pds1 could reflect biochemical regulation not recapitulated in the purified system. Alternatively, increased Pds1 transcription could replenish depleted Pds1; CDH1-m11 cells probably have unrestrained SBF activity, because SBF is inactivated by the Cdh1 target Clb2 (Amon et al., 1993), and PDS1 is in the SBF regulon (Spellman et al., 1998). Further, Clb2 promotes its own transcription as well as that of CDC20 (Amon et al., 1993; Zhu et al., 2000). Such transcriptional circuitry could help ensure the proper order and function of APC coactivators; delayed inactivation of Cdh1 during a normal cell cycle could result in greater transcription of the G1 and S phase cyclins that serve to inactive Cdh1, while inhibiting anaphase through Pds1 synthesis.
Mitotic Cyclins May Be Central Cdh1-APC Targets
The hyperpolarized bud growth phenotype of CDH1-m11 cells is likely a direct consequence of removal of mitotic cyclins, because restoration of Clb2 to CDH1-m11 cells eliminates this phenotype. This presumably occurs either because Clb2 directly promotes isotropic growth, or because Clb2 inhibits expression of genes such as the G1 cyclin CLN2 that directly drive polarized bud growth (Lew and Reed, 1993; Amon et al., 1994).
In spindle morphogenesis, Cdh1 acts at various thresholds, and probably acts on multiple targets, to prevent final successful anaphase. Mitotic cyclins are capable of restoring a separated SPB phenotype in the context of CDH1-m11; however, these spindles have abnormal tubulin fluorescence and a relatively frequent occurrence of more than two SPB foci. This suggests that balance between Cdh1 and mitotic cyclins permits specific steps such as SPB separation to occur, with multiple other interactions and couplings present to orchestrate specific aspects of spindle physiology, including tubulin dynamics and spindle maintenance. Complex dynamics have been described at the spindle midzone regulated by both APC-Cdh1 targets and net CDK phosphorylation (Higuchi and Uhlmann, 2005; Fridman et al., 2009; Khmelinskii et al., 2009).
Our strategy in this study, to first deregulate Cdh1 at the endogenous level and then to add back single Cdh1 targets by introducing undegradable alleles expressed at endogenous levels, allows accurate dissection of the mechanism of action of even a highly pleiotropic regulator such as Cdh1. In bud morphogenesis, the situation is simple: the hyperpolarized bud phenotype is essentially due to a single target, Clb2. Spindle morphogenesis is clearly much more complicated, but nevertheless, we are able to implicate mitotic cyclins as major regulators sufficient for significant spindle morphogenesis in the absence of other Cdh1 targets. With appropriate variations, this strategy should be applicable to dissection of the action of other complex regulators.
Supplementary Material
ACKNOWLEDGMENTS
We thank O. Cohen-Fix (National Institute of Diabetes and Digestive and Kidney Diseases), M. Hall (Purdue University), M. Hoyt (Johns Hopkins University), K. Nasmyth (University of Oxford), S. Reed (Scripps Research Institute), M. Schwab (University of Regensburg), P. Sorger (Harvard Medical School), and F. Uhlmann (Cancer Research UK) for strains and plasmids. B. Drapkin developed the OpenLab microscope automation used to obtain images of fixed cells. We thank S. Di Talia, B. Drapkin, Y. Lu, and all other members of the Cross laboratory for helpful and insightful discussions. This work was supported by U.S. Public Health Service grant GM-47238. J.A.R. was supported by National Institutes of Health Medical Scientist Training Program grant GM-07739.
Abbreviations used:
- APC
anaphase promoting complex
- CDK
cyclin-dependent kinase
- FOA
5-fluoroorotic acid
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0901) on January 20, 2010.
REFERENCES
- Amon A., Irniger S., Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Amon A., Tyers M., Futcher B., Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Bean J. M., Siggia E. D., Cross F. R. Coherence and timing of cell cycle start examined at single-cell resolution. Mol. Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Benanti J. A., Matyskiela M. E., Morgan D. O., Toczyski D. P. Functionally distinct isoforms of Cik1 are differentially regulated by APC/ C-mediated proteolysis. Mol. Cell. 2009;33:581–590. doi: 10.1016/j.molcel.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles J. F., Jaspersen S. L., Tinker-Kulberg R. L., Hwang L., Szidon A., Morgan D. O. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J. M., Kirschner M. W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Crasta K., Huang P., Morgan G., Winey M., Surana U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006;25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Lim H. H., Giddings T. H., Jr., Winey M., Surana U. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 2008;10:665–675. doi: 10.1038/ncb1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin B. J., Lu Y., Procko A. L., Timney B. L., Cross F. R. Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Mol. Syst. Biol. 2009;5:328. doi: 10.1038/msb.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A. E., Cantley L. C., Yaffe M. B. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M., Sullivan M., Morgan D. O. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol. Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I., Dahmann C., Surana U., Amon A., Nasmyth K., Goetsch L., Byers B., Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M., Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., Roof D. M. Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p. Proc. Natl. Acad. Sci. USA. 2001;98:12515–12520. doi: 10.1073/pnas.231212498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. C., Jeong D. E., Henderson J. T., Choi E., Bremmer S. C., Iliuk A. B., Charbonneau H. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J. Biol. Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. C., Warren E. N., Borchers C. H. Multi-kinase phosphorylation of the APC/C activator Cdh1 revealed by mass spectrometry. Cell Cycle. 2004;3:1278–1284. doi: 10.4161/cc.3.10.1153. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Hoyt M. A. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Hutti J. E., Cantley L. C., Morgan D. O. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell. 2007;25:689–702. doi: 10.1016/j.molcel.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K., Saunders W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. N., Park I., Ellingson E., Littlepage L. E., Pellman D. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 2001;154:85–94. doi: 10.1083/jcb.200102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M., van Drogen F., Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Charles J. F., Morgan D. O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Juang Y. L., Huang J., Peters J. M., McLaughlin M. E., Tai C. Y., Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Kramer E. R., Scheuringer N., Podtelejnikov A. V., Mann M., Peters J. M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. H., Goh P. Y., Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- Martinez J. S., Jeong D. E., Choi E., Billings B. M., Hall M. C. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol. Cell. Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapenko D., Burton J. L., Wang R., Solomon M. J. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm 1, regulation by proteolysis and Cdc28 phosphorylation. Mol. Cell. Biol. 2008;28:4653–4664. doi: 10.1128/MCB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K. E., Cohen-Fix O. The role of Cdh1p in maintaining genomic stability in budding yeast. Genetics. 2003;165:489–503. doi: 10.1093/genetics/165.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Murray A. W. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Schuyler S. C., Liu J. Y., Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Lutum A. S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Mocker D., Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Toth A., Galova M., Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B. R., Ng T. M., Matyskiela M. E., Carroll C. W., Morgan D. O., Toczyski D. P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. CDC20 and CDH 1, a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wäsch R., Cross F. R. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Woodbury E. L., Morgan D. O. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhu G., Spellman P. T., Volpe T., Brown P. O., Botstein D., Davis T. N., Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.