GbpD, a guanine exchange factor specific for Rap1, has been implicated in adhesion, cell polarity, and chemotaxis of Dictyostelium cells. Here it is shown that activated Rap1 directly binds to PI3K. The activation of PI3K by Rap1 and RasG regulates basal and chemoattractant-stimulated PIP3 levels and pseudopod formation.
Abstract
GbpD, a Dictyostelium discoideum guanine exchange factor specific for Rap1, has been implicated in adhesion, cell polarity, and chemotaxis. Cells overexpressing GbpD are flat, exhibit strongly increased cell-substrate attachment, and extend many bifurcated and lateral pseudopodia. Phg2, a serine/threonine-specific kinase, mediates Rap1-regulated cell-substrate adhesion, but not cell polarity or chemotaxis. In this study we demonstrate that overexpression of GbpD in pi3k1/2-null cells does not induce the adhesion and cell morphology phenotype. Furthermore we show that Rap1 directly binds to the Ras binding domain of PI3K, and overexpression of GbpD leads to strongly enhanced PIP3 levels. Consistently, upon overexpression of the PIP3-degradating enzyme PTEN in GbpD-overexpressing cells, the strong adhesion and cell morphology phenotype is largely lost. These results indicate that a GbpD/Rap/PI3K pathway helps control pseudopod formation and cell polarity. As in Rap-regulated pseudopod formation in Dictyostelium, mammalian Rap and PI3K are essential for determining neuronal polarity, suggesting that the Rap/PI3K pathway is a conserved module regulating the establishment of cell polarity.
INTRODUCTION
Chemotaxis or directional movement toward a chemical compound is an essential property of many cells (Van Haastert and Devreotes, 2004). It plays a role in diverse functions such as the sourcing of nutrients by prokaryotes, the formation of multicellular structures in protozoa, the tracking of bacterial infections by neutrophils, and the organization of the embryo in metazoa (Baggiolini, 1998; Campbell and Butcher, 2000; Crone and Lee, 2002; Iijima et al., 2002). The mechanism of chemotaxis is essentially identical in all eukaryotes, and because of its genetically tractability, the social amoebae Dictyostelium discoideum is often used as model organism to study chemotaxis (Devreotes and Zigmond, 1988; Van Haastert and Devreotes, 2004). During the vegetative state, Dictyostelium are single-celled amoeba that feed on bacteria. On starvation, cells undergo a tightly regulated developmental process in which they secrete and chemotax toward cAMP, resulting in multicellular fruiting bodies.
During random movement in buffer, Dictyostelium cells repeatedly extend and retract pseudopodia. In response to cAMP cells rapidly polarize due to actin filaments in the front that induce the formation of local pseudopodia and an acto-myosin–containing uropod at the back (Van Haastert and Devreotes, 2004). An important response for establishing cell polarity and chemotaxis is the formation and accumulation of phosphatidylinositol-3,4,5-triphosphate [PIP3] at the leading edge (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). This PIP3 gradient is achieved by the reciprocal localization of phosphatidylinositol 3-kinase (PI3K), which translocates to the leading edge and produces PIP3 by the phosphorylation of PI(4,5)P2 (phosphatidylinositol 4,5-bisphosphate), and PTEN, which catalyzes the reverse reaction and localizes at the back of the cell (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). Recent studies have shown that the PI3K pathway is not essential for chemotaxis, but rather that chemotaxis depends on several interconnecting pathways (Loovers et al., 2006; Hoeller and Kay, 2007; Takeda et al., 2007). Two studies identified phospholipaseA2 as a component of a chemotactic pathway that acts in parallel to the PI3K pathway (Chen et al., 2007; Van Haastert et al., 2007), whereas Veltman et al. (2008) have shown the presence of a third soluble guanylyl cyclase-dependent pathway in cells starved for longer times. Kamimura et al. (2008) recently described another PIP3-independent pathway regulating chemotaxis, involving activation of TorC2 and protein kinase B (PKB). However, although PI3K is not essential for chemotaxis, it is clear that it plays an important role in directional sensing, especially in shallow gradients (Takeda et al., 2007; Van Haastert et al., 2007).
Rap proteins belong to the Ras subfamily of small G-proteins and are involved in processes like adhesion, exocytosis, differentiation, and cell proliferation (Bos et al., 2001; Bos, 2005). Small G-proteins, acting as molecular switches, which cycle between an active GTP-bound and inactive GDP-bound state. Activation is regulated by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP, and inactivation is regulated by GTPase-activating proteins (GAPs) that stimulate the hydrolysis of bound GTP to GDP (Bourne et al., 1991). Recently, the completed assembly of the Dictyostelium genome has led to the identification of 14 Ras subfamily members, an unusually large number (Eichinger et al., 2005; Weeks, 2005). Thus far, six Ras subfamily proteins have been characterized and have been shown to be involved in a wide variety of processes including cell movement, polarity, cytokinesis, chemotaxis, macropinocytosis, and multicellular development (Chubb and Insall, 2001; Wilkins and Insall, 2001; Weeks, 2005; Kortholt and Van Haastert, 2008). RasC and RasG are the best characterized Dictyostelium Ras proteins; both are activated in response to cAMP (Kae et al., 2004), are involved in regulation of the cAMP relay, and are important for cAMP-dependent chemotaxis (Bolourani et al., 2006; Bolourani et al., 2008). Disruption of both rasC and rasG results in a total loss of cAMP-mediated signaling, suggesting that all cAMP signal transduction in early development is partitioned between pathways that use either RasC or RasG (Bolourani et al., 2006, 2008). The only thus far characterized Rap subfamily member, Rap1, is essential and is involved in proliferation, growth, adhesion, development, and regulation of the cytoskeleton (Rebstein et al., 1993; Seastone et al., 1999; Kang et al., 2002; Kortholt et al., 2006; Jeon et al., 2007b; Parkinson et al., 2009).
Previously we have characterized GbpD as a Rap-specific D. discoideum GEF important for adhesion, cell polarity, and chemotaxis. GbpD contains a CDC25-homology domain, a Ras exchange motif (REM) domain, a GRAM domain, and two cyclic nucleotide–binding (CNB) domains (Goldberg et al., 2002). Overexpression of GbpD results in an increased level of active Rap1 and subsequently in increased cell flattening, increased cell-substrate attachment, and severely reduced chemotaxis, as these cells extend many bifurcated and lateral pseudopodia (Bosgraaf et al., 2005; Kortholt et al., 2006). Phg2, a serine/threonine-specific kinase, directly interacts with active Rap1 via its Ras association (RA) domain and is necessary for GbpD/Rap1-regulated adhesion but is not essential for GbpD/Rap1-regulated cell polarity (Kortholt et al., 2006). Jeon et al. (2007a,b) showed that Phg2 mediates cell adhesion at the leading edge by regulating myosin II disassembly. In this study we characterized the role of GbpD/Rap1 in cell polarity and chemotaxis in more detail. Together our data indicate that a GbpD/Rap/PI3K pathway helps controlling pseudopod formation in Dictyostelium.
MATERIALS AND METHODS
Cell Culture
All cell lines were grown either on 9-cm dishes or in shaking culture containing HG5 medium (14.3 g/l of peptone, 7.15 g/l of glucose, 0.49 g/l of KH2PO4, and 1.36 g/l of Na2HPO4·H2O). To select for transformants with one of the extrachromosomal plasmids described below, cells were grown in HG-5 supplemented with 10 μg/ml G418 (Gibco BRL, Rockville, MD) and/or hygromycin B (Invitrogen). The pi3k1/2- null strain was obtained from the Dictyostelium Stock Center (DSC; Columbia University, New York, NY).
