A combination of in vivo and in vitro approaches were used to show how tropomyosin and myosin-II contribute to contractile ring assembly in fission yeast. Ring assembly is sensitive to changes in the cellular levels of myosin-II, and tropomyosin works to maximize myosin-II motor function during this process by stabilizing actomyosin interactions.
Abstract
Myosin-II (Myo2p) and tropomyosin are essential for contractile ring formation and cytokinesis in fission yeast. Here we used a combination of in vivo and in vitro approaches to understand how these proteins function at contractile rings. We find that ring assembly is delayed in Myo2p motor and tropomyosin mutants, but occurs prematurely in cells engineered to express two copies of myo2. Thus, the timing of ring assembly responds to changes in Myo2p cellular levels and motor activity, and the emergence of tropomyosin-bound actin filaments. Doubling Myo2p levels suppresses defects in ring assembly associated with a tropomyosin mutant, suggesting a role for tropomyosin in maximizing Myo2p function. Correspondingly, tropomyosin increases Myo2p actin affinity and ATPase activity and promotes Myo2p-driven actin filament gliding in motility assays. Tropomyosin achieves this by favoring the strong actin-bound state of Myo2p. This mode of regulation reflects a role for tropomyosin in specifying and stabilizing actomyosin interactions, which facilitates contractile ring assembly in the fission yeast system.
INTRODUCTION
Myosin-II is essential for actomyosin contractile ring function and cytokinesis in animal cells (De Lozanne and Spudich, 1987; Mabuchi and Okuno, 1977). Two myosin-II isoforms are found in fission yeast, Myo2p and Myp2p, both of which function in the contractile ring. Myo2p is essential for ring formation, cytokinesis, and growth (Kitayama et al., 1997; May et al., 1997; Balasubramanian et al., 1998), whereas Myp2p only becomes important for growth under stress conditions (Bezanilla et al., 1997; Motegi et al., 1997).
Fission yeast actomyosin ring assembly occurs in two stages (Naqvi et al., 1999; Motegi et al., 2000; Wu et al., 2003; Wu et al., 2006): 1) Mid1p/anillin (Chang et al., 1996; Sohrmann et al., 1996) directs the recruitment of Myo2p to the incipient division site as a band of cortical dots (“nodes”). 2) By the start of anaphase B, Myo2p nodes and actin filaments coalesce into a ring. Unlike the initial recruitment of Myo2p to the division site, ring formation relies on Myo2p motor function and actin filaments (Naqvi et al., 1999). Myp2p joins the ring after assembly is complete (Bezanilla et al., 2000). Ring constriction is then delayed until completion of chromosome segregation and mitotic exit (Wolfe and Gould, 2005).
Although a number of proteins are essential for contractile ring formation in fission yeast, it is largely unknown how these proteins operate during ring assembly. Careful analysis of node dynamics during the assembly phase led to the development of a “search, capture, pull, and release” model. Myo2p at one node was proposed to associate with actin filaments growing from another node, forming a network that gradually compacts into a tight ring driven by myosin-II motor activity (Vavylonis et al., 2008). Recent studies with mid1Δ cells have shown that rings can assemble in the absence of nodes, suggesting that the underlying molecular mechanisms driving assembly can overcome the loss of spatial organization when nodes are absent (Hachet and Simanis, 2008; Huang et al., 2008). Such a mechanism may be accounted for by the “leading cable” model of ring assembly (Arai and Mabuchi, 2002; Wong et al., 2002; Kamasaki et al., 2007; Mishra and Oliferenko, 2008; Roberts-Galbraith and Gould, 2008). In this model actin cables emanating from a single nucleation site are proposed to encircle the cell equator. The two models provide a framework for understanding ring assembly, and the next challenge lies in determining how the key cytokinesis proteins are regulated, and how they work with one another to facilitate ring assembly.
Myo2p motor activity is established at the division site by UCS (Unc-45-/Cro1p-/She4p–related) protein Rng3p. Rng3p promotes the stability and motility activity of Myo2p (Lord and Pollard, 2004; Lord et al., 2008) and is essential for converting Myo2p nodes into functional rings (Wong et al., 2000). However, Myo2p motor function and ring assembly may also rely on actin filament–based regulation. Far from bare, contractile ring actin filaments are associated with tropomyosin (Balasubramanian et al., 1992) and actin-bundling proteins (Eng et al., 1998; Wu et al., 2001; Takaine et al., 2009). Tropomyosins are dimeric coiled-coil proteins that bind along the length of actin filaments and regulate the function of a number of actin-binding proteins (Cooper, 2002). In addition to regulating association with myosins (Ostap, 2008), tropomyosins regulate filament depolymerization and severing by cofilin and gelsolin (Fattoum et al., 1983; Bernstein and Bamburg, 1992; Ono and Ono, 2002; Nakano and Mabuchi, 2006; Fan et al., 2008), inhibit Arp2/3-mediated growth of branched actin filaments (Blanchoin et al., 2001), and promote rates of formin-mediated actin polymerization (Wawro et al., 2007; Skau et al., 2009).
In complex eukaryotes multiple isoforms of tropomyosin are expressed in the same cell, localizing to distinct compartments (Martin and Gunning, 2008). The ability of tropomyosins to differentially regulate key actin-binding proteins implies a role in the functional specification of different actin structures in the cell. Cdc8p, the sole tropomyosin in fission yeast, is essential for contractile ring assembly (Balasubramanian et al., 1992). Cdc8p localizes to actin cables spanning the cell length, contractile rings, and (to a lesser degree) actin patches (Balasubramanian et al., 1992; Arai et al., 1998; Skoumpla et al., 2007). Its distribution may influence the function of the different myosins operating at these structures.
In this study we used a combination of in vivo and in vitro techniques to understand how myosin-II functions in contractile ring assembly. We utilized a haploid strain engineered to carry two copies of myo2 to assess how changes in Myo2p levels influences ring dynamics and the function of other cytokinesis proteins. We find that tropomyosin maximizes Myo2p motor function by influencing myosin-II kinetics to stabilize the actomyosin interactions critical for contractile ring assembly. Our work provides mechanistic insight into the process of actomyosin ring assembly in fission yeast, and is consistent with an emerging role for tropomyosin isoforms in specifying myosin function at given actin structures in cells.
MATERIALS AND METHODS
Microscopy
A Nikon TE2000-E2 inverted microscope with motorized fluorescence filter turret and a Plan Apo 60× (1.45 NA) objective was used to capture DIC and epifluorescence cell images (Melville, NY). Fluorescence utilized an EXFO X-CITE 120 illuminator (Nikon). NIS Elements software was used to control the microscope (Nikon), two Uniblitz shutters (Vincent Associates, Rochester, NY), a Photometrics CoolSNAP HQ2 14-bit camera (Tucson, AZ), and autofocusing. Time-lapse movies typically monitored contractile ring assembly and dynamics by capturing yellow fluorescent protein (YFP)/green fluorescent protein (GFP) fluorescence every 2 min for 2 h at room temperature. Autofocusing was performed on the differential interference contrast (DIC) channel before captures. Cell suspensions (3 μl) were mounted on flat 30-μl media pads (solidified by 1% agarose) prepared on the slide surface. A combination of petroleum jelly, lanolin, and paraffin (1:1:1) was used to seal slides and coverslips. To simultaneously track spindle pole bodies (Sad1p-GFP) and rings (YFP-Myo2p/Rlc1p-GFP), a z-stack of six images (taken every 0.75 μm, spanning the depth of the cell) was collected every 2–3 min for 90 min using the YFP or GFP filter. Cells were grown in EMM-Ura− medium and mounted on EMM-Ura− agarose pads when comparing ring dynamics in 1x and 2xYFP-myo2 strains expressing excess Rlc1p (from a multicopy plasmid). Images were captured using Nikon ND software and analysis of contractile ring assembly time, dwell time, and constriction rate was performed using ImageJ (http://rsb.info.nih.gov/ij/), Microsoft Excel (Redmond, WA), and KaleidaGraph software (Synergy Software, Reading, PA).
Fluorescence microscopy was used to estimate YFP-Myo2p levels in contractile rings (Wu and Pollard, 2005). Fluorescence intensities of contractile rings in YFP-myo2 cells were compared side-by-side with those in 2xYFP-myo2 cells in images derived from mixed samples. To differentiate between the two strains, one harbored an integrated Sad1p-GFP fusion marking spindle pole bodies. Images were maximum projections derived from z-stacks (0.5 μm) spanning the depth of the cells (4 μm). Final ring intensity measurements were corrected for cytoplasmic or out-of-focus signal by blanking with an identical area measurement taken from the cytoplasm, a distance away from the ring. Analysis was performed using ImageJ software.
