Abstract
Background
Plants engineered for abiotic stress tolerance may soon be commercialized. The engineering of these plants typically involves the manipulation of complex multigene networks and may therefore have a greater potential to introduce pleiotropic effects than the simple monogenic traits that currently dominate the plant biotechnology market. While research on unintended effects in transgenic plant systems has been instrumental in demonstrating the substantial equivalence of many transgenic plant systems, it is essential that such analyses be extended to transgenic plants engineered for stress tolerance. Drought-tolerant Arabidopsis thaliana were engineered through overexpression of the transcription factor ABF3 in order to investigate unintended pleiotropic effects. In order to eliminate position effects, the Cre/lox recombination system was used to create control plant lines that contain identical T-DNA insertion sites but with the ABF3 transgene excised. This additionally allowed us to determine if Cre recombinase can cause unintended effects that impact the transcriptome.
Results
Microarray analysis of control plant lines that underwent Cre-mediated excision of the ABF3 transgene revealed only two genes that were differentially expressed in more than one plant line, suggesting that the impact of Cre recombinase on the transcriptome was minimal. In the absence of drought stress, overexpression of ABF3 had no effect on the transcriptome, but following drought stress, differences were observed in the gene expression patterns of plants overexpressing ABF3 relative to control plants. Examination of the functional distribution of the differentially expressed genes revealed strong similarity indicating that unintended pathways were not activated.
Conclusions
The action of ABF3 is tightly controlled in Arabidopsis. In the absence of drought stress, ectopic activation of drought response pathways does not occur. In response to drought stress, overexpression of ABF3 results in a reprogramming of the drought response, which is characterized by changes in the timing or strength of expression of some drought response genes, without activating any unexpected gene networks. These results illustrate that important gene networks are highly regulated in Arabidopsis and that engineering stress tolerance may not necessarily cause extensive changes to the transcriptome.
Background
Drought is a major abiotic stress that limits crop productivity [1]. Climate change models predict an increase in summer drying in the midlatitudes, which could contribute to an increase in the number of episodes of drought [2,3]. Engineering plants with enhanced tolerance of abiotic stresses such as drought is a major objective of plant biotechnology that is expected to be commercialized in the near future [4,5]. Tolerance to abiotic stress may be achieved through the modification of endogenous plant pathways, often by manipulating important regulatory proteins such as transcription factors. Altering the level of expression of key transcription factors involved in abiotic stress pathways has been shown to enhance tolerance to various abiotic stresses in Arabidopsis [6-9] as well as in important crop species such as rice [10-12], maize [13], and alfalfa [14].
Traits involving tolerance to abiotic stresses are considered to be more complex than those that are currently commercialized due to the large number of genes and pathways that may be affected. Furthermore, the interaction between plants and the environment is an intricate, continuous process that has been difficult to characterize, further adding to the complexity of manipulating abiotic stress tolerance traits. The increased complexity of these traits may correspond with a greater potential for unintended effects to occur in transgenic plants.
In transgenic systems, two different types of unintended effects are generally known to occur [15]. Position effects are attributed to the insertion of a transgene at a particular locus in the genome and the resulting interference this might cause. These effects will vary with the site of integration and will therefore be unique to each independent plant line. Position effects can be easily eliminated by screening for plant lines that have no or little position effects. In contrast, pleiotropic effects are independent of the site of transgene insertion and are the sum of all the phenotypic effects caused by expression of the transgene. While some of these may be the intended trait, others may occur through unexpected interactions of the gene with plant processes and constitute the unintended pleiotropic effects. These effects are of greater interest since they are more difficult to eliminate and more likely to create safety issues.
Engineering more complex traits such as abiotic stress tolerance in plants through the manipulation of transcription factors may uncover cryptic properties of the transcription factor that could produce some of the unintended pleiotropic effects. Many transcription factors are part of large families that have complex evolutionary histories [16,17]. These families typically arise through gene duplications followed by functional divergence in separate expression domains or through the acquisition of new functions. These processes often result in functional redundancies within the families that can be difficult to detect. Furthermore, some transcription factors may retain ancestral functions that are sometimes only revealed by altering the normal pattern of expression. Therefore the manipulation of transcription factors in engineering complex traits such as abiotic stress tolerance may be likely to produce unintended pleiotropic effects.