Aggregation Test
Dictyostelium cells were grown to an amount of 2 × 107 cells per 9-cm dish. Cells were collected, washed two times in 10 mM phosphate buffer (PB), pH 6.5, and suspended in 500 μl of PB. Subsequently cells were placed on nonnutrient agar plates (15.0 g/l agar in PB), and pictures were taken after 24 h with an Olympus DP10 camera (Melville, NY).
Construction of Plasmids
For expression of GbpD in Dictyostelium the previously described MB74HYG GbpD vector and MB74GbpD-GFP were used (Bosgraaf et al., 2005). The PIP3 detector PHcracGFP and GbpD were expressed from a single vector. For this the pleckstrin homology (PH) domain of CRAC was subcloned in the previously described shuttle vector pDM329 (Veltman et al., 2009a). Subsequently the expression cassette with the PH domain fused at its C-terminus to GFP was excited by NgoMIV and ligated into the unique NgoMIV site of MB74HYG GbpD. For expression and purification of the RBD domain of PI3K2 (amino acids 757-918) the encoding DNA fragment was cloned in pGEX-4T3 (Pharmacia, Piscataway, NJ) leading to an N-terminal glutathione S-transferase (GST) fusion proteins. The RBD domain of PI3K2 was amplified using the forward primer (5′- CAGGATCCATGGGTCATATTCAGGCAGAGTTG-3′) and reverse primer (5′-CTGCGGCCGCTTAATAGTCATAGTCCATTAATGAG-3′) with Dictyostelium cDNA as template. The fragment was digested with BamHI and NotI and cloned in pGEX4T-3. The expression vector was checked by sequencing. For expression of RalGDS-RBD (amino acids 786-884) in Dictyostelium, the encoding DNA fragment was amplified using the forward primer (5′-GGATCCAAAAAATGAGCTCCGCGCTGCCGCTC-3′) and reverse primer (5′-TCTAGACCGCTTCTTGAGGACAAAGTCATAG-3′). The BamHI/XbaI fragment was cloned in the BglII-SpeI site of a modified MB74GFP vector that contained a hygromycin-resistance cassette (Bosgraaf et al., 2005), resulting in the plasmid pDM115-RalGDS-RBD-GFP. For the regulated expression of Rap1G12V and Rap1S17N the previously described tetracycline-controlled inducible expression system was used (Veltman et al., 2009b). The Rap1 mutants were amplified using the forward primers RapG12VF (5′GGATCCAAAATGCCTCTTAGAGAATTCAAAATCGTCGTTTTAGGTTCAGTTGGTGTAGGTAAATCTGCTTTG-3′) and Rap1S17NF (5′-GGATCCAAAATGCCTCTTAGAGAATTCAAAATCGTCGTTTTAGGTTCAGTTGGTGTAGGTAAATCTGCTTTG-3′) in combination with the reverse primer RapR (5′- GACTAGTTTACAATAAAGCACATTTTG-3′). Subsequently the BamHI-SpeI fragments were cloned into the BglII-SpeI site of a pDM309 vector (Veltman et al., 2009b). RasGS17N and RasGG12V were expressed under the previously described folate-repressible discoidin promoter (Khosla et al., 1996). For expression and purification of the C-terminal-truncated Rap1 (amino acids 1-169) and RasG, the previous described constructs were used (Kortholt et al., 2006). The plasmid WF38 expressing the PH domain of CRAC fused to enhanced green fluorescent protein (eGFP) and a plasmid expressing PTEN-GFP were kindly provided by Dr. P. Devreotes ((Johns Hopkins University).
Protein Preparation
The GST fusion constructs were expressed in Bl21(DE3)codonplus-RIL cells (Stratagene, La Jolla, CA). The cells were grown in standard I medium (Merck, Schwalbach, Germany) containing 50 mg/ml ampicillin and 25 mg/ml chloramphenicol, induced at an OD600 of 0.8 with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), and incubated overnight at 25°C. After protein production, the cells were pelleted (15 min, 4000 × g and 4°C), washed in 0.9% NaCl, and resuspended in lysis buffer (5 mM 1,4-dithioerythritol [DTE], 50 mM NaCl, 5 mM MgCl2, and 30 mM Tris HCl, pH 7.9). To inhibit protein degradation, 1 mM of phenylmethylsulfonylfluoride (PMSF) was added. Cells were lysed in a Microfluidizer (Microfluidics, Newton, MA), and 0.1 μg/ml DNAseI was added. Lysates were cleared by centrifugation (45 min, 100,000 × g at 4°C), and fusion proteins were purified using a reduced glutathione (GSH) affinity column (Pharmacia). Rap1 (amino acid 1-169) and RasG were eluted from the column as GST fusion proteins in lysis buffer containing 20 mM glutathione. For PI3K2-RBD the GST tag was cleaved on the column using 300 U of thrombin (Serva, Paramus, NJ), followed by elution of the protein in lysis buffer. Isolated proteins were analyzed using SDS-page and the protein concentration was determined by the method of Bradford (Bio-Rad, Hercules, CA).
Confocal Analysis and Chemotaxis Assays
Chemotaxis toward cAMP was tested using micropipettes filled with 10−4 M cAMP applied to a field of aggregation competent cells with an Eppendorf femtotip (Hamburg, Germany) at a pressure of 25 hPa. Cells were starved in PB for 6–8 h, resuspended in PB, and monitored by phase-contrast microscopy. The motile behavior of cells in spatial gradients of cAMP was analyzed using computer-assisted methods previously described (Soll, 1999). Briefly, images were recorded every 10 s during 15 min. The contour of the cell and the position of the cell centroid were determined at each time point for ∼25 cells at a distance of 50–100 μm from the pipette.
Adhesion Assay
To quantify the strength of cell adhesion to the surface, we used a previously published protocol (Fey et al., 2002) with a few modifications. Briefly, cells were grown on HG-5 medium in six-well plates (Nunc, Wiesbaden-Biebrich, Germany) to a maximum of 70% confluence. The medium was refreshed and incubated for 1 h. The plates were then rotated on a shaker at 150 rpm, and after 1 h a sample of 150 μl of the medium was taken. The remaining medium was removed and replenished with fresh HG-5, and the remaining adhering cells were detached by repeated pipetting. The number of cells in the two samples was determined in triplicate using a hemocytometer. The number of nonadhered cells in the first sample was divided by the total number of cells (first plus second sample) to yield the percentage of loosen-adhered cells.
Guanine Nucleotide Dissociation Inhibition Measurements
For the guanine nucleotide dissociation inhibition (GDI) measurements, Rap1 was loaded by incubating in ammonium sulfate-buffer (200 mM (NH4)2SO4, 50 mM DTE, 10 μM ZnCl2, pH 7.6) containing alkaline phosphatase (2 U/mg protein) and a 1.5 M excess of the fluorescence GTP analogue mGppNHp (John et al., 1990). The protein–nucleotide complex was separated from unbound nucleotides by size exclusion chromatography (Superdex 75 16/20, Pharmacia). The affinity between PI3K2-RBD and the indicated G-proteins was determined using the inhibition of the mGppNHp release as described (Herrmann et al., 1996). Briefly, mGppNHp-loaded G-proteins were incubated in the presence of varying concentrations of PI3K2-RBD and a 200-fold excess of unlabeled GppNHp in assay buffer at 25°C. The decay in fluorescence was measured in a Spex1 spectrofluorometer (Spex Industries, Edison, NJ), with excitation and emission wavelengths of 366 and 450 nm, respectively. The observed rate constants (kobs) were single exponential fitted using the program Grafit (Erithacus Software, West Sussex, United Kingdom). The dependence of the observed rate constants on the effector concentration was fitted to yield the dissociation constant (Kd) as described (Herrmann et al., 1996).