Fluorescence recovery after photobleaching (FRAP) experiments used confocal laser scanning microscopy with a Zeiss LSM 510 META system equipped with an argon laser, META detector, and a Plan Apo 100× (1.4 NA) objective (Thornwood, NY). Cells were mounted on 1% agarose pads (as described above) before microscopy at room temperature. A region of interest (ROI) was selected on YFP-Myo2p contractile rings for directed bleaching. Photobleaching iterations were performed briefly at high laser power, resulting in ∼90–100% signal loss. Signal recovery was monitored by time-lapse analysis at low laser power with images collected every 3 s after bleach until the recovery signal reached a plateau (∼1 min). The LSM 510 software (version 4.2) was used to collect images and perform data analysis (see the Figure 3B legend). Recovery curves of YFP-Myo2p signal versus time were plotted and fit using KaleidaGraph software (Synergy).
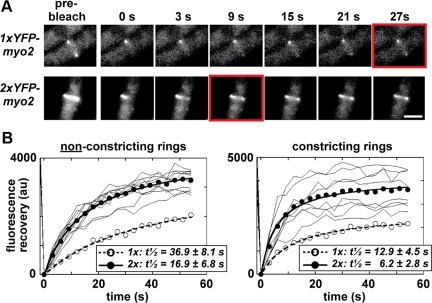
Figure 3.
Doubling Myo2p cellular levels increases its exchange rate at contractile rings. (A) FRAP was used to measure YFP-Myo2p exchange rates in contractile rings using confocal laser scanning fluorescence microscopy. Micrographs compare recovery of YFP-Myo2p fluorescence in nonconstricting rings from 1x (top panels) and 2xYFP-myo2 (bottom panels) cells. Panels on the far left show prebleached rings that were subsequently bleached at a region of interest (ROI). Subsequent panels chart recovery of signal at the ring (0–27 s after bleach). Red boxes indicate the point when recovery is half-maximal (t1/2). Cells were grown in YE5S media at 25°C before imaging at ambient temperature. White bar, 4 μm. (B) Plots charting FRAP at ROIs on nonconstricting and constricting rings from 2xYFP-myo2 cells. Fluorescence intensities measured before bleach (−1.5 s) and after bleach (every 3 s, 0–60 s) are plotted. Individual ROI traces (thin lines) are shown (n = 8–10) along with an average fit (•, thick line). Datasets for each trace were corrected for additional bleaching encountered during time-lapse imaging by a control ROI (derived from an unbleached ring in the same field of cells). To facilitate curve fitting, zero signal intensity was set for each trace by subtracting residual YFP-Myo2p signal (detected at the first time point after bleach, 0 s) from all trace values. Mean YFP-Myo2p recovery curves from FRAP experiments performed in parallel with 1xYFP-myo2 cells (Sladewski et al., 2009) are included on each plot (○, dashed line). t1/2 values represent the mean generated from the fits of each individual FRAP experiment.
In Vitro Motility Assays
We used motility assays based on an established protocol (Kron and Spudich, 1986) with 0.4–100 μg/ml Myo2p delivered into motility chambers. Fluorescent actin stocks (5 μM) were polymerized by addition of 50 mM KCl and 1 mM MgCl2 (with or without 2.5 μM Cdc8p), incubated for 30 min, and then labeled with 5 μM rhodamine phalloidin for 30 min. After Myo2p was adhered to the surface of a nitrocellulose-coated coverslip for 10 min, the chamber was washed as follows: a) three times with motility buffer (25 mM imidazole, pH 7.4, 50 mM KCl, 1 mM EGTA, 4 mM MgCl2, 2 mM DTT) plus 0.5 mg/ml BSA; b) three times with motility buffer alone; c) twice with motility buffer containing vortexed (30 s) unlabeled actin filaments (1 μM); d) three times with motility buffer plus 1 mM ATP; e) twice with motility buffer plus 25 nM rhodamine phalloidin-labeled actin filaments (±Cdc8p), and oxygen scavengers (50 μg/ml catalase, 130 μg/ml glucose oxidase, and 3 mg/ml glucose); f) twice with motility buffer plus 20 mM DTT, 0.5% methyl-cellulose, and scavengers; and g) twice with motility buffer plus 20 mM DTT, 0.5% methyl-cellulose, 1.5 mM ATP, and scavengers. Washes (e–g) included 2 μM Cdc8p for all assays using Cdc8p-bound filaments. Experiments testing the effects of ADP on Myo2p motility used 0.4 mM ATP and indicated concentrations of ADP in the final wash buffer. Filaments were observed at room temperature by epifluorescence microscopy and recorded at 2-s intervals. ImageJ software was used to calculate filament velocities from time-lapse series (n = ∼50 filaments).
ATPase Assays
Actin-activated Myo2p ATPase assays were carried out at room temperature in 2 mM Tris-HCl, pH 7.2, 10 mM imidazole, 60–165 mM KCl, 0.1 mM CaCl2, 3 mM MgCl2, 2 mM ATP, and 1 mM DTT with 20 nM Myo2p and 0–100 μM actin/actin-Cdc8p filaments. Malachite green was used to quantitate Pi release (Henkel et al., 1988). Controls omitting Myo2p were used for each sample, and basal activity (detected in controls lacking actin) was subtracted to derive actin-activated ATPase rates. Curves were fit to Michaelis-Menten kinetics using KaleidaGraph software.
Actin Filament Cosedimentation Assays
Myo2p was incubated with 10 μM actin (or Cdc8p-saturated actin) filaments in 60 mM KCl, 10 mM imidazole, pH 7.0, 1 mM DTT, 8 mM MgCl2, and 8 mM ADP (with 0.5–2 mM ATP). Samples were ultracentrifuged at 4°C in a TL100 rotor (Beckman Instruments, Fullerton, CA) at 120,000 × g for 60 min. Supernatants were recovered and the actin filament pellets were resuspended in an equal volume of G buffer (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT). Pellet and supernatant samples were boiled in SDS-PAGE sample buffer, run on 10% SDS-PAGE gels, and stained with Coomassie blue.
Duty Ratio Measurements
Duty ratios (percentage of time myosin spends in the strong actin-bound state per ATP hydrolysis cycle) for Myo2p with actin versus actin-Cdc8p filaments were determined using two different approaches. First, the equation: v = d/ton (Huxley, 1990) was used, where v = motility rate (nm/ms; see Table 4), d = Myo2p step size (nm), and ton = time Myo2p spends in the strong actin-bound state per ATP hydrolysis cycle (ms). Given that the Myo2p amino acid sequence indicates a similar sized light chain-binding domain (with 2 IQ motifs) to that of skeletal muscle myosin, we assume Myo2p step-size equals the ∼10 nm of muscle myosin-II (Finer et al., 1994). The VMAX values for Myo2p derived from curves in Figure 6A (see Table 4) indicate the time taken for a single ATP hydrolysis cycle (210/287 ms +/− Cdc8p). Duty ratio was estimated by dividing ton values derived from maximal motility rates by the total cycle time: 14.5/287 = 0.051 (5.1%) without Cdc8p and 28.6/204 = 0.140 (14.0%) with Cdc8p.
Table 4.
Effect of tropomyosin on the biochemical properties of Myo2p
| Property | No Cdc8p | +Cdc8p |
|---|---|---|
| In vitro motility (μm · s−1)a | ||
| Myo2p | 0.69 ± 0.04 | 0.35 ± 0.02 |
| Control: myosin-Va HMM | 0.22 ± 0.03 | 0.23 ± 0.02 |
| Actin-activated Mg2+-ATPaseb | ||
| Km (μM) | 20.4 ± 2.9 | 3.0 ± 0.8 |
| VMAX (s−1)c | 3.5 ± 0.2 | 4.9 ± 0.2 |
| ton (ms)d | 14.5 | 28.6 |
| Duty ratio (%)e | ||
| i) | 5.1 | 14.0 |
| ii) | 3.2 ± 0.3 | 5.8 ± 0.4 |
a n = 50 filaments. Rates (±SD) derived using 32 μg/ml myosin (plus excess calmodulin for myosin-Va HMM).
b Values are derived from curves in Figure 6A (±SE).
c ATPase activity: molecules of ATP hydrolyzed per motor per second.
d Values calculated using the equation: v = d/ton, where v = motility rate, and d = step size (assumed to be 10 nm).
e Values represent i) the percentage of time ton is occupied within an ATPase cycle time (derived from VMAX values) and ii) experimental values derived from curves in Figure 8D (±SE).
Figure 6.
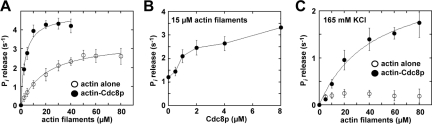
Tropomyosin promotes Myo2p's actin-activated ATPase activity. Actin-activated Mg2+-ATPase activity of purified Myo2p was measured as a function of actin (or actin-Cdc8p) concentration. Basal Myo2p ATPase activities (detected from control reactions lacking actin/actin-Cdc8p) and background Pi from actin or actin-Cdc8p (determined from controls lacking myosin) were subtracted from all measurements. Myo2p was included in reactions at a final concentration of 20–30 nM. Plots were generated from average values obtained from three to four different datasets using at least two independent preparations of Myo2p, actin, and Cdc8p. Curves were fit to Michaelis Menten kinetics using KaleidaGraph software. KCl concentration in reactions was 60 mM unless otherwise stated. (A) Myo2p activity in the presence of actin filaments versus Cdc8p-saturated filaments. Actin filaments were preincubated with Cdc8p at a concentration of actin to Cdc8p molecules of 2:1. (B) Myo2p activity as a function of Cdc8p concentration. Actin was preincubated with a range of Cdc8p concentrations and included in reactions at a final concentration of 15 μM. (C) As in A, only reactions included a higher concentration of KCl (165 mM). Actin filaments were preincubated with Cdc8p at a concentration of actin to Cdc8p at 6:1.