The use of non-targeted global profiling technologies, such as microarray analysis, to identify unintended effects in plant systems has proven an effective means of determining the "substantial equivalence" of a transgenic plant to its non-transgenic counterpart. Such approaches have been used to investigate unintended effects in a number of transgenic plant systems [18-27]. To date, these studies have primarily focused on simple, monogenic traits such as those that are currently commercially grown. As transgenic crops with more complex traits involving the modification of endogenous plant pathways will soon be entering the market, it is important to extend these analyses to investigate the potential for unintended pleiotropic effects in such systems.
In order to understand the extent and kinds of unintended effects that could be induced in transgenic plants engineered for complex traits, we conferred drought tolerance on Arabidopsis thaliana by overexpressing the transcription factor ABF3. This system targets drought resistance, a trait that will likely enter the market in the near future. Since transcription factors ultimately function by altering the levels of expression of target genes, we investigated unintended effects using microarray analysis to survey global gene expression profiles. In order to eliminate position effects in our analysis and focus on the pleiotropic unintended effects, we employed the Cre/lox system to excise the ABF3 transgene from the site of insertion, leaving behind the selectable marker, to create control plant lines. Without the ABF3 transgene, the pleiotropic effects will be absent but the site of integration is still interrupted by the selectable marker such that position effects are maintained in these lines.
ABF3 belongs to the ABF/AREB subfamily of bZIP transcription factors which consists of thirteen members in Arabidopsis. Several members have been shown to function in ABA signalling either during seed maturation or in response to stress [28]. These factors can bind to ABA-response elements (ABREs), cis-regulatory elements found in the promoters of many ABA- and stress-responsive genes [29-31]. In addition to drought tolerance, overexpression of ABF3 confers tolerance to salt, cold, heat, and oxidative stresses, suggesting that it regulates multiple abiotic stress pathways in Arabidopsis [7,32]. Three other ABF/AREB transcription factors are predicted to function in ABA-dependent stress signalling based on expression profiling and overexpression studies. Expression of ABF1 is induced by cold-treatment [29]. ABF2/AREB1 is induced by salt-treatment as well as dehydration but not cold and overexpression confers tolerance to a wide range of abiotic stresses, including salt, drought, heat, and oxidative stress [29,31,33]. Interestingly, ABF2/AREB1 also appears to function in glucose signalling as well as in the regulation of seedling growth [33]. ABF4/AREB2 is expressed in response to cold, drought, and salt and overexpression renders plants tolerant to drought and salt [7,29,31]. Therefore, ABF3 likely shares some redundant functions with other members of the ABF/AREB subfamily.
Plant response to drought involves changes in the expression patterns of a large number of genes [34-36] and, in addition to members of the ABF/AREB family, a number of other transcription factors have been identified that play a role in the drought response in Arabidopsis. These include the AP2/ERF transcription factors DREB2A, DREB2B, and CBF4 [8,37,38], AtMYB2, which functions in concert with AtMYC2 [39-41], and the NAC family transcription factors ANAC19, ANAC055/ATNAC3, and ANAC072/RD26 [42], which at least partially function in concert with a zinc finger homeodomain protein ZFHD1 [43]. Therefore, while overexpression of ABF3 affects one of the key drought response pathways, it is not the only pathway mediating the drought response at the gene expression level.
The use of the Cre/lox system to create control lines also created an opportunity to examine the effects of the Cre/lox system on the transcriptome. Site-specific recombination technologies can be used to excise selectable markers or other undesirable genetic elements and can also be used to direct site-specific integration of transgenes [44]. While many studies have employed Cre-mediated recombination in plant systems with no apparent unintended effects [45-48], other studies have observed a range of abnormal phenotypes including growth defects, leaf chlorosis, delayed flowering, and male sterility [49,50]. In tobacco plants transformed with a chloroplast targeted Cre recombinase, recombination was observed involving cryptic lox sites in the plastid genome, but invariably the second lox site was located within the transgene [51-53]. These recombinations could result in deletions of up to 147 kb, but they did not cause any deleterious effects in the plants [51-53]. Studies in animal systems have similarly revealed that Cre recombinase can have unintended effects, often leading to chromosomal aberrations [54-57].
These studies suggest that the Cre/lox system has the potential to cause unintended effects in plant systems, by mediating recombination with cryptic lox sites that may be present in the genome resulting in large deletions. Such cryptic lox sites are difficult to identify since they may deviate substantially from conventional loxP sites [52] and not all of the unintended effects may produce readily apparent phenotypic abnormalities, so studying the unintended effects of Cre recombinase using a non-targeted approach such as microarray analysis is essential for establishing the utility and safety of this technology.