GST-Ras-binding Domain Pulldown Assay
The GST-Ras-binding domains (RBDs) of PI3K1–3 and Byr2 were expressed in Escherichia coli and purified as described previously (Kae et al., 2004). After cAMP pulsing, AX-2 cells were harvested by centrifugation and resuspended at a density of 2 × 107 cells/ml in PB containing 1 mM caffeine. After 30 min, aliquots (2 ml) of cell suspension were stimulated by addition of cAMP to 15 μM. Cell suspensions (350 μl) were lysed at the indicated times by mixing with an equal volume of 2× lysis buffer (20 mM sodium phosphate, pH 7.2, 2% Triton X-100, 20% glycerol, 300 mM NaCl, 20 mM MgCl2, 2 mM EDTA, 2 mM Na3VO4, 10 mM NaF, containing two tablets of Roche Complete protease inhibitor per 50 ml buffer; Mannheim, Germany). The lysates were centrifuged for 10 min, and the protein concentrations of the supernatants were determined using the Bio-Rad protein assay from 400 μg of lysate protein was then incubated with 100 μg of GST-RBD on glutathione-Sepharose beads (Amersham Biosciences, Freiburg, Germany) at 4°C for 1 h. The glutathione-Sepharose beads were harvested by centrifugation and washed three times in 1× lysis buffer. Gel loading buffer (50 μl of 1× SDS) was then added to the pelleted beads, and the suspension boiled for 5 min. Samples were subjected to SDS-PAGE, and Western blots probed with anti-Rap1– or anti-RasG–specific antibody.
PKB/Akt Phosphorylation Assays
PKB/Akt phosphorylation assays were performed as previously described (Lim et al., 2001). Briefly, 100-μl aliquots of cAMP-stimulated cell suspension were removed at the indicated time points and immediately lysed as described above for the RBD pulldown assays. Protein samples (10 μg) were fractionated by SDS-PAGE, blotted onto nitrocellulose, and probed with the phospho-threonine antibody (Cell Signaling Technologies, Beverly, MA).
RESULTS
PI3K Is Essential for GbpD/Rap1-mediated Cell Protrusions and Adhesion
Previously the strong GbpDOE phenotype was used to screen for downstream targets of GbpD and Rap1 in Dictyostelium (Kortholt et al., 2006). GbpD was overexpressed in mutants defected in adhesion or cell polarity, and Phg2, a serine/threonine-specific kinase, was identified as a Rap1 effector necessary for adhesion but not for cell polarity and chemotaxis (Kortholt et al., 2006).
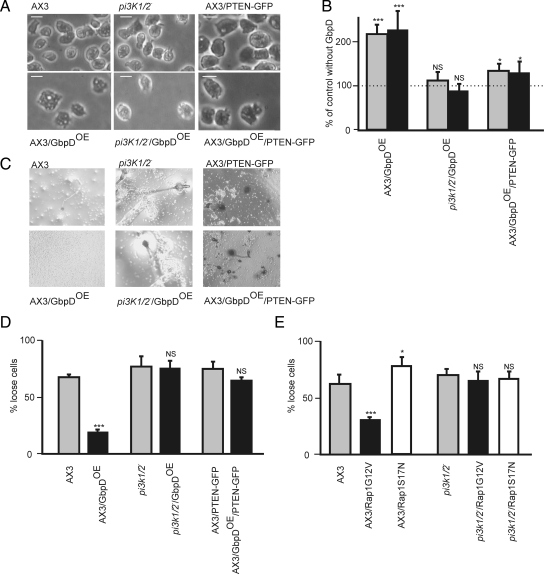
PIP3 is known to be a strong inducer of pseudopod formation (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). To investigate the possible role of PIP3 in the increased extension of pseudopodia by GbpDOE cells, we overexpressed GbpD in cells lacking PI3K1/PI3K2 (pi3k1/2- null). Disruption of PI3K1/PI3K2, results in cells that have a strongly reduced amount of PIP3, a 90% reduced AKT/PKB activation in response to cAMP, defects in polarity, and a reduced chemotaxis speed (Zhou et al., 1998; Funamoto et al., 2001, 2002). First, the morphology of the mutants was analyzed microscopically. Wild-type cells overexpressing GbpD (AX3/GbpDOE) extended more pseudopodia and were very large and flat compared with wild-type cells (AX3; Figure 1A.). Confocal fluorescence microscopy revealed that GFP-tagged GbpD was expressed in pi3k1/2- null cells at levels approximately equal to that of wild-type cells (data not shown). Although expressing similar amounts of GbpD, pi3k1/2- null cells overexpressing GbpD (pi3k1/2−/GbpDOE) did not show an increased amount of pseudopodia or a flattened cell morphology (Figure 1, A and B). Furthermore, starved pi3k1/2-null and pi3k1/2-null/GbpDOE cells, although defective in cell polarity, are able to aggregate and showed normal development, whereas AX3/GbpDOE cells were unable to aggregate (Figure 1C).
Figure 1.
Cell morphology and adhesion of pi3k1/2-null cells overexpressing GbpD. To investigate the role of PI3K in the GbpD pathway, GbpD was expressed in wild-type (AX3) and pi3k1/2-null cells and also in cells overexpressing the PIP3-degrading enzyme PTEN. (A) Phase-contrast images of wild-type, pi3k1/2-null, and PTEN-GFP–expressing cells, grown in HG-5 medium. Scale bar, 10 μm. (B) Quantification of contact area (gray) and number of protrusions (black) in AX3, pi3k-null and AX3/PTEN-GFP cells without (control) or with overexpression of GbpD was performed as described (Bosgraaf et al., 2005). Data are presented as percentage of GbpDOE cells relative to its control; values are means; error bars, SDs; n = 10 cells. (C) Aggregation of wild-type (AX3) and mutant cells on agar after 24 h of starvation. (D and E) Cell attachment was determined by measuring the percentage of nonadherent wild-type and mutant vegetative cells after 1 h of rotation. Results are the means; error bars, SDs; n = 3 independent experiments. The effect of GbpDOE is significant; **p < 0.001; *p < 0.05; NS, not significant.
Second, the adhesive capacities of the cell lines were tested via an adhesion assay. As shown in Figure 1D, 77% of the pi3k1/2-null cells were detached from the substratum after 1 h of shaking, a slightly higher percentage than that for wild-type cells (68%). AX3/GbpDOE cells showed a large increase in cell-substrate adhesion, because only 18% of the cells were detached after 1 h of shaking. In contrast, cell attachment in pi3k1/2−/GbpDOE cells is similar to that for pi3k1/2-null cells (76% detached). These results suggest that PI3K is essential for GbpD-mediated adhesion. To investigate the role of Rap1 in a more direct way, we expressed constitutive active (Rap1G12V) and dominant negative (Rap1S17N) mutants in wild-type cells and pi3k1/2-null cells. Cell attachment is substantially enhanced in AX3/Rap1G12V cells and is reduced in AX3/Rap1S17N cells as previously described (Figure 1E; Jeon et al., 2007b). Consistent with an essential role for PI3K in GbpD/Rap1-mediated adhesion, expression of Rap1G12V or Rap1S17N does not alter cell-substratum attachment of pi3k1/2-null cells.