Second, we used an experimental approach to estimate duty ratio (Uyeda et al., 1990; Harris and Warshaw, 1993). Rates of Myo2p-driven actin versus actin-Cdc8p filament motility were measured at five myosin concentrations (0.8, 1.6, 3.2, 6.4, and 16 μg/ml Myo2p). At these concentrations motility rates steadily decline from the maximum rate (see Figure 7A). Ultimately, motility rates are plotted versus the number of myosin heads available to a given actin filament. Numbers of heads per square micrometer nitrocellulose were calculated based on concentrations of Myo2p used and the chamber dimensions. We assume that 50% of myosin entered into chambers attaches to nitrocellulose (given efficiency of Myo2p binding was ∼50–60% at low Myo2p concentrations by Western blot analysis) and that all heads within a 26-nm-wide band can interact with a passing actin filament (Uyeda et al., 1990). Numbers of heads available to a given actin filament were based on the surface density of Myo2p and filament length. Thus, a 1-μm-long filament associated with a surface containing 100 heads/μm2 would have access to (26/1000 × 100) 2.6 heads. Data from plots of filament velocity versus number of Myo2p heads available were fit to the equation: V = (a × VMAX) × (1 − [1 − f]N), where V is actin filament velocity, VMAX is maximum filament velocity, f is duty ratio, N is total number of motors capable of interacting with the actin filament, and (a × VMAX) relates to the ability to move filaments at low motor density. Fitting by nonlinear regression allowed (a × VMAX) and f to be determined as parameters of the fit.
Figure 7.
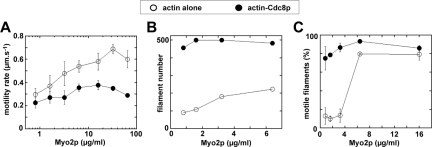
Tropomyosin reduces the speed but increases the efficiency of Myo2p-driven in vitro motility. Epifluorescence microscopy was used to generate time-lapse movies tracking movement of rhodamine-labeled actin filaments across the surface of Myo2p coated coverslips. Data presented was derived from independent experiments using two different preparations of Myo2p. (A) Average rates of Myo2p-driven actin/actin-Cdc8p filament gliding as a function of the Myo2p concentration adhered to coverslips. (n = 50). (B) The number of actin versus actin-Cdc8p filaments bound to coverslip surfaces coated with low concentrations of Myo2p. Total number of filaments from three 130-μm2 fields (from three independent experiments) are plotted at each concentration. (C) Percentage of actin (vs. actin-Cdc8p) filaments bound to the coverslip surface undergoing motility as a function of Myo2p concentration. (n = 270–300 filaments).
Supplementary Material
Yeast strain and plasmid lists are provided in Supplementary Tables S1 and S2. Additional details regarding yeast strains, plasmid constructions, and protein purifications are provided in the Supplementary Material. Supplementary Figures S1 and S2 (relating to contractile ring dynamic measurements in Figure 5), Figure S3 (actin-binding activity of fission yeast-purified Cdc8p), and Movies 1 and 2 (showing Myo2p motility ± Cdc8p) are also provided here.
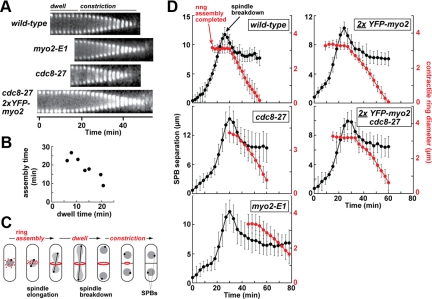
Figure 5.
Myo2p motor function and tropomyosin promote contractile ring assembly. (A) Representative kymographs comparing contractile ring lifetimes and dynamics in wild-type (YFP-Myo2p), myo2-E1 (Rlc1p-GFP), cdc8-27 (YFP-Myo2p), and cdc8-27 2xYFP-myo2 cells. Kymographs were generated from time-lapse fluorescence microscopy images (as detailed in Figure 2A) from cell montages (shown in Supplementary Figure S1). (B) A plot showing the correlation between contractile ring assembly and dwell times derived from values listed in Table 2. (C) Schematic of a wild-type fission yeast cell illustrating the timing of contractile ring (red) assembly, dwell, and constriction phases relative to spindle elongation and SPB (black) separation during mitosis. Nuclei are shown in gray. Diameters of assembled rings are monitored in relation to SPB position to determine the timing of contractile ring dynamics in relation to progression through mitosis. (D) Average SPB separation distances (black) in relation to contractile ring diameter (red) were monitored in cells over time. Plots were generated from analysis of time-lapse images (six z-sections) recorded every 2–3 min by fluorescence microscopy of SPBs (Sad1p-GFP) and rings (Rlc1p-GFP for wild-type, cdc8-27, and myo2-E1 strains; YFP-Myo2p for the 2x strains). Rlc1p-GFP was used instead of 1xYFP-Myo2p because its signal was much brighter and relatively resistant to the photobleaching incurred during collection of 1xYFP-myo2 z-stack movies. With respect to ring diameters, the horizontal lines start upon completion of ring assembly and reflect the dwell phase before constriction (diagonal lines). For each strain shown, n = 25–30 cells. Ring diameter and corresponding SPB separation measurements for each cell were collectively aligned in average plots at the point in time when rings initiate constriction. The average time represented in the plots spanning SPB duplication (time zero) to spindle breakdown (defined as peak SPB separation) reflects the mean of the dataset (rounded-up to the nearest time point). Likewise, the length of ring dwell time plotted represents the mean.
RESULTS
Actomyosin Contractile Ring Dynamics Rely on an Optimal Concentration of Myo2p
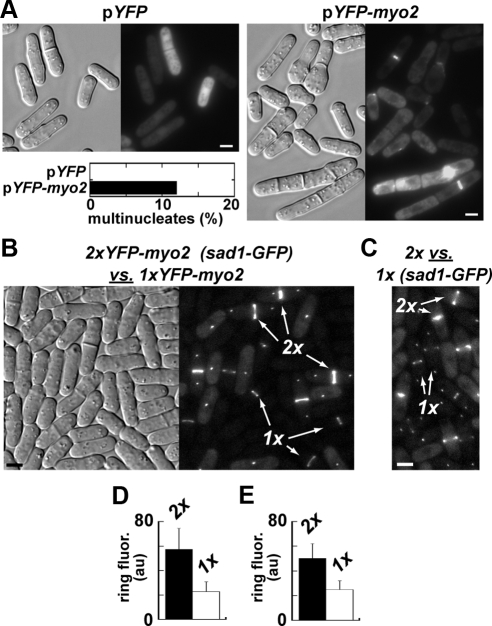
Contractile rings possess a remarkably uniform concentration of Myo2p, which gradually increases as rings constrict (Wu and Pollard, 2005). We wanted to test whether this uniform accumulation of Myo2p reflected the emergence of an optimal concentration that (once reached) facilitates Myo2p function in ring assembly. We initially took a standard approach to heightening Myo2p cellular levels using plasmid-based expression (from the myo2 promoter). This led to obvious defects in contractile ring integrity and cytokinesis in a subpopulation of cells (Figure 1A). The relatively high and inconsistent levels of Myo2p expressed in these cells, along with the aforementioned cytokinesis defects, made it difficult to accurately test whether contractile ring dynamics are fine-tuned by Myo2p cellular levels. Thus, we used a more subtle approach to alter Myo2p levels. Myo2p expression in haploids was doubled by integrating a second copy of myo2 into the genome. Fluorescence intensity measurements of YFP were used to compare Myo2p levels at contractile rings in cells with one copy (1xYFP-myo2) versus two copies (2xYFP-myo2) of myo2 (Figure 1, B–E). For comparisons, mixtures of 1x and 2xYFP-myo2 cells were imaged together. To distinguish one from the other, one of the two strains was marked by a spindle pole body (SPB) marker (Figure 1, B and C). Doubling myo2 copy number led to about a twofold increase in Myo2p levels at the ring (Figure 1, D and E, Table 1).
Figure 1.