In this study, we performed microarray analysis on Arabidopsis plants engineered to be drought-tolerant through overexpression of the transcription factor ABF3 with the goal of identifying unintended pleiotropic effects. The results suggest that overexpression of ABF3 has a minimal impact on the transcriptome, with differences in the gene expression pattern only detectable in response to drought and then being suggestive of transcriptional reprogramming as opposed to the activation of novel pathways. In addition, we examined the impact of Cre recombinase on the transcriptome to detect any unintended effects of this technology and found that it had minimal effects on gene expression patterns in plants following transgene excision.
Results
Phenotype of plants overexpressing ABF3
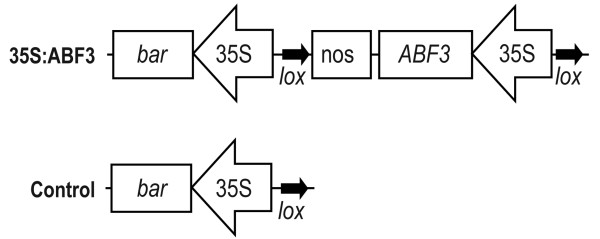
A construct was created containing the CaMV 35S promoter followed by the coding sequence of ABF3 and then the nopaline synthase transcriptional terminator, all of which was flanked by two loxP sites (Figure 1A). This construct was transformed into Arabidopsis thaliana to generate 35S:ABF3 plants. Fifty-nine independent transformants were recovered and three high-expressing lines containing single insertions were selected for further analysis (35S:ABF3-48, -57, and -59).
Figure 1.
35S:ABF3 and control T-DNA regions. Schematic map of the 35S:ABF3 and control T-DNA regions. ABF3, coding region of ABF3 gene; 35S, CaMV 35S promoter; nos, nopaline synthase transcriptional terminator; bar, bialaphos resistance; lox, loxP sites.
Control plant lines were generated by excising the 35S:ABF3 transgene, leaving only the selectable marker transgene at the site of insertion (Figure 1B). This was achieved by crossing 35S:ABF3 plants with plants expressing the Cre recombinase gene and then backcrossing the progeny to wild-type Arabidopsis to remove the Cre recombinase gene, generating three control lines (Control-48, -57, and -59). The loss of both the ABF3 transgene and the Cre recombinase transgene was confirmed by PCR (Additional file 1).
Previously, Arabidopsis plants overexpressing ABF3 were found to be more tolerant to drought conditions, which could at least partially be attributed to a lower rate of transpiration [7]. To confirm that the 35S:ABF3 lines show a similar phenotype, the transpiration rate was determined. Excised leaves from 4-week-old 35S:ABF3 plants left at room temperature for 1 day lost less fresh weight than both wild-type and control plants, indicating a lower rate of transpiration (Figure 2A). In addition, 35S:ABF3 plants showed a mild growth retardation that became more evident as plants matured (Figure 2B, C), which is also consistent with previous results [7].
Figure 2.
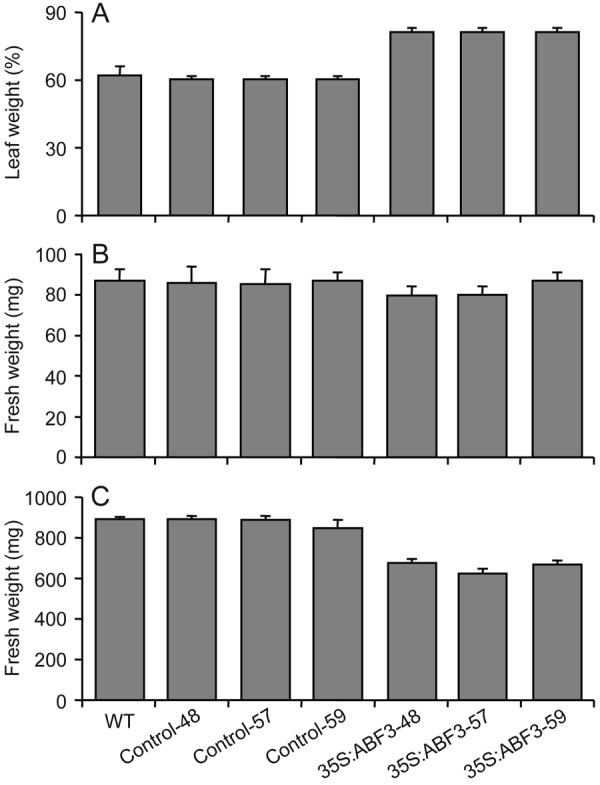
Phenotype of 35S:ABF3 plants. (A) Transpiration rate of leaves from 35S:ABF3, control, and wild-type plants. Leaves from 4-week-old plants were excised and left at room temperature for one day and the percentage of leaf weight remaining was measured (n = 5). For each measurement, five leaves were pooled. Error bars indicate standard deviation. (B, C) Growth retardation of 35S:ABF3 plants. Three-day-old seedlings were transplanted to fresh MS plates and fresh weight was measured after 1 (B) (n = 6) or 4 weeks (C) (n = 5) of growth. For each measurement, eight (B) or 5 (C) seedlings were pooled. Error bars indicate standard deviation.