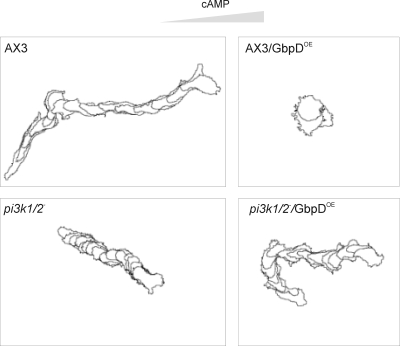
Thirdly we analyzed the chemotaxis properties of the mutants. When a pipette filled with cAMP is placed in the surrounding of starved wild-type cells, cells rapidly respond and chemotax persistently toward the source of cAMP, as depicted in Figure 2. pi3k1/2-null cells exhibit good chemotaxis toward the pipette (Funamoto et al., 2002). Overexpression of GbpD in wild-type cells strongly inhibits chemotaxis as these cells showed little movement toward the pipette. In contrast pi3k1/2−/GbpDOE cells exhibit a chemotaxis response that is nearly identical to the response of the parent pi3k1/2−cell line (Figure 2).
Figure 2.
Chemotaxis assay. To analyze the chemotaxis behavior of wild-type (AX3), pi3k1/2−, AX3/GbpDOE and pi3k−/GbpDOE strains, cells were starved and pulsed for 6–8 h, resuspended in PB, and monitored by phase-contrast microscopy. Cells were stimulated with a micropipette containing 10−4 M cAMP, from the right. The contours of the cells are shown at 1-min interval for AX3, pi3k1/2−, and pi3k1/2−/GbpDOE and at 5-min intervals for AX3/GbpDOE for a total of 15 min.
Overexpression of GbpD in wild type affects cell morphology, adhesion, and chemotaxis. The results presented here show that none of these effects are observed upon overexpression of GbpD in pi3k1/2-null cells or addition of the PI3K-inhibitor LY294002 (data not shown), indicating that PI3K is essential for all these GbpD-mediated signaling pathways.
Enhanced PIP3 Levels in GbpDOE Cells
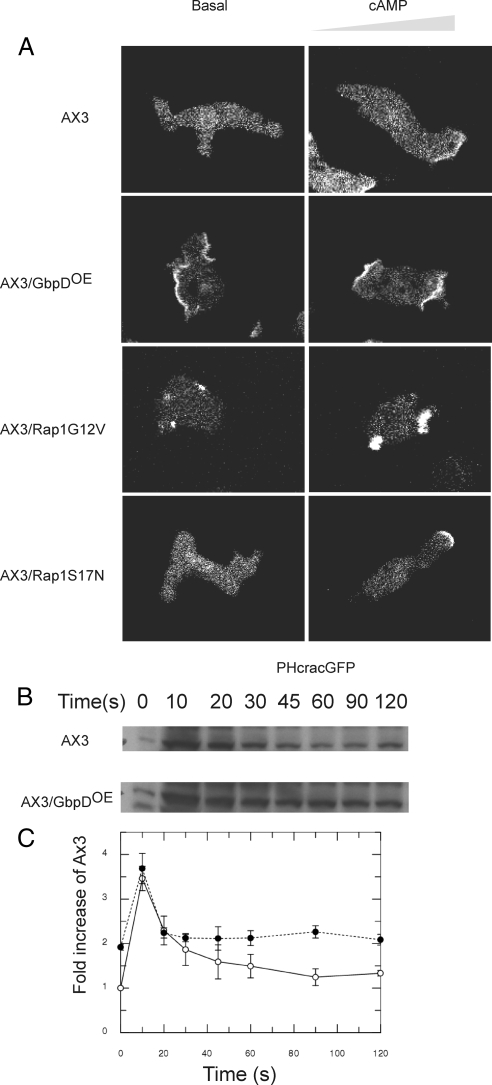
To investigate if GbpD is able to regulate PI3K in vivo, the effect of GbpD overexpression on PIP3 levels was measured by expressing the PIP3 detector PHcracGFP in GbpDOE cells (Parent et al., 1998). Nonstimulated, differentiated AX3/PHcracGFP-expressing cells moved in random directions and showed an evenly distributed PHcrac localization in the cytosol (Figure 3A, Movie 1). On the contrary, nonstimulated GbpDOE/PHcracGFP cells made multiple and broad PIP3 patches along the entire plasma membrane, from which multiple pseudopodia are extended (Figure 3A, Movie 2).
Figure 3.
Effect of GbpD expression on PI3K activity. (A) To investigate the effect of GbpDOE on PIP3 levels, the PIP3 detector PHcracGFP was expressed in wild-type, GbpDOE, RapG12VOE and RapS17NOE cells. Confocal images are shown for unstimulated cells (left) and for cells stimulated with cAMP from a micropipette on the right (right). Movies 1 and 2 for wild-type and GbpDOE cells, respectively, are available as Supplementary Information. (B) PKB phosphorylation in wild-type and GbpDOE cells. Cells were cAMP pulsed and then stimulated with 1 μM cAMP. Samples were removed at the times indicated and lysed directly into SDS loading buffer. Samples were subjected to SDS-PAGE and analyzed by Western blotting by probing with a phospho-threonine–specific antibody. The indicated ∼51-kDa protein corresponds to the phosphorylation of PKB/Akt. The blot shown is representative of three independent experiments. (C) Quantification of PKB phosphorylation for AX3 (○) and GbpDOE (•) cells. Data are presented as fold increase relative to AX3 cells before stimulation.
Both AX3/PHcracGFP and GbpDOE/PHcracGFP cells were stimulated with cAMP. Similar to previous investigations (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003), introduction of a pipette filled with cAMP in the surroundings of wild-type cells induced a strong translocation of PHcracGFP from the cytosol to the leading edge (Figure 3A, Movie 1). Pseudopodia are extended from PHcracGFP-containing areas of the plasma membrane, and cells move persistently toward the pipette. On the contrary, GbpDOE/PHcracGFP cells make multiple and broad PIP3 patches along the entire plasma membrane. Cells extend multiple, functional pseudopodia at the front and at lateral sides and showed little movement toward the pipette (Figure 3A, Movie 2). In Dictyostelium the activity of Akt/PKB is transiently stimulated in response to cAMP, in a PIP3-dependent manner (Meili et al., 1999; Lim et al., 2005). Akt/PKB is activated by phosphorylation at a conserved threonine residue, which can be detected by using a phospho-threonine specific antibody (Lim et al., 2005; Kortholt et al., 2007). Wild-type and GbpDOE cells were stimulated with cAM, and cell lysates were analyzed by Western blotting. In lysates from wild-type cells, a 51-kDa protein was transiently phosphorylated, peaking at 10 s after stimulation (Figure 3, B and C). GbpDOE cells show about twofold higher basal level of active PKB; the response to cAMP is similar as in wild type, but it recovers to the elevated basal activity.
These experiments show that GbpDOE cells have enhanced PIP3 formation. Consistently, cells expressing dominant active RapG12V also make broad PIP3 patches and show poor chemotaxis (Figure 3A). These data suggest a stimulatory role for GbpD and Rap1 in the regulation of PI3K activity.