Artificially altering Myo2p expression levels in the cell. (A) Cell morphology (DIC images, left) and YFP/YFP-Myo2p localization (fluorescence images, right) in wild-type fission yeast cells harboring pYFP (empty vector) or pYFP-myo2. Cells were grown in EMM-Leu− media at 32°C. Elevated expression of Myo2p from a multicopy plasmid leads to obvious morphological defects in cytokinesis in ∼12% of cells. The plot summarizes cytokinesis phenotypes scored as multinucleate cells (3+ nuclei/cell) by fluorescence microscopy after treating cells with Hoechst stain to mark nuclei. n = 300 cells. (B) Comparison of Myo2p levels at contractile rings in a mixed population of cells carrying one (1xYFP-myo2) or two (2xYFP-myo2) copies of myo2. The two strains were grown separately in YE5S medium before mixing and imaging by light microscopy using DIC (left) and YFP (right) filters. The SPB marker Sad1p-GFP was incorporated into the genome of the 2xYFP-myo2 strain to differentiate it from the 1x strain. Examples of each strain are indicated (arrows). (C) Fluorescent image comparing 2x and 1x strains. As in B, except the Sad1p marker was engineered into the 1x strain. (D and E) Plots show relative YFP-Myo2p fluorescence intensity in contractile rings from corresponding 2x versus 1xYFP-myo2 strain pairs indicated in B (D) and C (E). For each strain average intensity values were derived from 30 rings from images of mixed populations.
Table 1.
Relative concentrations of Myo2p at contractile rings as a function of myo2 copy number
a Local concentration (±SD) of YFP-Myo2p at mature, nonconstricting contractile rings based on fluorescence intensity measurements (relative to those of 1xYFP-myo2 haploid strains) presented in Figure 1. n = 60 rings.
b Final average fluorescence intensity values for YFP-Myo2p in rings of the 1xYFP-myo2 strain were set to 20 μM, the established local concentration of Myo2p in nonconstricting rings in YFP-myo2 cells (Wu and Pollard, 2005).
The impact of doubling Myo2p levels on cytokinesis was assessed by comparing the phenotypes of 1xYFP-myo2 and 2xYFP-myo2 cells. Both strains showed normal cell morphology and exhibited no obvious defects in cytokinesis (Figure 1, B and C). We next compared contractile ring performance. Four different properties of the ring were measured: assembly: time taken for Myo2p to compact into a ring after its appearance as a broad band of nodes; dwell: time from completion of ring assembly until initiation of constriction; constriction: change in ring circumference over time; and lifetime: time from completion of ring assembly to completion of constriction. Our analysis revealed that rings assembled about twofold quicker in the 2xYFP-myo2 strain (Table 2). These rings had a ∼1.5 longer lifetime than those in the 1xYFP-myo2 strain, which was reflected by a ∼1.5-fold longer dwell time and ∼1.5 slower rate of constriction (Figure 2A, Table 2).
Table 2.
Effect of myo2 copy number and mutations on contractile ring dynamics
| Strain | Ring property (±SD)a |
|||
|---|---|---|---|---|
| Assembly (min) | Dwell (min) | Constriction (μm/min) | Lifetime (min) | |
| YFP-myo2b | 17.2 ± 1.6 | 13.3 ± 2.2 | 0.44 ± 0.02 | 38.3 |
| (17.4 ± 2.8) | (12.1 ± 2.4) | (0.40 ± 0.05) | ||
| 2xYFP-myo2b | 8.9 ± 2.3 | 20.7 ± 3.0 | 0.31 ± 0.05 | 56.2 |
| (12.3 ± 3.5) | (21.8 ± 2.7) | (0.26 ± 0.03) | ||
| YFP-myo2 cdc8-27 | 22.4 ± 3.8 | 6.2 ± 3.1 | 0.46 ± 0.08 | 30.1 |
| 2xYFP-myo2 cdc8-27 | 14.9 ± 3.0 | 19.7 ± 4.9 | 0.30 ± 0.06 | 56.4 |
| rlc1-GFP | 17.9 ± 3.1 | 15.0 ± 3.1 | 0.40 ± 0.05 | 42.5 |
| rlc1-GFP cdc8-27 | 23.1 ± 3.6 | 10.5 ± 3.1 | 0.38 ± 0.06 | 39.4 |
| rlc1-GFP myo2-E1 | 26.7 ± 5.0 | 7.9 ± 3.3 | 0.21 ± 0.04 | 60.3 |
a Assembly: time taken for Myo2p to compact into a mature ring after its appearance as a broad band of nodes; dwell: time from completion of ring compaction until initiation of constriction; constriction: change in ring circumference over time. n = 30–50 rings/strain. Dwell times and constriction initiation were discerned by charting ring diameter over time for each ring. Individual constriction rates were derived from slopes of constricting ring circumferences. Lifetimes are the sum of the average dwell and constriction times. Constriction time was estimated by dividing a representative contractile ring circumference (11 μm) by the average constriction rate. All cells were grown at 25°C, and time-lapse microscopy was carried out at room temperature.
b Values shown in parentheses for the YFP-myo2 and 2xYFP-myo2 strains were generated in the presence of excess Rlc1p (expressed from a multicopy plasmid). The difference in ring constriction rates in the YFP-myo2 and 2xYFP-myo2 strains was significant: p = < 0.0001 (p = 0.0002 with excess Rlc1p).
Figure 2.
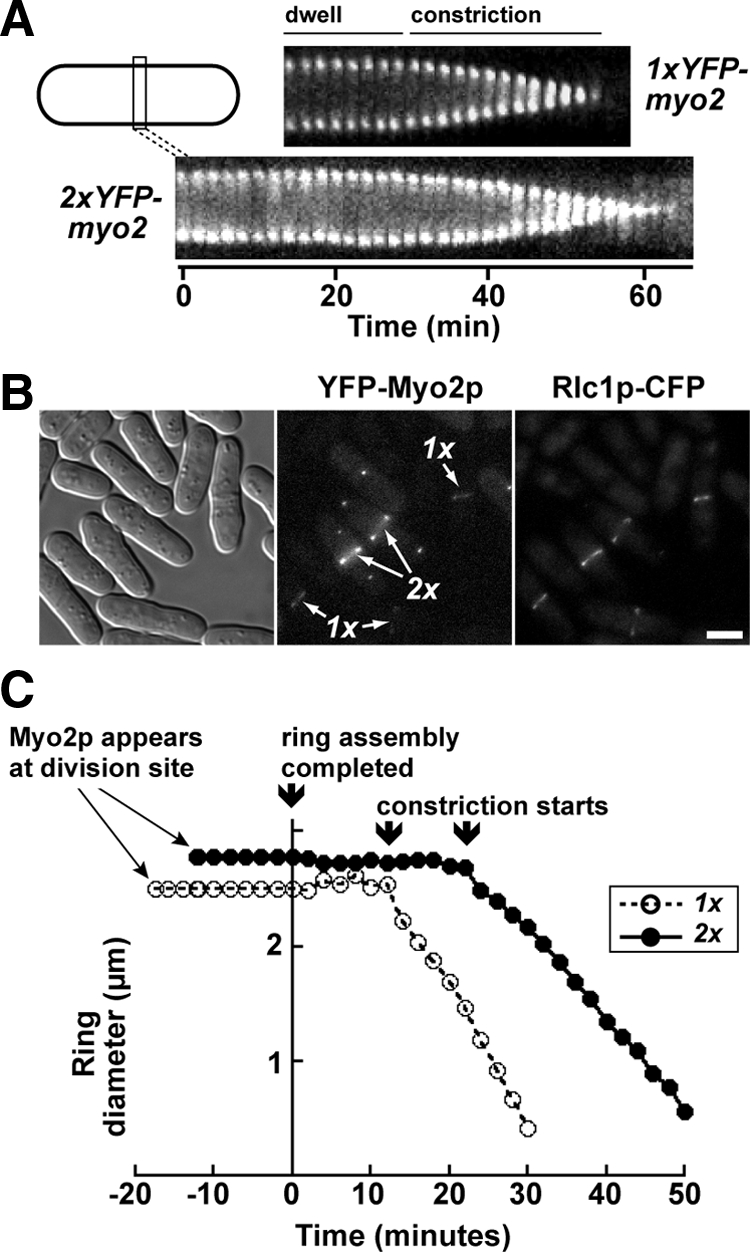
Doubling Myo2p expression levels influences contractile ring dynamics. (A) Kymographs comparing contractile ring lifetime and dynamics in representative 1x and 2xYFP-myo2 cells. Each kymograph is made up of a series of thin slices centered on the contractile ring using images captured every 2 min. Slice height, 4.4 μm. Kymographs start from the point at which rings have assembled, spanning dwell and constriction phases. Kymographs are aligned based on the point at which constriction begins. (B) Comparing the relative levels of Rlc1p at contractile rings in 1x and 2xYFP-myo2 cells. Colocalization of YFP-Myo2p and Rlc1p-CFP in representative cells was performed by epifluorescence microscopy. 1x and 2xYFP-myo2 strains were grown separately before mixing and imaging by light microscopy using DIC (left), YFP (center), and CFP (right) filters. The SPB marker Sad1p-GFP was incorporated into the genome of the 2xYFP-myo2 strain to differentiate it from the 1x strain. (C) Plot charting the average assembly times, dwell times, and constriction rates for contractile rings in 1x and 2xYFP-myo2 cells (n = 20) harboring excess Rlc1p (expressed from a multicopy plasmid). Horizontal lines to the left of the y-axis represent the average assembly time. On assembly ring diameters were measured through dwell and constriction phases and then collectively aligned at the point in time when rings initiate constriction and averaged. The dwell phases (horizontal line to right of y-axis) reflect the mean of the dataset (rounded up to the nearest 2-min time point).