Identification of position effects and impact of Cre recombinase on the transcriptome
Microarray analysis of control plant lines was performed to identify position effects as well as to determine if use of the Cre/lox system can cause unintended effects in plant systems. Position effects should be specific to each of the three control lines, as each line is expected to have a unique insertion site. In contrast, unintended effects resulting from excision at cryptic lox sites are more likely to affect independent plant lines similarly, since cryptic sites that may be present in the Arabidopsis genome will equally be found in each plant line.
Using a P-value cut-off of 0.05, only a small number of genes were differentially expressed in control lines compared to wild-type plants (Table 1). In the Control-48 line, only a single gene was differentially expressed. In the Control-57 and -59 lines, 10 and 4 genes were differentially expressed and two of these, rps7 (AtCg00900/AtCg01240) and rps12.1 (transplice part 1 of 2) (AtCg00065), were common to both lines. These genes could represent either position effects or natural background variation in gene expression. The rps7 and rps12.1 genes that were differentially expressed in both Control-57 and -59 lines may reflect unintended effects of the Cre/lox system.
Table 1.
Genes differentially expressed (P < 0.05) in control lines following Cre-mediated excision of the ABF3 transgene
| Line | Probe Set ID | AGI | Annotation | M | P |
|---|---|---|---|---|---|
| 48 | 267442_at | At2g19080 | Metaxin-related | 0.68 | 1.22 × 10-2 |
| 57 | 244939_at | AtCg00065 | Ribosomal protein s12 (Transplice part 1 of 2) (rps12.1)* | 2.58 | 9.11 × 10-4 |
| 244992_s_at | AtCg00900/AtCg01240 | Ribosomal protein s7 (rps7)* | 1.50 | 2.97 × 10-2 | |
| 245608_at | At4g14350 | Protein kinase family protein | -0.99 | 5.86 × 10-3 | |
| 247137_at | At5g66210 | Calcium-dependent protein kinase family protein (CDPK28) | -1.20 | 3.35 × 10-2 | |
| 247754_at | At5g59080 | Expressed protein | 1.33 | 2.20 × 10-2 | |
| 248812_at | At5g47330 | Palmitoyl protein thioesterase family protein | 2.34 | 7.24 × 10-3 | |
| 252053_at | At3g52400 | Syntaxin, putative (SYP122) | -1.09 | 3.60 × 10-2 | |
| 255655_at | At4g00980 | Zinc knuckle (CCHC-type) family protein | 1.39 | 4.60 × 10-6 | |
| 259072_at | At3g11700 | Fasciclin-like arabinogalactin protein 18 precursor (FLA18) | -0.73 | 2.29 × 10-3 | |
| 259348_at | At3g03770 | Leucine-rich repeat transmembrane protein kinase, putative | 0.91 | 1.82 × 10-2 | |
| 59 | 244939_at | AtCg00065 | Ribosomal protein s12 (Transplice part 1 of 2) (rps12.1)* | 2.50 | 1.52 × 10-3 |
| 244992_s_at | AtCg00900/AtCg01240 | Ribosomal protein s7 (rps7)* | 1.51 | 2.69 × 10-2 | |
| 246727_at | At5g28010 | Bet v I allergen family protein | 2.02 | 1.10 × 10-5 | |
| 262719_at | At1g43590 | Transposable element gene | 1.29 | 1.30 × 10-2 | |
M is log2 fold change and P is FDR-adjusted P-value for Student's t-test.
* indicates genes encoded by the chloroplast genome.
Effect of ABF3 overexpression on the transcriptome in the absence of drought
Microarray analysis of 35S:ABF3 plants was performed to identify unintended effects resulting from overexpression of a transcription factor in the absence of stress. Expression of ABF3 in the absence of stress is generally very low but it is rapidly induced in response to ABA [7,29]. Overexpression of ABF3 would therefore be expected to initiate those pathways that are typically activated in response to stress via ABA-mediated signalling. This has been observed for other transcription factors and is generally used as a means of identifying targets of that particular transcription factor [39,42,58].