Subcellular Distribution of Activated Rap1 Coincides with PI3K Activation and PIP3 Formation
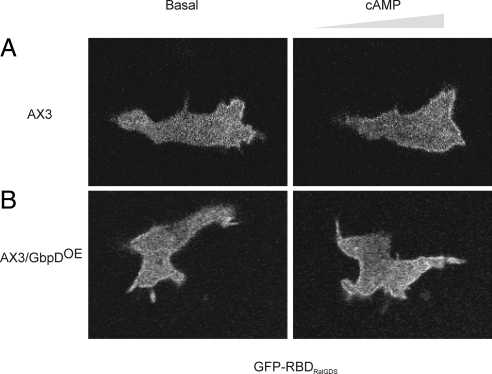
To examine the localization of Rap1 activation, a GFP-fused RalGDS-RBD construct (GFP-RBDRalGDS) was expressed in AX3 and AX3/GbpDOE cells. Previously it has been shown that GFP-RBDRalGDS only interacts with the GTP-bound form of Rap and can be used as a reporter to study dynamic changes in Rap activation in human cells (Bivona et al., 2004). Like PHcracGFP, GFP-RBDRalGDS is mainly localized in the cytosol of nonstimulated wild-type cells (Figure 4A, Movie 3), whereas in GbpDOE cells multiple and broad patches of GFP-RBDRalGDS along the entire plasma membrane were observed (Figure 4B). Because GbpD does not activate RasC or RasG, these results indicate that the GFP-RBDRalGDS probe can be used to visualize Rap activation, although it may not be completely specific. As shown in the right panel of Figure 4A, introduction of a pipette filled with cAMP in the surroundings of wild-type cells resulted in the rapid translocation of cytosolic GFP-RBDRalGDS to the membrane, similar to the translocation of the PIP3 detector PHcracGFP (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). Jeon et al. (2007) previously presented a similar localization of activated Rap in wild-type cells to that presented here. On the contrary, GbpDOE cells make multiple and broad GFP-RBDRalGDS patches along the entire plasma membrane (Figure 4B), similar to the distribution of PHcracGFP. These data show that the localization of Rap1 activation coincides with the localization of PI3K activation and PIP3 formation.
Figure 4.
Subcellular localization of Rap1 activation. To analyze the subcellular localization of Rap1 activation, the GFP-RBDRalGDS reporter was expressed in AX3 (A) and AX3/GbpDOE (B). Confocal images are shown for unstimulated cells (left) and for cells stimulated with cAMP from a micropipette on the right (right).
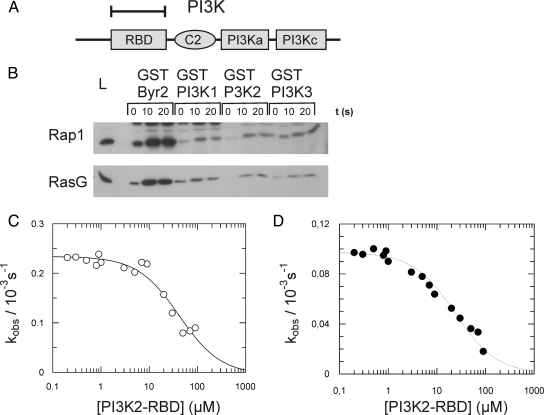
Rap1 and RasG Bind to the RBD Domain of PI3K
PI3K consist of an N-terminal membrane-targeting region, an RBD, a C2 lipid-binding domain, a PI3K catalytic association domain (PI3Ka), and a PI3K catalytic domain (PI3Kc; Figure 5A). RBD domains have an ubiquitin fold and interact tightly only with the GTP-bound but not with the GDP-bound conformation of Ras-like proteins (Herrmann, 2003). Previous studies have shown that the RBD of PI3K is essential for its activation and has been shown to interact weakly with RasD and RasC and strongly with RasG (Funamoto et al., 2002; Kae et al., 2004). To further investigate the binding specificity, pulldown experiments with the RBD domain of PI3K1–3 were performed. As shown in Figure 5B, all three PI3K RBD domains are able to bind activated Rap1 and RasG with similar properties. Jeon et al. (2007b) showed that Rap1 is rapidly activated upon cAMP stimulation. Consistent with these results, more active Rap1 was detected in cell lysates stimulated with cAMP than in lysates from unstimulated cells (Figure 5B). To further characterize the PI3K/Rap1 and PI3K/RasG interaction, in vitro GDI assays were performed. The interaction of the GTP-bound G-protein with an effector protein stabilizes the interaction between the G-protein and the nucleotide. This stabilization results in decreased dissociation of the nucleotide from the G-protein/nucleotide/effector complex (Herrmann et al., 1996). Incubating G-protein loaded with mGppNHp, a hydrolysis-resistant fluorescent GTP analogue, with an excess of unlabeled GppNHp results in the exchange of mGppNHp for GppNHp. This exchange is monitored as decay in fluorescence, from which the rate constant kobs is calculated. The addition of increasing concentrations of the effector results in a concentration dependent decrease of kobs. The affinity (Kd) between the G-protein and the effector is determined from this dependency (Herrmann et al., 1996). For the PI3K/Rap1 and PI3K/RasG interaction we determined a Kd of 40 and 24 μM, respectively (Figure 5, C and D). This affinity is similar to that described for the interaction between Rap1 and Phg2 (Kortholt et al., 2006) and the interaction of human Rap with several effectors (Wohlgemuth et al., 2005). Together our data show that both in vivo and vitro Rap1 and RasG bind with approximately the same affinity and specificity to PI3K.
Figure 5.
Interaction between the RBD domain of PI3K and Rap1. (A) Schematic showing the domain composition of PI3K. The bracket indicates the isolated fragment. RBD, Ras-binding domain; PI3Ka, PI3K catalytic association domain; PI3Kc, PI3K catalytic domain. (B) To determine the ability of Rap1 and RasG to bind to the RBD domains of PI3K1, PI3K2, and PI3K3, pulldown experiments were performed. At the indicated time points after cAMP stimulation (s), the amounts of activated Rap1 (top) and RasG (bottom) that bound to the indicated GST-RBD constructs were determined, as described in Materials and Methods. In lane 1 the total amount of G-protein in the lysate is shown (L). The previously described RBD domain of Schizosaccharomyces pombe Byr2 was used as a control for activity of the G-proteins (lanes 2–4). The dissociation rate of mGpp(NH)p from Rap1 (C) and RasG (D) was measured in the presence of varying concentration of PI3K2-RBD. These data were used to calculate the observed rate constants, kobs. The kobs values were plotted against the indicated effector concentration of PI3K-RBD. The addition of increasing concentrations of the effector results in a concentration dependent decrease of kobs. These data were used to calculate the dissociation constant of the Rap1-GTP/PI3K-RBD and RasG-GTP/PI3K-RBD complex, yielding a Kd of 40 and 24 μM, respectively.
The GbpDOE Phenotype Is Lost upon Overexpression of the PIP3-degrading Enzyme PTEN
The data presented in the previous sections suggest that GbpD/Rap regulated PIP3 formation is important for cell morphology, adhesion, and chemotaxis. If PIP3 formation is essential for GbpD mediated signaling, expression of the PIP3 degrading enzyme PTEN should interfere with GbpD-mediated processes. To investigate this hypothesis, PTEN was expressed in wild-type (AX3) and GbpDOE cells. Expression of PTEN in wild-type cells did not influence cell morphology (Figure 1A) or adhesive capacity (Figure 1D). In contrast, the strong phenotype of GbpDOE cells was lost upon expression of PTEN. Overexpression of GbpD in AX3 cells induced a ∼125% increase of the surface contact area and the number of protrusions (Figure 1B). Overexpression of PTEN-GFP in these cells (AX3/GbpDOE/PTEN-GFP) resulted in a pronounced reduction of the GbpDOE-induced increase of the surface contact area and the number of protrusions (Figure 1B). Furthermore, the strong substrate attachment of GbpDOE cells (only 18% of cells detached from the substratum after 1-h shaking) is almost completely lost upon expression of PTEN (64% detached cells; Figure 4D). Finally, although AX3/GbpDOE cells fail to aggregate upon starvation, overexpression PTEN in these cells allows cell aggregation (Figure 1C). These data reveal that the PIP3-degrading enzyme PTEN largely reverses all the phenotypic defects induced by overexpression of GbpD.