Although the estimated cellular concentration of the essential light chain Cdc4p (4.75 μM) is about sixfold in excess of Myo2p plus Myp2p (0.45 + 0.38 = 0.83 μM), the regulatory light chain Rlc1p (0.60 μM) is limiting (Wu and Pollard, 2005). Thus, in theory there is not enough Rlc1p available to cover both Myp2p and the excess Myo2p in the 2xYFP-myo2 strain. A lack of Rlc1p on the Myo2p heavy chain may account for the altered contractile ring dynamics of the 2xYFP-myo2 strain. Alternatively, Rlc1p levels may simply equilibrate with myosin-II levels in cells and (provided Myo2p is not grossly overexpressed) maintain full coverage of Myo2p in the 2x strain. To distinguish between these two possibilities, we compared Myo2p and Rlc1p levels in 1xYFP-myo2 rlc1-CFP and 2xYFP-myo2 rlc1-CFP strains (Figure 2B). Although YFP-Myo2p levels at contractile rings were clearly different in the two strains, levels of Rlc1p-CFP were essentially the same suggesting that Rlc1p is limiting and that excess YFP-Myo2p in the 2x strain is lacking Rlc1p. However, analysis of rings in the presence of excess Rlc1p (expressed from a multicopy plasmid) revealed that a lack of light chains was not responsible for the difference in ring dynamics observed between the 1xYFP-myo2 and 2xYFP-myo2 strains (Figure 2C; Table 2).
Cellular Concentration Regulates Myo2p Exchange Rates at Contractile Rings
We performed FRAP experiments to test whether the concentration of Myo2p influences the rate at which it exchanges at the ring. Analysis of the 1xYFP-myo2 strain indicated an average half-time (t1/2) for Myo2p recovery of ∼37 s in nonconstricting rings which becomes shorter (∼13 s) during constriction (Figure 3, A and B; Sladewski et al., 2009). Compared with 1xYFP-myo2 rings Myo2p exchanged about twofold faster in both nonconstricting and constricting rings of the 2xYFP-myo2 strain (Figure 3, A and B). Thus, doubling Myo2p levels speeds up the rate it moves in and out of the ring.
Collectively, FRAP (Figure 3) and ring dynamics (Figure 2, A and C, and Table 2) measurements with the 1x and 2xYFP-myo2 strains indicate that the cellular level of Myo2p influences actomyosin ring performance and function.
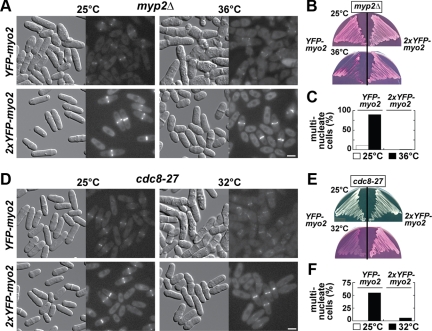
Doubling Myo2p Levels Suppresses Cytokinesis Defects Associated with Compromised Myosin-II and Tropomyosin Function
Given the increased lifetime exhibited by the 2xYFP-myo2 rings, we wanted to test whether doubling Myo2p expression exacerbated cytokinesis defects associated with mutations in other contractile ring proteins. We found 2xYFP-myo2 interacted genetically with a myp2Δ null and a temperature-sensitive cdc8-27 mutant. Myp2p is the nonessential myosin-II, whereas Cdc8p is the sole tropomyosin in fission yeast. In each case doubling Myo2p expression suppressed cytokinesis defects associated with these mutants. Temperature-sensitive cytokinesis defects of a myp2Δ strain were no longer evident (Figure 4, A–C), whereas obvious cytokinesis defects associated with cdc8-27 mutants grown at 32°C (but not at 36°C) were absent in the 2xYFP-myo2 background (Figure 4, D–F). The ability of excess levels of Myo2p to suppress a myp2 null is understandable because Myp2p and Myo2p provide the motor activity for the actomyosin ring. However, until now, no direct role for Cdc8p in Myo2p function has been reported. The fact that a two-fold increase in Myo2p levels can rescue cytokinesis in a cdc8 mutant suggests these two proteins work together and share a common goal.
Figure 4.
Doubling Myo2p cellular levels suppresses cytokinesis defects in myp2Δ and cdc8-27 cells. (A–C) The influence an extra copy of myo2 imparts on the temperature-sensitive phenotype of a myp2Δ strain was assessed by comparing cell morphology and contractile ring integrity (A), growth (B), and cytokinesis defects (C) in myp2Δ YFP-myo2 versus myp2Δ 2xYFP-myo2 strains. (A) Cells grown at 25 and 36°C were imaged and processed in identical manner using DIC (left) and YFP (right) filters by light microscopy. Bar, 4 μm. (B) Cells were grown at 25 and 36°C on YE5S plates containing phloxin B, which labels dead cells (dark pink) over live cells (light pink). (C) Cells grown in YE5S media at 25 and 36°C were treated with Hoechst stain to mark nuclei and cytokinesis phenotypes scored as described in Figure 1A. (D–F) As in A–C, only the temperature-sensitive cdc8-27 mutation was used in place of the myp2Δ mutation. Phenotypes were compared after growth at 25 versus 32°C. Bar, 4 μm.
Myo2p Motors and Tropomyosin Promote Actomyosin Ring Assembly
Time-lapse analysis of contractile ring dynamics was undertaken to explore whether Myo2p and Cdc8p share a role in ring function and to understand how increased Myo2p levels rescue Cdc8p function. We found that temperature-sensitive Myo2p motor (myo2-E1) and cdc8-27 mutants exhibited similar defects in ring dynamics. Rings took longer to assemble and had shorter dwell times compared with wild-type (Figure 5A; Table 2). However, although cdc8-27 rings had normal rates of constriction (after growth at 25°C), myo2-E1 rings constricted about twofold slower than wild-type cells (Figure 5A; Table 2). Even at a semipermissive temperature (30°C) where many cdc8-27 cells fail to form a ring and exhibit cytokinesis phenotypes, this mutant still had a constriction rate (0.39 ± 0.09 μm/min; n = 25) similar to wild type (0.40 ± 0.08 μm/min; n = 20). A shorter dwell time meant that the average contractile ring lifetime for the cdc8-27 strain was shorter than that of a wild-type strain (Figure 5A; Table 2). For the most part, ring defects associated with myo2-E1 and cdc8-27 mutants contrasted with those exhibited by the 2xYFP-myo2 strain (where ring assembly is quicker, dwell times longer, and ring lifetime extended; Table 2). Suppression in the cdc8-27 2xYFP-myo2 strain (Figure 4, D–F) was reflected by shorter, wild-type-like assembly times, with dwell times and constriction rates being more characteristic of the 2xYFP-myo2 strain (Figure 5A, Table 2).
Based on our analysis of ring dynamics, dwell time is inversely proportional to assembly time (Table 2, Figure 5B). In this case, the short dwell times in the myo2-E1 and cdc8-27 mutants are a result of delayed assembly, whereas the long dwell times in the 2xYFP-myo2 strain reflect premature assembly. However, at this stage we cannot rule out that these altered dwell times are unrelated to assembly and instead reflect defects in the timing of constriction. To differentiate between these two possibilities, we compared the timing of ring assembly and constriction relative to SPB position and mitotic progression, as illustrated in Figure 5C (and as exemplified by representative wild-type, cdc8-27, and myo2-E1 cells; Figure S2). Ring assembly is completed later in the cell cycle of cdc8-27 and myo2-E1 mutants compared with wild-type cells (Figure 5D). On average wild-type rings complete assembly ∼8 min before spindle breakdown, whereas cdc8-27 and myo2-E1 rings completed assembly ∼1 min before and ∼6 min after spindle breakdown respectively (Table 3). Therefore, shorter dwell times are a consequence of delayed ring assembly and are not due to premature constriction. In fact, the initiation of ring constriction was clearly delayed in myo2-E1 cells (Figure 5D, Table 3).
Table 3.
Cell cycle timing of contractile ring assembly andconstriction
| Strain | Time relative to spindle breakdown (min)a |
|
|---|---|---|
| Assembly complete | Constriction initiates | |
| Wild typeb | −8.3 ± 2.7 | +5.7 ± 2.0 |
| cdc8-27 | −1.3 ± 2.9 | +8.9 ± 3.3 |
| myo2-E1b | +5.6 ± 6.2 | +15.4 ± 6.7 |
| 2xYFP-myo2 | −14.0 ± 3.2 | +6.7 ± 2.9 |
| cdc8-27 2xYFP-myo2 | −14.1 ± 5.2 | +8.7 ± 4.0 |
a Values represent the mean (±SD) generated from analysis of rings and SPBs in 30 cells. Values correspond to the average datasets provided in Figure 5C. Spindle breakdown is defined as the time when SPB separation peaks.
b Contractile rings in these strains visualized by Rlc1p-GFP.