Three independent plant lines containing the 35S:ABF3 transgene were compared to the corresponding control plant lines from which the 35S:ABF3 transgene was excised. Based on microarray analysis, using a cut-off of P < 0.05, only a small number of genes (7, 1, and 8) were differentially expressed in the three 35S:ABF3 transgenic plant lines (Table 2). The only gene that was differentially expressed in all three plant lines was ABF3.
Table 2.
Genes differentially expressed (P < 0.05) in 35S:ABF3 transgenic plants
| Line | Probe Set ID | AGI | Annotation | M | P |
|---|---|---|---|---|---|
| 48 | 244964_at | AtCg00580 | PSII cytochrome b559 (psbE)* | 0.73 | 1.18 × 10-2 |
| 245608_at | At4g14350 | Protein kinase family protein | -1.54 | 1.27 × 10-6 | |
| 253263_at | At4g34000 | ABA-responsive elements-binding factor 3 (ABF3) | 3.35 | 2.09 × 10-11 | |
| 253399_at | At4g32850 | Nuclear poly(A) polymerase (nPAP) | 0.7 | 2.22 × 10-2 | |
| 256683_at | At3g52220 | Expressed protein | -0.47 | 4.43 × 10-2 | |
| 256940_at | At3g30720 | Expressed protein | 1.68 | 3.25 × 10-2 | |
| 262719_at | At1g43590 | Transposable element gene | 2.02 | 3.12 × 10-6 | |
| 57 | 253263_at | At4g34000 | ABA-responsive elements-binding factor 3 (ABF3) | 4.44 | 2.58 × 10-14 |
| 59 | 244992_s_at | AtCg00900/AtCg01240 | Ribosomal protein s7 (rps7)* | -1.68 | 4.80 × 10-3 |
| 244996_at | AtCg00160 | Ribosomal protein s2 (rps2)* | -1.28 | 9.05 × 10-3 | |
| 245018_at | AtCg00520 | Protein required for photosystem I assembly and stability (ycf4)* | -1.22 | 9.22 × 10-3 | |
| 245019_at | AtCg00530 | Hypothethical protein (ycf10/cemA)* | -1.34 | 1.48 × 10-2 | |
| 245020_at | AtCg00540 | Cytochrome f apoprotein (petA)* | -1.21 | 1.25 × 10-2 | |
| 253263_at | At4g34000 | ABA-responsive elements-binding factor 3 (ABF3) | 2.04 | 1.35 × 10-6 | |
| 261888_at | At1g80800 | Pseudogene, 40S ribosomal protein S12 (rps12B) | -0.92 | 2.31 × 10-2 | |
| 264683_at | At1g65580 | Inositol or phosphatidylinositol phosphatase/FRAGILE FIBRE 3 (FRA3) | -0.71 | 2.34 × 10-2 | |
M is log2 fold change and P is FDR-adjusted P-value for Student's t-test.
* indicates genes encoded by the chloroplast genome.
Drought response of 35S:ABF3 plants
Overexpression of ABF3 in Arabidopsis confers enhanced drought tolerance [7]. Since ABF3 is a transcription factor, it is likely that this is achieved at the level of gene expression. Therefore, microarray analysis should provide insight into the mechanism of drought tolerance. Since the expression pattern of ABF3 during the drought response is achieved by constitutive overexpression from the CaMV 35S promoter, it is possible that unintended effects may also be generated downstream. By comparing the transcriptional profile of 35S:ABF3 plants to control plants, it may be possible to identify these unintended effects. Two time points were examined in order to consider both early and late responses to drought. For this analysis, the Control-48 and 35S:ABF3-48 lines were selected as the Control-48 lines showed the smallest number of differentially expressed genes (Table 1).
Following 2 h of drought stress, using a cut-off of P < 0.05 and a fold-change of 2, 508 genes were differentially expressed in the control plants exposed to drought stress compared to unstressed control plants and 468 genes were differentially expressed in 35S:ABF3 plants exposed to drought stress compared to unstressed 35S:ABF3 plants. Following 24 h of drought stress, 592 genes were differentially expressed in control plants exposed to drought stress compared to unstressed control plants and 797 genes were differentially expressed in 35S:ABF3 plants exposed to drought stress compared to unstressed 35S:ABF3 plants.