DISCUSSION
Overexpression of GbpD in wild-type cells results in a very strong phenotype; cells have a flat morphology and exhibit enhanced adhesion, many protrusions, and impaired chemotaxis (Kortholt et al., 2006). In our previous study we showed that GbpD is a GEF specific for Rap1. In a screen for effectors, Phg2 was identified as a Rap1 effector necessary for adhesion (Kortholt et al., 2006). Overexpression of GbpD in phg2-null cells still resulted in a flattened cells with many protrusions and defective chemotaxis (Figure S1), indicating that Dictyostelium cells have at least two Rap1 effectors: Phg2 mediating cell substrate attachment and another effector mediating cell polarity (Kortholt et al., 2006). In this study we demonstrate that PI3K is the second Rap1 effector in Dictyostelium and is essential for the adhesion, cell morphology, and chemotaxis phenotypes of GbpDOEcells.
GbpD/Rap1-mediated PIP3 Production
Dictyostelium contains eight putative PI3K family members (Janetopoulos et al., 2005). It has been shown that PI3K1 and PI3K2 contribute most to the regulated PI3K activity (Huang et al., 2003). We show here that GbpD/Rap1 activates PI3K, which is essential for GbpD/Rap1-mediated signaling, because 1) GbpDOE cells contain strongly enhanced basal and cAMP-stimulated level of PIP3; 2) the strong GbpDOE phenotype is lost in pi3k1/2-null cells and upon overexpression of the PIP3 degradating enzyme PTEN; 3) expression of Rap1G12V or Rap1S17N does not alter cell-substratum attachment of pi3k1/2-null cells; 4) Rap1 both in vivo and vitro directly interacts with the RBD domain of PI3K; and 5) GFP-RBDRalGDS, the detector of Rap1-GTP, colocalizes with PI3K and PIP3 at the sides of enhanced pseudopod formation.
Rap1 Regulates PI3K Activation in Conjunction with Ras Proteins
Recent studies on the regulation of Dictyostelium PI3K by small G-proteins have suggested that RasG is the main regulator of PI3K signaling (Bolourani et al., 2006). GbpDOE cells, like cells overexpressing constitutive active RasG12V, have enhanced basal and cAMP-stimulated PIP3 formation. GbpDOE cells do not exhibit elevated levels of RasG-GTP or RasC-GTP (Kortholt et al., 2006). In addition, the characteristic GbpDOE phenotype, including the elevated levels of PIP3, is still present in rasC−/GbpDOE, rasG−/GbpDOE, rasC−/rasG−/GbpDOE, and RasGS17N/GbpDOE cells (unpublished data; Kortholt et al., 2006), but is completely lost upon expression of dominant negative Rap1S17N (Figure S1). These results indicate that Rap1 does not activate PI3K indirectly, by activating RasC or RasG. Binding studies with different PI3K RBD domains show that Rap1 and RasG bind with approximately the same affinity and specificity to PI3K. Together these data suggest that RasG-GTP and Rap1-GTP may independently activate PI3K. These two proteins may have a different contribution in PI3K activation for basal movement, cAMP-stimulation chemotaxis, and feedback mechanisms, which we will discuss in more detail below.
Rap1 Regulates Basal Pseudopod Formation in Conjugation with Ras Proteins
A recent study by Sasaki et al. (2007) showed that a Gβ-independent PI3K/Ras pathway is important for the regulation of random cell movement and the extension of F-actin projections in the absence of extracellular stimuli. Gβ− cells exhibit normal random movement and show spontaneous Ras activation and PIP3 accumulation at sites of actin protrusion. In contrast, wild-type cells treated with LY294002 or pi3k1/2/3-null cells lack spontaneous Ras activation and show defects in random movement. Active RasG is localized at sites of actin protrusion, and rasG-null cells have decreased random cell movement, suggesting that RasG is one of the Ras isoforms that plays a role in the Ras/PI3K regulatory circuit (Tuxworth et al., 1997; Sasaki and Firtel, 2006; Sasaki et al., 2007). However, because rasG-null cells only show modest defects in random movement, most likely other Ras/Rap isoforms are also involved. GbpDOE cells moving randomly in buffer make multiple and broad PIP3 patches along the entire plasma membrane, from which multiple pseudopodia are extended, a phenotype similar to that described for cells expressing membrane-bound PI3K2 (Funamoto et al., 2002; Sasaki et al., 2007). Furthermore, randomly moving gbpD-null cells are, like pi3k1/2/3-null cells, much rounder and extend fewer pseudopodia than wild-type cells. Therefore, our data suggest that Rap1 is the second Ras isoform involved in regulation of random cell movement. The GbpD/Rap1/PI3K pathway, which regulates local activation of PIP3 and the basal formation of pseudopodia, apparently escalates in GbpDOE cells leading to many intense PIP3 patches and multiple pseudopodia. Because, nonstimulated wild-type cells do not extend multiple pseudopodia and show an even distribution of PHcrac, there has to be down-regulation or inactivation of the GbpD/Rap pathway in wild-type cells after its initial activation. GbpD/Rap signaling could be blocked at several steps, including inhibition of GbpD activity, inactivation of Rap-GTP levels by a yet unidentified Rap-specific GAP protein, or reduced PI3K activity by, for instance, depletion of PI(4,5)P2 levels. One of the proteins involved in the down-regulation of the Rap/PI3K loop could be RapGAP1 (Jeon et al., 2007a). Deletion of RapGAP1 results in a phenotype similar to that described for cells overexpressing GbpD or RapG12V, and the localization of RapGAP1 is regulated by F-actin.
Role of cAMP-stimulated Rap1 Activation in Chemotaxis
In response to chemoattractant stimulation, PIP3 accumulates at the leading edge of chemotaxing cells (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). The localized production of PIP3 is mediated by PI3K at the leading edge and the PIP3-degradating enzyme PTEN in the back of the cell (Parent et al., 1998; Funamoto et al., 2002; Huang et al., 2003). The RBD domain of PI3K is essential for its activation by extracellular cAMP (Funamoto et al., 2002). We have shown that cAMP mediates Rap1 activation at the leading edge, that Rap1-GTP binds to the RBD domain of PI3K, and that the Rap1-GTP detector GFP-RBDRalGDS colocalizes with the formed PIP3, suggesting that Rap1 may also mediate chemoattractant stimulation of PI3K. It was previously shown that in a cell line lacking RasC and RasG, cAMP-mediated PI3K activation is almost completely absent (Bolourani et al., 2006). Furthermore, since the defect in PI3K signaling is much more severe in rasG-null cells than in rasC-null cells, and the translocation of the PIP3 detector PHcracGFP to the leading edge is almost absent in cells expressing dominant negative RasGS17N (Figure S1), RasG seems to be the main regulator of PI3K signaling. Also Rap1 activity seems to be predominantly regulated by RasG because Rap1 activation is totally abolished in rasC−/rasG− and rasG−cells, but only slightly reduced in rasC− cells (Bolourani et al., 2008). On cAMP stimulation, Ras is activated with levels peaking at 3–6 s, and PI3K translocates to the membrane with similar kinetics, whereas PHcrac is translocated with slightly slower kinetics (Sasaki and Firtel, 2006). This is consistent with the model that Ras-driven PI3K production initiates PIP3 formation. On the contrary, the kinetics of Rap1 activation are significantly more slowly, with levels peaking at 6–9 s (Jeon et al., 2007b). Because of these slower kinetics, it is unlikely that Rap1 activation is necessary for the initial PI3K activation. Consistently, cells expressing dominant negative Rap1S17N are capable of producing PIP3 at the leading edge and can chemotax persistently toward cAMP (Figure 2). However Rap1 signaling could be involved in amplifying and stabilizing the initial PIP3 gradient. Our data and previous studies by Jeon et al., (2007a,b) suggest that Rap1 regulates leading-edge formation through two mechanisms: it regulates PIP3 formation by activation of PI3K, and it activates Phg2, which controls leading edge myosin II disassembly thereby facilitating actin-mediated leading edge protrusions.