All cells were grown at 25°C, and time-lapse microscopy was carried out at room temperature.
The differences in constriction initiation times in wild-type vs. mutants were significant (p = 0.0006 for cdc8-27; p = < 0.0001 for myo2-E1).
We found that, consistent with the cell cycle analysis of myo2 and cdc8 mutants, the longer dwell times of 2xYFP-myo2 rings reflect premature assembly, which was completed ∼6 min earlier than in the 1xYFP-myo2 strain (Figure 5D, Table 3). The ability of elevated Myo2p levels to suppress cdc8-27 mutants (Figure 4, D–F) was reflected in the timing of ring assembly (Figure 5D, Table 2) that saw cdc8-27 2xYFP-myo2 rings form ∼13 min earlier than those in a cdc8-27 background (Figure 5D, Table 3). Rings in the cdc8-27 background initiated constriction at the same time, irrespective of whether one or two copies of Myo2p were present (Figure 5D, Table 3). In summary, defects in the Myo2p motor or Cdc8p delay ring assembly, although doubling Myo2p cellular levels, speeds up ring assembly, which in turn suppresses delayed ring assembly in cdc8-27 mutants.
Tropomyosin Enhances Myo2p ATPase Activity
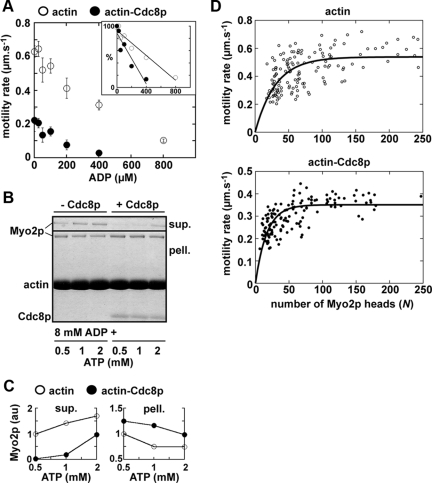
The observation that increased Myo2p expression suppresses the cdc8-27 mutant implies a role for Cdc8p in maintaining effective Myo2p function in vivo. We therefore undertook in vitro studies to determine whether Cdc8p directly influences Myo2p function. Myo2p was purified as previously described (Lord and Pollard, 2004). Functional Cdc8p was overexpressed in fission yeast and purified from cell extracts (Skau et al., 2009; Skoumpla et al., 2007; Figure S3).
We used actin-activated ATPase assays to test whether association of Cdc8p with actin filaments influences Myo2p activity. Actin decorated with Cdc8p increased the activity of Myo2p (Figure 6A). The effect was most prominent at lower actin concentrations leading to a shift in the Michaelis Menten kinetics (Km) reflecting a seven-fold increase in apparent affinity for actin (Table 4). The VMAX was increased ∼1.4-fold (Figure 6A, Table 4). Cdc8p promoted Myo2p activity in a concentration-dependent manner, with full occupancy of Cdc8p on actin being required for maximal activation (Figure 6B). An increase in ionic strength led to diminished Myo2p activity (Figure 6C). However, Cdc8p restored Myo2p activity at high salt (Figure 6C) indicative of a difference in the nature of the actomyosin interaction when actin is associated with Cdc8p.
Tropomyosin Maximizes Myo2p-driven Actin Filament Gliding
In vitro motility assays were used to test how Cdc8p affected Myo2p motor function. Although Cdc8p enhances Myo2p activity in ATPase assays (Figure 6), association of Cdc8p with actin filaments reduced motility rates about twofold over a wide range of Myo2p concentrations (Figure 7A, Supplementary Movie 1). However, Myo2p's ability to bind actin filaments was greatly enhanced by Cdc8p in this assay. When Myo2p concentration was reduced, the number of actin filaments associated with Myo2p at the coverslip surface was much higher in the presence of Cdc8p (Figure 7B, Supplementary Movie 2). Myo2p eventually failed to support any motility when applied to chambers at 0.4 μg/ml, yet motility was maintained when Cdc8p was present. Furthermore, at lower Myo2p concentrations (0.8–3.2 μg/ml) most Cdc8p-bound filaments are motile, whereas the relatively few filaments apparent in the absence of Cdc8p were mostly nonmotile (Figure 7C), presumably nonspecifically adhered to the surface. Because about five-fold more filaments (of which ∼80% are motile) are apparent in the presence of Cdc8p, compared with its absence (where only ∼15% of filaments are motile), the number of motile filaments increases >25-fold when Cdc8p is bound to actin.
Although surfaces in our motility chambers are blocked after attachment of Myo2p, we wanted to rule out the possibility that reduced rates of Myo2p motility were due to drag caused by Cdc8p simultaneously interacting with actin filaments and the coverslip surface. We used myosin-Va as a control here given that this motor is not thought to be regulated by tropomyosin (Wang et al., 2000). The fact that myosin-Va exhibited identical rates of motility in the presence and absence of Cdc8p (Table 4) confirms that the effect of Cdc8p on Myo2p motility is specific.
Tropomyosin Favors the Strong Actin-bound State of Myo2p
Although ATPase and motility assays indicate Cdc8p promotes actomyosin interactions, Myo2p's motility rate was consistently slower in the presence of Cdc8p (Figure 7A). At saturating concentrations of myosin-II, motility is limited by the relative length of time the motor occupies the strong actin-bound ADP state (Siemankowski et al., 1985; Wang et al., 2000). Thus, we wished to test whether Myo2p's increased actin affinity and reduced motility speed reflected a preference for the ADP-bound state when associated with Cdc8p-bound actin.
We first measured Myo2p properties as a function of ADP:ATP ratios. We found that Myo2p motility was more sensitive to increasing concentrations of ADP in the presence of Cdc8p (Figure 8A). Linear fits from plots of relative motility rate versus ADP concentration indicated that 422 μM ADP was needed to reduce Myo2p motility by 50% in the absence of Cdc8p, whereas only 186 μM ADP was needed when actin was saturated with Cdc8p (Figure 8A). Conversely, we used actin filament cosedimentation assays to test the ability of increasing concentrations of ATP to release Myo2p from a strong actin-bound (ADP) state (Figure 8B). As ATP concentration was increased, Myo2p shifted from the strong-bound (pellet) to the weak-bound (supernatant) state both in the presence and absence of Cdc8p. However, this shift was more pronounced in the absence of Cdc8p (Figure 8, B and C). Collectively, the data suggests that Myo2p prefers the strong actin-bound state when Cdc8p is bound to filaments.
Figure 8.
Tropomyosin increases the duty ratio of Myo2p. (A) Average rates of Myo2p-driven actin/actin-Cdc8p motility as a function of the ADP concentration in the running buffer. ATP at 400 μM was included in the running buffer throughout (n = 50, from two independent experiments). Inset, motility rates are plotted as percentages (where 100% represents the rate measured in the absence of ADP). Linear fits compare the ADP sensitivity of Myo2p motility with and without Cdc8p. (B) SDS-PAGE gel summarizing the distribution of Myo2p in supernatant and pellet fractions after cosedimentation with actin versus actin-Cdc8p filaments. All samples included 8 mM ADP and the indicated concentrations of ATP. (C) Plots summarizing results in B. Densitometry of the Myo2p bands was used to compare relative levels of Myo2p in supernatant and pellet fractions. For each plot the value 1 represents Myo2p levels found in the 0.5 mM ATP samples lacking Cdc8p. (D) Plots of actin filament motility rate versus the number of Myo2p heads available to interact with the filament for actin (left) and actin-Cdc8p (right); n = 150. The number of Myo2p heads available was determined based on actin filament length, the concentration of Myo2p bound to the coverslip surface, and the estimated reach of a myosin head (Uyeda et al., 1990). Curve fits were used to determine the duty ratio of Myo2p with and without Cdc8p (Table 4).
We measured Myo2p's duty ratio (percentage of time spent in the strong actin-bound state per ATP hydrolysis cycle) with and without Cdc8p using two different approaches. First, motility rates were used to derive ton values (Table 4). Knowing ton and the VMAX of the actin-activated ATP hydrolysis allowed us to calculate duty ratios of ∼5% without Cdc8p, versus ∼14% with Cdc8p (Table 4). Second, duty ratios were determined by performing motility assays at low concentrations of Myo2p to measure the relationship between motility rate and actin filament length (Uyeda et al., 1990; Harris and Warshaw, 1993). Curve fits from plots shown in Figure 8D determined duty ratios of ∼3% without Cdc8p, versus ∼6% with Cdc8p (Table 4). Therefore, Cdc8p enhances the affinity of Myo2p for actin by increasing the amount of time the motor spends in the strong actin-bound state, reflected by a ∼2–3-fold increase in duty ratio.
DISCUSSION
The rate of ring assembly is sensitive to the cellular concentration of Myo2p. Cdc8p associates with actin filaments and maximizes Myo2p motor function by promoting actomyosin interactions. Thus, successful actomyosin interactions and contractile ring assembly relies both on the establishment of effective concentrations of active Myo2p motors and the emergence of Cdc8p-bound actin filaments at the division site.