In order to compare the profile of genes expressed in the two plant lines in response to drought stress, functional categorizations of the differentially expressed genes were obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) and compared. Although the profile of genes differentially expressed in control and 35S:ABF3 plants show some differences, overall they show a similar pattern of distribution among the different functional classes (Additional file 2). The greatest difference between 35S:ABF3 and control plant lines in the percentage of genes belonging to a particular functional category was 1.3% for the 'other metabolic processes' category at the 2 h time point. This suggests that the overall functional response of 35S:ABF3 and control plant lines at the gene expression level was similar.
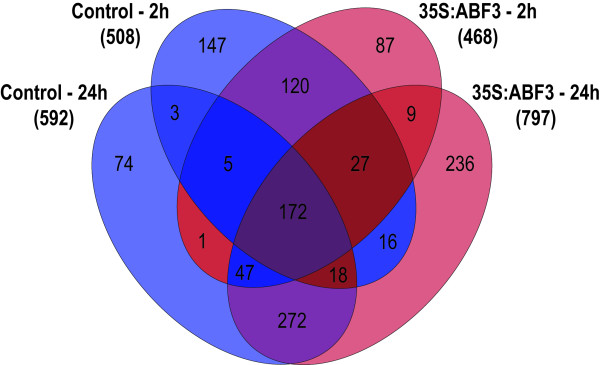
In total, 1234 genes were differentially expressed in at least one plant line during at least one time point. The overlap in genes expressed in the two plant lines at the two different time points is depicted with a four-way Venn diagram in Figure 3. These genes can be subdivided into three categories. There are 564 genes that were differentially expressed in both control and 35S:ABF3 lines at the same time points suggesting that they were commonly regulated in both lines. There are 407 genes that are regulated differently in control and 35S:ABF3 plant lines that show an enhanced response in the 35S:ABF3 line. Finally, there are 263 genes that are regulated differently in control and 35S:ABF3 plant lines that show an attenuated response in the 35S:ABF3 line. In the latter two categories, these genes are either uniquely differentially expressed in one line or the other, or they are differentially expressed in one line at one time point but not in the other line at that time point. Since only two time points were examined, it is difficult to determine if the observed differences in the two lines are due to differences in the timing of gene expression or in the magnitude of gene expression or some combination of both of these factors. Additional file 3 contains a list of all of the differentially expressed genes found in each of the three categories.
Figure 3.
Overlap of differentially expressed genes in 35S:ABF3 and control plants in response to drought. Four-way Venn diagram showing the overlap of differentially expressed genes in 35S:ABF3 and control plant lines in response to drought at 2 h and 24 h. Regions corresponding to genes that are regulated commonly in both plant lines are shaded in purple. Regions corresponding to genes that show enhanced regulation in 35S:ABF3 plants are shaded in red and those that show attenuated regulation in 35S:ABF3 plants are shaded in blue.
In order to confirm the microarray results, RT-PCR was performed on 32 genes from the enhanced and attenuated categories. Twenty-nine of the examined genes exhibited expression patterns that were consistent with the microarray results, confirming the reliability of the microarray data (Figure 4). Amplification of one gene, At2g22760, produced two bands, one corresponding to the expected size of 526 bp. The primers for this gene were designed around an intron and the size of the second band is similar to the 664 bp that would be expected if the intron were not spliced, suggesting that this band may represent a splice variant.
Figure 4.