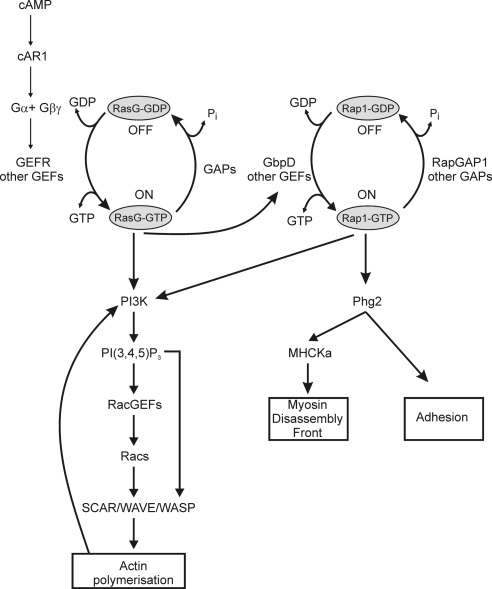
The data of this report allow for a refinement of our previous published model of the Rap1 pathway in Dictyostelium mediating adhesion and cell polarity (Kortholt et al., 2006; Figure 6). GbpD is one of the GEFs that catalyze the exchange of GDP for GTP thereby activating the Rap1 protein. The mechanism of regulation of GbpD remains unclear. Although containing cyclic nucleotide binding domains, radioactive cyclic nucleotide binding assays have revealed no detectable binding of cAMP or cGMP to GbpD (Bosgraaf et al., 2002). In support of these data, it has been reported that activity of GbpD is not dependent on intracellular cAMP or cGMP (Kortholt et al., 2006). Because lipid dot-blot assays showed that the regulatory domain of GbpD can bind various lipids (unpublished observations), it is possible that lipids are involved in the regulation of GbpD activity. The targets of Rap1 are Phg2 and PI3K. Phg2, a serine/threonine kinase, is essential for Rap1 regulated adhesion (Kortholt et al., 2006) and controls leading edge myosin II disassembly (Jeon et al., 2007a,b). Rap1 directly interacts with PI3K via its RBD and regulates local activation of PIP3 and the basal formation of pseudopodia. cAMP-mediated Rap1 activation is predominantly regulated by RasG through the activation of a so far unidentified GEF (Bolourani et al., 2008). RasG is the main regulator of cAMP-stimulated PI3K activation, whereas Rap1 amplifies and stabilize the initial PIP3 gradient.
Figure 6.
Model of the Rap1 pathway in Dictyostelium regulating adhesion and cell morphology. Dictyostelium Rap1 switches between an active GTP–bound and inactive GDP–bound state. GbpD is one of the GEFs that catalyze the exchange of GDP for GTP, thereby activating the Rap1 protein. Phg2, a serine/threonine kinase, is necessary for Rap1-regulated adhesion. PI3K directly interacts with active Rap1, thereby regulating PIP3 formation and subsequently adhesion and basal cell morphology. Binding of cAMP to the cAR1 receptor results in the rapid activation of GEFs and subsequently Ras at the leading edge. RasG activation is essential for regulating PIP3 signaling and subsequently essential for amplifying the initial signal, establishing cell polarity and actin polymerization at the leading edge. Rap1 activation in response to cAMP occurs downstream of RasG. Rap1 may regulate leading edge formation through two mechanisms: it amplifies and stabilizes the initial PIP3 gradient, and it activates Phg2, which directly or indirectly phosphorylates MHCKa that controls leading edge myosin II disassembly thereby facilitating actin-mediated leading edge protrusions.
Mammalian Rap, like Dictyostelium Rap1, is involved in processes such as phagocytosis, adhesion, and chemotaxis (Bos et al., 2001). The interaction between mammalian Rap and PI3K is not completely clear and seems to be cell type specific. In some cells, such as human embryonic kidney cells (HEK 293) and thyroid cells (Tsygankova et al., 2001; Mei et al., 2002), Rap activation increases PI3K activity, whereas in PCCL3 and B-cells (Lou et al., 2002; Christian et al., 2003) activation of Rap leads to inhibition of PI3K. In terms of cell polarity and cell migration, mammalian Rap plays an essential role in the determination of neuronal polarity (Schwamborn and Puschel, 2004). In addition, mammalian PI3K is essential for axon specification and probably acts both upstream and downstream of Rap (Shi et al., 2003; Schwamborn and Puschel, 2004), a situation similar to that for Rap1 regulated pseudopod formation in Dictyostelium.
In summary, PI3K is the Rap1 effector in Dictyostelium necessary for Rap1-regulated adhesion and cell polarity. We propose a role for PIP3 and GbpD in the basal formation of pseudopodia, whereas RasC and RasG may be more important during cAMP stimulation. The Rap pathway that regulates pseudopod formation in Dictyostelium is reminiscent of the pathway that determines neuronal polarity, suggesting that the Rap/PI3K pathway is a conserved module regulating the establishment of cell polarity.
Supplementary Material
ACKNOWLEDGMENTS
The pi3k1/2-null strain used in this study was obtained from the Dictyostelium stock center. A.K. is a research fellow of the Alexander von Humboldt foundation. H.R. was supported by the Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) and is recipient of the Otto-Hahn-Medaille of the Max-Planck-Gesellschaft. The studies were partially supported by a grant from the Canadian Institutes of Health Research to G.W.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0177) on January 20, 2010.