Role of Myo2p and Tropomyosin in Actomyosin Ring Assembly and Function
Doubling Myo2p expression levels led to premature contractile ring assembly, slower ring constriction, and faster Myo2p exchange at the ring. Our findings suggest that ring dynamics are fine-tuned by an optimal concentration of Myo2p which in a wild-type cell balances timely ring assembly with optimal rates of constriction. The slower rate of constriction might be explained by myosin-II's role in actin filament turnover at contractile rings (Guha et al., 2005; Murthy and Wadsworth, 2005). Higher levels of Myo2p could lead to less actin filament sliding and more filament destabilization resulting in nonproductive contractility and slower ring constriction.
In addition to promoting ring assembly and faster rates of Myo2p exchange at rings, increased Myo2p levels completely suppressed cytokinesis phenotypes associated with myp2Δ mutants lacking the nonessential myosin-II Myp2p. This finding indicates that Myo2p levels at rings are uniformly lower in wild-type cells (given they do not compensate for loss of Myp2p). Based on our results, relatively low levels of myosin-II appear important to delay the precise timing of ring assembly, but increased levels may become important once rings have matured, dictating recruitment of Myp2p (or the reliance on additional Myo2p in a myp2Δ background).
Analysis of contractile ring dynamics in myo2-E1 and cdc8-27 mutants indicated that Myo2p motor activity and Cdc8p support efficient rates of ring assembly. The role of the Myo2p motor in driving ring compaction is consistent with a recent study showing that ring assembly is delayed in mutants with compromised Myo2p motor function (Coffman et al., 2009). Doubling Myo2p levels suppressed cytokinesis defects of cdc8-27 cells by promoting ring assembly. This finding implies that Cdc8p plays a role in maximizing actomyosin interactions in vivo. Although suppression was evident in cdc8-27 cells grown at 32°C, it was not sustained at 36°C. Thus, either there is not enough Myo2p in the 2xYFP-myo2 strain to support suppression at the restrictive temperature, or Cdc8p has other essential roles whose function cannot be rescued by simply elevating Myo2p levels.
Unlike ring assembly, constriction rates were normal in cdc8-27 cells grown at 25°C (or 30°C). In contrast, even at 25°C constriction in myo2-E1 mutants was much slower than wild-type. Thus, Cdc8p's ability to regulate Myo2p appears be critical in establishing actomyosin interactions during ring assembly, as opposed to maintaining them during constriction. Once rings are formed their subsequent constriction may also rely on further enhancement of Myo2p motor activity by other regulators. For example, activation by the UCS protein may be more critical later given that Rng3p only begins to concentrate at rings once they have matured (Lord and Pollard, 2004). Phosphorylation of regulatory light chain Rlc1p may also contribute to Myo2p activity during constriction given this modification promotes maximal rates of Myo2p motility and optimal rates of ring constriction (Sladewski et al., 2009).
Regulation of Myo2p Motor Activity by Tropomyosin
Association of actin with Cdc8p increases the actin-activated ATPase activity of Myo2p, reflected by a reduced Km and an increased VMAX. The presence of Cdc8p resulted in about a two-fold drop in the rate of Myo2p-driven actin filament gliding in motility assays, although the number of filaments undergoing motility was much greater in the presence of Cdc8p. The ability of Cdc8p to increase the Myo2p duty ratio suggests it may promote effective actomyosin interactions by favoring Pi release and occupancy of the strong actin-bound state. Because myosin-II motility is limited by the strong bound state (Siemankowski et al., 1985), increasing the relative time Myo2p spends in this state (per ATP hydrolysis cycle) limits the speed at which it translocates individual actin filaments in motility assays, despite an increase in the overall numbers of productive actomyosin interactions.
The ability of Cdc8p to promote actomyosin interactions does not reflect the ability of Cdc8p to bind Myo2p. Such a role for tropomyosin would be unprecedented and we found no evidence of this. Cdc8p's ability to promote filament binding by Myo2p in actin cosedimentation assays was sensitive to ADP-ATP ratios, suggesting Cdc8p mediates its effects indirectly by influencing the ratio of weak- (ATP) to strong- (ADP) bound actomyosin states.
Tropomyosin increases the motility rate of skeletal muscle myosin about threefold (Umemoto and Sellers, 1990). Here regulation of actomyosin interactions depends on myosin concentration. At low myosin concentrations actomyosin interactions are inhibited by tropomyosin, whereas this effect is reversed as myosin concentration increases owing to its ability to cooperatively bind and “activate” tropomyosin-bound filaments (Lehrer, 1994; Swartz et al., 1996; VanBuren et al., 1999; Strand et al., 2001). However, unlike nonmuscle cells, tropomyosins in striated muscle function specifically with troponin to regulate myosin in a Ca2+-dependent manner. Previous work with Cdc8p revealed that it inhibits initial associations between actin and skeletal muscle myosin S1 forms owing to its ability to occupy the closed conformation on actin filaments (Skoumpla et al., 2007), which regulates interactions between actin and muscle myosin (McKillop and Geeves, 1993). However, our experiments with Myo2p are consistent with Cdc8p promoting actomyosin interactions, as opposed to gating access to the filament.
Relating Cdc8p to Tropomyosin Function in Nonmuscle Cells
Although our work highlights the role of tropomyosin in promoting actomyosin ring assembly in fission yeast, the molecular mechanism we define is probably relevant in higher eukaryotes, where regulation of nonmuscle myosins by tropomyosin is not well understood. Overall, our findings are reminiscent of some previous studies with nonmuscle myosin-II and tropomyosins in higher eukaryotes. Overexpression of the human low-molecular-weight (LMW) Tm5NM1 in rat cortical neuronal cells increases stress fiber formation and leads to recruitment of myosin-IIA at these structures (Bryce et al., 2003). LMW tropomyosins also appear to be important for actomyosin ring function and cytokinesis in transformed cells. For example, in cultured astrocytes and low-grade astrocytoma tumor cells high-molecular-weight (HMW) tropomyosins localize at contractile rings, whereas in high-grade tumor cells HMW tropomyosin expression is lost and LMW Tm5a and 5b take over at contractile rings (Hughes et al., 2003). Forced expression of human LMW Tm5 and a chimera Tm5/3 (made from Tm5 and Tm3) in Chinese hamster ovary cells speeds up cell division and cytokinesis (Eppinga et al., 2006). The ability of these tropomyosins to accelerate cytokinesis may reflect their ability to promote myosin-II motor function given that purified Tm5 and Tm5/3 were previously found to enhance the actin-activated ATPase activity of smooth muscle myosin-II in vitro (Novy et al., 1993).
As with Myo2p and Cdc8p, budding yeast myosin-II (Myo1p) also exhibits slower rates of in vitro motility in the presence of tropomyosin Tpm2p (Huckaba et al., 2006). However, this reduced rate of motility was found to be due to Tpm2p's ability to inhibit myosin-II-actin binding (Huckaba et al., 2006).
Implications for the Regulation of Contractile Ring Function
A duty ratio of at least 50% is needed to support processive walking of a double-headed myosin molecule along an actin filament. Duty ratios of ∼4% (minus Cdc8p) and ∼10% (plus Cdc8p) indicate that Myo2p is nonprocessive. Based on these values, a Myo2p ensemble of at least 25 molecules would be needed to support effective motility of actin filaments in the absence of Cdc8p. The minimum number of molecules required in this ensemble drops to 10 when Cdc8p is present. Thus, in order to support effective actomyosin interactions during ring assembly, Myo2p must either self assemble into filaments or use a scaffold to tightly cluster. The nodes may provide such scaffolds.
How do our findings impinge on current models of ring assembly? In the search, capture, pull, and release model (Vavylonis et al., 2008; Coffman et al., 2009), formin-mediated growth of new actin filaments from nodes provides tracks that Myo2p in other nodes tug on to affect ring compaction. Doubling Myo2p levels most likely increases the concentration of Myo2p motors available at nodes, which allows faster ring assembly by increasing the frequency of capture (Myo2p-based motility), leading to greater force production within the actin network and faster ring compaction. The association of Cdc8p with growing actin filaments in these networks would have a similar effect, which would explain why doubling Myo2p levels suppresses defects in ring assembly in a cdc8 mutant. In the leading cable model (Arai and Mabuchi, 2002; Kamasaki et al., 2007) an increase in the number of effective actomyosin interactions promoted by Cdc8p (or by doubling Myo2p levels) would favor formation of the actomyosin bundle/cable (Hachet and Simanis, 2008; Huang et al., 2008; Mishra and Oliferenko, 2008). This is presumably a prerequisite for this model given that Myo2p motor function is ultimately essential for ring assembly (Naqvi et al., 1999; Wong et al., 2000).