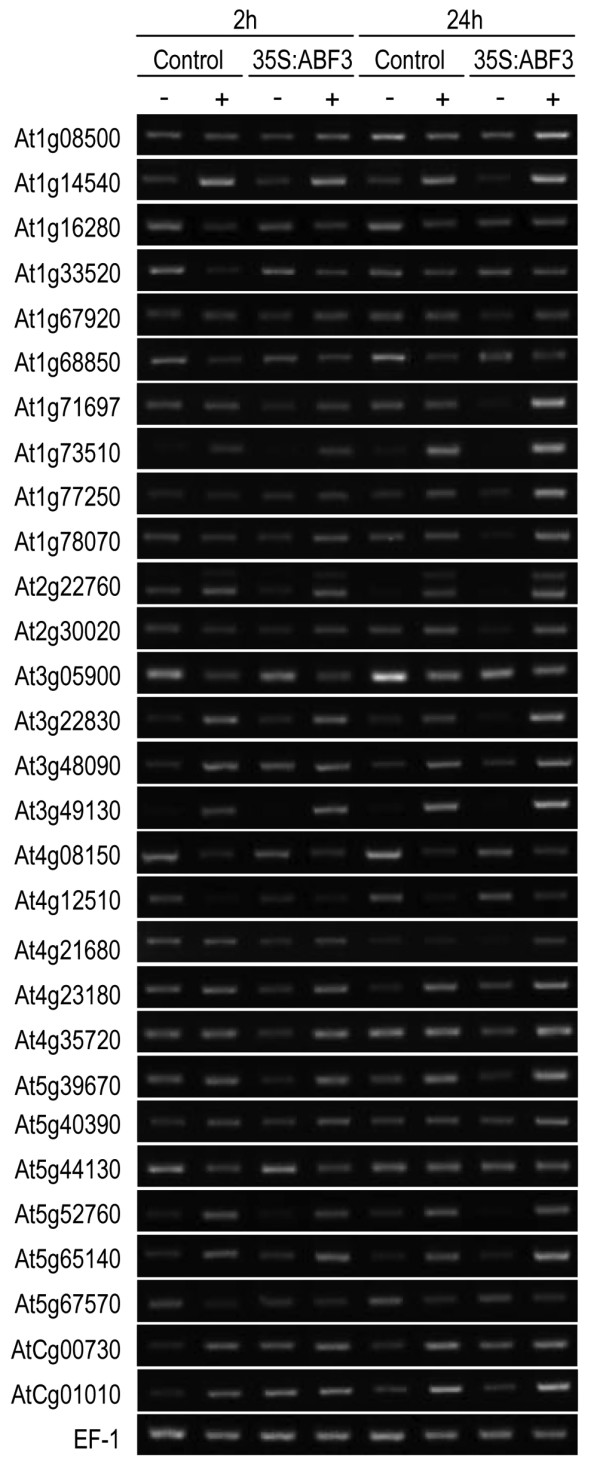
RT-PCR confirmation of microarray data. RT-PCR was performed using RNA from unstressed (-) or drought stressed (+) control and 35S:ABF3 plants at either the 2 h or 24 h time point. Genes showing either enhanced or attenuated regulation in 35S:ABF3 plants in response to drought were amplified using gene-specific primers. Elongation factor 1-α (EF-1) was amplified as an internal control.
Genes commonly regulated in both 35S:ABF3 and control lines
There are 564 genes that are commonly regulated in both 35S:ABF3 and control lines. Of these, 172 show differential expression in both lines at both time points, 120 are only differentially expressed at 2 h and 272 are only differentially expressed at 24 h.
A number of genes in this category are known to act in pathways that are upstream or independent of ABF3, which is consistent with their common pattern of regulation in both 35S:ABF3 and control plant lines (Table 3). Since ABF3 is an ABA-dependent transcription factor, ABA biosynthesis should occur upstream of ABF3 activity. Consistent with this, AtNCED3 (At3g14440), which encodes a 9-cis-epoxycarotenoid dioxygenase enzyme involved in ABA biosynthesis, was upregulated in both lines at both time points in response to drought treatment as was the gene CYP707A3 (At5g45340), which encodes a cytochrome P450 monooxygenase involved in ABA catabolism.
Table 3.
Genes commonly regulated in 35S:ABF3 and control plant lines that act in pathways upstream or independent from ABF3
| 2 h | 24 h | |||||
|---|---|---|---|---|---|---|
| Probe Set ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| ABA Metabolism | ||||||
| 257280_at | At3g14440 | AtNCED3 | 3.653 | 4.141 | 2.385 | 2.834 |
| 248964_at | At5g45340 | CYP707A3 | 2.007 | 2.192 | 3.177 | 3.399 |
| DREB Transcription Factors | ||||||
| 250781_at | At5g05410 | DREB2A | 3.945 | 4.672 | 4.755 | 4.602 |
| 256430_at | At3g11020 | DREB2B | 1.376 | 1.562 | 1.94 | 1.987 |
| NAC Transcription Factors | ||||||
| 260203_at | At1g52890 | ANAC019 | 3.945 | 4.409 | 4.264 | 4.608 |
| 258395_at | At3g15500 | ANAC055/AtNAC3 | 3.088 | 3.258 | 2.297 | 2.557 |
| 253872_at | At4g27410 | ANAC072/RD26 | 2.602 | 3.183 | 2.567 | 2.594 |
Values are log2 fold changes.