REFERENCES
- Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- Bivona T. G., Wiener H. H., Ahearn I. M., Silletti J., Chiu V. K., Philips M. R. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G. B., Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell. 2006;17:4543–4550. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G. B., Weeks G. Rap1 activation in response to cAMP occurs downstream of ras activation during Dictyostelium aggregation. J. Biol. Chem. 2008;283:10232–10240. doi: 10.1074/jbc.M707459200. [DOI] [PubMed] [Google Scholar]
- Bos J. L. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Bos J. L., de Rooij J., Reedquist K. A. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., Van Haastert P.J.M. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L., Waijer A., Engel R., Visser A. J., Wessels G., Soll D., Van Haastert P. J. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J. Cell Sci. 2005;118:1899–1910. doi: 10.1242/jcs.02317. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The Gtpase superfamily—conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Campbell J. J., Butcher E. C. Chemokines in tissue-specific and microenvironment-specific lymphocyte homing. Curr. Opin. Immunol. 2000;12:336–341. doi: 10.1016/s0952-7915(00)00096-0. [DOI] [PubMed] [Google Scholar]
- Chen L., Iijima M., Tang M., Landree M. A., Huang Y. E., Xiong Y., Iglesias P. A., Devreotes P. N. PLA(2) and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian S. L., Lee R. L., Mcleod S. J., Burgess A. E., Li A.H.Y., Dang-Lawson M., Lin K.B.L., Gold M. R. Activation of the Rap GTPases in B lymphocytes modulates B cell antigen receptor-induced activation of Akt but has no effect on MAPK activation. J. Biol. Chem. 2003;278:41756–41767. doi: 10.1074/jbc.M303180200. [DOI] [PubMed] [Google Scholar]
- Chubb J. R., Insall R. H. Dictyostelium: an ideal organism for genetic dissection of Ras signalling networks. Biochim. Biophys. Acta. 2001;1525:262–271. doi: 10.1016/s0304-4165(01)00111-8. [DOI] [PubMed] [Google Scholar]
- Crone S. A., Lee K. F. The bound leading the bound: target-derived receptors act as guidance cues. Neuron. 2002;36:333–335. doi: 10.1016/s0896-6273(02)01009-7. [DOI] [PubMed] [Google Scholar]
- Devreotes P. N., Zigmond S. H. Chemotaxis in eukaryotic cells—a focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J. A., Glockner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P., Stephens S., Titus M. A., Chisholm R. L. SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Funamoto S., Milan K., Meili R., Firtel R. A. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 2001;153:795–809. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. M., Bosgraaf L., Van Haastert P.J.M., Smith J. L. Identification of four candidate cGMP targets in Dictyostelium. Proc. Natl. Acad. Sci. USA. 2002;99:6749–6754. doi: 10.1073/pnas.102167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C. Ras-effector interactions: after one decade. Curr. Opin. Struct. Biol. 2003;13:122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Herrmann C., Horn G., Spaargaren M., Wittinghofer A. Differential interaction of the Ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- Hoeller O., Kay R. R. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Huang Y. E., Iijima M., Parent C. A., Funamoto S., Firtel R. A., Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P-3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M., Huang Y. E., Devreotes P. Temporal and spatial regulation of chemotaxis. Dev. Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Jeon T. J., Lee D. J., Lee S., Weeks G., Firtel R. A. Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J. Cell Biol. 2007a;179:833–843. doi: 10.1083/jcb.200705068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon T. J., Lee D. J., Merlot S., Weeks G., Firtel R. A. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 2007b;176:1021–1033. doi: 10.1083/jcb.200607072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., Goody R. S. Kinetics of interaction of nucleotides with nucleotide-free H-Ras P21. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- Kae H., Lim C. J., Spiegelman G. B., Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P., Devreotes P. N. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 2008;18:1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R. J., Kae H., Ip H., Spiegelman G. B., Weeks G. Evidence for a role for the Dictyostelium Rap1 in cell viability and the response to osmotic stress. J. Cell Sci. 2002;115:3675–3682. doi: 10.1242/jcs.00039. [DOI] [PubMed] [Google Scholar]
- Khosla M., Spiegelman G. B., Weeks G. Overexpression of an activated rasG gene during growth blocks the initiation of Dictyostelium development. Mol. Cell. Biol. 1996;16:4156–4162. doi: 10.1128/mcb.16.8.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A., King J. S., Keizer-Gunnink I., Harwood A. J., Van Haastert P. J. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol. Biol. Cell. 2007;18:4772–4779. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A., Rehmann H., Kae H., Bosgraaf L., Keizer-Gunnink I., Weeks G., Wittinghofer A., Van Haastert P. J. Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J. Biol. Chem. 2006;281:23367–23376. doi: 10.1074/jbc.M600804200. [DOI] [PubMed] [Google Scholar]
- Kortholt A., Van Haastert P. J. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 2008;20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Lim C. J., Spiegelman G. B., Weeks G. RasC is required for optimal activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 2001;20:4490–4499. doi: 10.1093/emboj/20.16.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. J., Zawadzki K. A., Khosla M., Secko D. M., Spiegelman G. B., Weeks G. Loss of the Dictyostelium RasC protein alters vegetative cell size, motility and endocytosis. Exp. Cell Res. 2005;306:47–55. doi: 10.1016/j.yexcr.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Loovers H. M., Postma M., Keizer-Gunnink I., Huang Y. E., Devreotes P. N., Van Haastert P. J. Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: a quantitative in vivo analysis by inhibition of PI3-kinase. Mol. Biol. Cell. 2006;17:1503–1513. doi: 10.1091/mbc.E05-09-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L. G., Urbani J., Ribeiro-Neto F., Altschuler D. L. cAMP inhibition of Akt is mediated by activated and phosphorylated Rap1b. J. Biol. Chem. 2002;277:32799–32806. doi: 10.1074/jbc.M201491200. [DOI] [PubMed] [Google Scholar]
- Mei F. C., Qiao J. B., Tsygankova O. M., Meinkoth J. L., Quilliam L. A., Cheng X. D. Differential signaling of cyclic AMP—opposing effects of exchange protein directly activated by cyclic AMP and cAMP-dependent protein kinase on protein kinase B activation. J. Biol. Chem. 2002;277:11497–11504. doi: 10.1074/jbc.M110856200. [DOI] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T. B., Ma H., Firtel R. A. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C. A., Blacklock B. J., Froehlich W. M., Murphy D. B., Devreotes P. N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Parkinson K., Bolourani P., Traynor D., Aldren N. L., Kay R. R., Weeks G., Thompson C. R. Regulation of Rap1 activity is required for differential adhesion, cell-type patterning and morphogenesis in Dictyostelium. J. Cell Sci. 2009;122:335–344. doi: 10.1242/jcs.036822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebstein P. J., Weeks G., Spiegelman G. B. Altered morphology of vegetative amoebae induced by increased expression of the Dictyostelium discoideum ras-related gene rap1. Dev. Genet. 1993;14:347–355. doi: 10.1002/dvg.1020140504. [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Firtel R. A. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N., Firtel R. A. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J. C., Puschel A. W. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat. Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Seastone D. J., Zhang L. Y., Buczynski G., Rebstein P., Weeks G., Spiegelman G., Cardelli J. The small M-r Ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell. 1999;10:393–406. doi: 10.1091/mbc.10.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. H., Jan L. Y., Jan Y. N. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Soll D. R. Computer-assisted three-dimensional reconstruction and motion analysis of living, crawling cells. Comput. Med. Imaging Graphics. 1999;23:3–14. doi: 10.1016/s0895-6111(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Takeda K., Sasaki A. T., Ha H. J., Seung H. A., Firtel R. A. Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 2007;282:11874–11884. doi: 10.1074/jbc.M610984200. [DOI] [PubMed] [Google Scholar]
- Tsygankova O. M., Saavedra A., Rebhun J. F., Quilliam L. A., Meinkoth J. L. Coordinated regulation of Rap1 and thyroid differentiation by cyclic AMP and protein kinase A. Mol. Cell. Biol. 2001;21:1921–1929. doi: 10.1128/MCB.21.6.1921-1929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth R. I., Cheetham J. L., Machesky L. M., Spiegelmann G. B., Weeks G., Insall R. H. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J., Keizer-Gunnink I., Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol. 2007;177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P.J.M., Devreotes P. N. Chemotaxis: Signalling the way forward. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Akar G., Bosgraaf L., Van Haastert P. J. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009a;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Keizer-Gunnik I., Van Haastert P. J. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D. M., Keizer-Gunnink I., Haastert P. J. An extrachromosomal, inducible expression system for Dictyostelium discoideum. Plasmid. 2009b;61:119–125. doi: 10.1016/j.plasmid.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Weeks G. The small GTPase superfamily. In: Loomis Wiliam F., Kuspa Adam., editors. Dictyostelium Genomics. Norwich, United Kingdom: Horizon Bioscience; 2005. pp. 173–210. [Google Scholar]
- Wilkins A., Insall R. H. Small GTPases in Dictyostelium: lessons from a social amoeba. Trends Genet. 2001;17:41–48. doi: 10.1016/s0168-9525(00)02181-8. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth S., Kiel C., Kramer A., Serrano L., Wittinghofer F., Herrmann C. Recognizing and defining true Ras binding domains 1: biochemical analysis. J. Mol. Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Zhou K. M., Pandol S., Bokoch G., Traynor-Kaplan A. E. Disruption of Dictyostelium P13K genes reduces [P-32]phosphatidylinositol 3,4 bisphosphate and [P-32]phosphatidylinositol trisphosphate levels, alters F-actin distribution and impairs pinocytosis. J. Cell Sci. 1998;111:283–294. doi: 10.1242/jcs.111.2.283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.