Supplementary Material
ACKNOWLEDGMENTS
We thank Joe Clayton for performing motility assays with myosin-Va HMM and Jessica Johnson, Jake Mink, and Matt Sammons for assistance with data analysis. We thank Mohan Balasubramanian (Temasek Life Sciences Laboratory, Singapore) for the myo2-E1 strain. We are indebted to Marilyn Wadsworth and Doug Taatjes from the University of Vermont Microscopy Imaging Facility for their assistance with FRAP experiments. We are grateful to the Lowey and Trybus labs (University of Vermont) for access to equipment and providing us with myosin-Va HMM. We thank Alex Hodges, Susan Lowey, Tom Pollard, Kathy Trybus, and David Warshaw for comments on the manuscript. Work in the Lord laboratory is funded by a New Research Initiative Award from the University of Vermont and a Scientist Development Grant (0835236N) from the American Heart Association.
Abbreviations used:
- FRAP
fluorescence recovery after photobleaching
- ROI
region of interest
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0852) on January 28, 2010.
REFERENCES
- Arai R., Mabuchi I. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 2002;115:887–898. doi: 10.1242/jcs.115.5.887. [DOI] [PubMed] [Google Scholar]
- Arai R., Nakano K., Mabuchi I. Subcellular localization and possible function of actin, tropomyosin and actin-related protein 3 (Arp3) in the fission yeast Schizosaccharomyces pombe. Eur J. Cell Biol. 1998;76:288–295. doi: 10.1016/S0171-9335(98)80007-1. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Helfman D. M., Hemmingsen S. M. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature. 1992;360:84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., McCollum D., Chang L., Wong K. C., Naqvi N. I., He X., Sazer S., Gould K. L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. W., Bamburg J. R. Actin in emerging neurites is recruited from a monomer pool. Mol. Neurobiol. 1992;6:95–106. doi: 10.1007/BF02780546. [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Forsburg S. L., Pollard T. D. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Wilson J. M., Pollard T. D. Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr. Biol. 2000;10:397–400. doi: 10.1016/s0960-9822(00)00420-6. [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T. D., Hitchcock-DeGregori S. E. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Bryce N. S., et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Woollard A., Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 1996;109(Pt 1):131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Coffman V. C., Nile A. H., Lee I. J., Liu H., Wu J. Q. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A. Actin dynamics: tropomyosin provides stability. Curr. Biol. 2002;12:R523–R525. doi: 10.1016/s0960-9822(02)01028-x. [DOI] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Eng K., Naqvi N. I., Wong K. C., Balasubramanian M. K. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr. Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- Eppinga R. D., Li Y., Lin J. L., Lin J. J. Tropomyosin and caldesmon regulate cytokinesis speed and membrane stability during cell division. Arch. Biochem. Biophys. 2006;456:161–174. doi: 10.1016/j.abb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fan X., Martin-Brown S., Florens L., Li R. Intrinsic capability of budding yeast cofilin to promote turnover of tropomyosin-bound actin filaments. PLoS One. 2008;3:e3641. doi: 10.1371/journal.pone.0003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattoum A., Hartwig J. H., Stossel T. P. Isolation and some structural and functional properties of macrophage tropomyosin. Biochemistry. 1983;22:1187–1193. doi: 10.1021/bi00274a031. [DOI] [PubMed] [Google Scholar]
- Finer J. T., Simmons R. M., Spudich J. A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Guha M., Zhou M., Wang Y. L. Cortical actin turnover during cytokinesis requires myosin II. Curr. Biol. 2005;15:732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Hachet O., Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. E., Warshaw D. M. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]
- Henkel R. D., VandeBerg J. L., Walsh R. A. A microassay for ATPase. Anal. Biochem. 1988;169:312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M. K. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba T. M., Lipkin T., Pon L. A. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J. Cell Biol. 2006;175:957–969. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. A., Cooke-Yarborough C. M., Chadwick N. C., Schevzov G., Arbuckle S. M., Gunning P., Weinberger R. P. High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia. 2003;42:25–35. doi: 10.1002/glia.10174. [DOI] [PubMed] [Google Scholar]
- Huxley H. E. Sliding filaments and molecular motile systems. J. Biol. Chem. 1990;265:8347–8350. [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C., Sugimoto A., Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron S. J., Spudich J. A. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S. S. The regulatory switch of the muscle thin filament: Ca2+ or myosin heads? J. Muscle Res. Cell Motil. 1994;15:232–236. doi: 10.1007/BF00123476. [DOI] [PubMed] [Google Scholar]
- Lord M., Pollard T. D. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Sladewski T. E., Pollard T. D. Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells. Proc. Natl. Acad. Sci. USA. 2008;105:8014–8019. doi: 10.1073/pnas.0802874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I., Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J. Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Gunning P. Isoform sorting of tropomyosins. Adv. Exp. Med. Biol. 2008;644:187–200. doi: 10.1007/978-0-387-85766-4_15. [DOI] [PubMed] [Google Scholar]
- May K. M., Watts F. Z., Jones N., Hyams J. S. Type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil. Cytoskelet. 1997;38:385–396. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- McKillop D. F., Geeves M. A. Regulation of the interaction between actin and myosin subfragment 1, evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Oliferenko S. Cytokinesis: catch and drag. Curr. Biol. 2008;18:R247–R50. doi: 10.1016/j.cub.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Motegi F., Nakano K., Kitayama C., Yamamoto M., Mabuchi I. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 1997;420:161–166. doi: 10.1016/s0014-5793(97)01510-x. [DOI] [PubMed] [Google Scholar]
- Motegi F., Nakano K., Mabuchi I. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 2000;113(Pt 10):1813–1825. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- Murthy K., Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi N. I., Eng K., Gould K. L., Balasubramanian M. K. Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J. 1999;18:854–862. doi: 10.1093/emboj/18.4.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy R. E., Sellers J. R., Liu L. F., Lin J. J. In vitro functional characterization of bacterially expressed human fibroblast tropomyosin isoforms and their chimeric mutants. Cell Motil. Cytoskelet. 1993;26:248–261. doi: 10.1002/cm.970260308. [DOI] [PubMed] [Google Scholar]
- Ono S., Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap E. M. Tropomyosins as discriminators of myosin function. Adv. Exp. Med. Biol. 2008;644:273–282. doi: 10.1007/978-0-387-85766-4_20. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith R. H., Gould K. L. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- Siemankowski R. F., Wiseman M. O., White H. D. ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc. Natl. Acad. Sci. USA. 1985;82:658–662. doi: 10.1073/pnas.82.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau C. T., Neidt E. M., Kovar D. R. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol. Biol. Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoumpla K., Coulton A. T., Lehman W., Geeves M. A., Mulvihill D. P. Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 2007;120:1635–1645. doi: 10.1242/jcs.001115. [DOI] [PubMed] [Google Scholar]
- Sladewski T. E., Previs M. J., Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol. Biol. Cell. 2009;20:3941–3952. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Fankhauser C., Brodbeck C., Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Strand J., Nili M., Homsher E., Tobacman L. S. Modulation of myosin function by isoform-specific properties of Saccharomyces cerevisiae and muscle tropomyosins. J. Biol. Chem. 2001;276:34832–34839. doi: 10.1074/jbc.M104750200. [DOI] [PubMed] [Google Scholar]
- Swartz D. R., Moss R. L., Greaser M. L. Calcium alone does not fully activate the thin filament for S1 binding to rigor myofibrils. Biophys J. 1996;71:1891–1904. doi: 10.1016/S0006-3495(96)79388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M., Numata O., Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–3131. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto S., Sellers J. R. Characterization of in vitro motility assays using smooth muscle and cytoplasmic myosins. J. Biol. Chem. 1990;265:14864–14869. [PubMed] [Google Scholar]
- Uyeda T. Q., Kron S. J., Spudich J. A. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- VanBuren P., Palmiter K. A., Warshaw D. M. Tropomyosin directly modulates actomyosin mechanical performance at the level of a single actin filament. Proc. Natl. Acad. Sci. USA. 1999;96:12488–12493. doi: 10.1073/pnas.96.22.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J. Q., Hao S., O'Shaughnessy B., Pollard T. D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wang F., Chen L., Arcucci O., Harvey E. V., Bowers B., Xu Y., Hammer J. A., 3rd, Sellers J. R. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J. Biol. Chem. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- Wawro B., Greenfield N. J., Wear M. A., Cooper J. A., Higgs H. N., Hitchcock-DeGregori S. E. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46:8146–8155. doi: 10.1021/bi700686p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wong K. C., D'Souza V. M., Naqvi N. I., Motegi F., Mabuchi I., Balasubramanian M. K. Importance of a myosin II-containing progenitor for actomyosin ring assembly in fission yeast. Curr. Biol. 2002;12:724–729. doi: 10.1016/s0960-9822(02)00790-x. [DOI] [PubMed] [Google Scholar]
- Wong K. C., Naqvi N. I., Iino Y., Yamamoto M., Balasubramanian M. K. Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J. Cell Sci. 2000;113(Pt 13):2421–2432. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Bahler J., Pringle J. R. Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell. 2001;12:1061–1077. doi: 10.1091/mbc.12.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Q., Kuhn J. R., Kovar D. R., Pollard T. D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Pollard T. D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Sirotkin V., Kovar D. R., Lord M., Beltzner C. C., Kuhn J. R., Pollard T. D. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.