Several transcription factors have been identified that mediate drought response pathways independent from the ABF3 pathway. Members of the DREB family of transcription factors function in abiotic stress signalling and members of the DREB2 subfamily are known to function in ABA-independent drought stress signalling [59]. DREB2A (At5g05410) and DREB2B (At3g11020), as expected, were upregulated in response to drought in both plant lines at both time points (Table 3). In addition, DREB1B/CBF1 (At4g25490) and DREB1C/CBF2 (At4g25470) were also upregulated in both lines at both time points and DDF1 (At1g12610) was upregulated at 24 h (Additional file 3). While the DREB1 subfamily is primarily associated with cold stress signalling [59], there is some evidence that members also function in dehydration stress [8,38,60,61].
Three NAC transcription factors also function in a drought signalling pathway that is independent from ABF3 [42]. ANAC19 (At1g52890), ANAC055/ATNAC3 (At3g15500), and ANAC072/RD26 (At4g27410) are all expressed at both time points in both plant lines (Table 3). Another six members of the NAC family of transcription factors (At1g01720, At1g69490, At1g77450, At2g02450, At3g49530, and At5g63790) are also commonly regulated in both 35S:ABF3 and control plant lines (Additonal file 3).
Also in this category are many genes that are known to be regulated in response to drought stress. This includes genes involved in the biosynthesis of osmolytes, late embryogenesis abundant proteins (LEA), kinases, phosphatases, and transcription factors, as well as several different types of transporters (Table 4). This further suggests that some drought signalling pathways are unaffected by ABF3 overexpression
Table 4.
Examples of typical drought-responsive genes regulated similarly in 35S:ABF3 and control plant lines
| 2 h | 24 h | |||||
|---|---|---|---|---|---|---|
| Probe Set ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| Osmolyte metabolism | ||||||
| 251505_at | At3g59050 | Polyamine oxidase 3 | 1.365 | 1.409 | ||
| 266072_at | At2g18700 | Trehalose-phosphatase/synthase 11 (AtTPS11) | 1.173 | 1.290 | ||
| 254321_at | At4g22590 | Trehalose-6-phosphate phosphatase, putative | 2.262 | 2.652 | ||
| 250467_at | At5g10100 | Trehalose-6-phosphate phosphatase, putative | 1.716 | 1.560 | ||
| LEA class proteins | ||||||
| 250648_at | At5g06760 | LEA group 1 domain-containing protein | 2.281 | 2.823 | 2.537 | 3.013 |
| 262128_at | At1g52690 | LEA protein, putative | 3.087 | 3.667 | ||
| 267261_at | At2g23120 | Expressed protein | 1.651 | 1.688 | ||
| 266392_at | At2g41280 | LEA protein M10 | 2.258 | 2.473 | ||
| 252988_at | At4g38410 | Dehydrin, putative | 2.692 | 2.516 | ||
| Kinases | ||||||
| 252592_at | At3g45640 | MAPK, putative (MPK3) | 1.681 | 1.772 | 1.259 | 1.651 |
| 258682_at | At3g08720 | Serine/threonine protein kinase (PK19) | 1.160 | 1.383 | ||
| 250673_at | At5g07070 | CBL-interacting protein kinase 2 (CIPK2) | 1.408 | 1.261 | 1.551 | 1.827 |
| 254996_at | At4g10390 | Protein kinase family protein | 2.177 | 1.763 | ||
| 249361_at | At5g40540 | Protein kinase, putative | 2.768 | 3.066 | ||
| 248821_at | At5g47070 | Protein kinase, putative | 1.547 | 1.487 | ||
| Phosphatases | ||||||
| 247957_at | At5g57050 | Abscisic acid-insensitive 2 (ABI2) | 1.617 | 2.228 | ||
| 247723_at | At5g59220 | Protein phosphatase 2C, putative | 1.675 | 2.115 | ||
| 251259_at | At3g62260 | Protein phosphatase 2C, putative | 2.533 | 2.710 | 2.902 | 2.440 |
| 253323_at | At4g33920 | Protein phosphatase 2C family protein | 1.385 | 1.615 | 1.767 | 1.779 |
| Transcription factors | ||||||
| 250582_at | At5g07580 | Ethylene-responsive element-binding family protein | -1.878 | -1.668 | ||
| 265452_at | At2g46510 | Basic helix-loop-helix family protein | 1.918 | 2.460 | ||
| 247509_at | At5g62020 | Heat shock transcription factor B2A (AtHSFB2A) | 1.133 | 1.135 | 1.594 | 1.900 |
| 251272_at | At3g61890 | Homeobox-leucine zipper protein 12 (HB-12) | 2.421 | 2.536 | 1.914 | 2.554 |
| 260237_at | At1g74430 | Myb family transcription factor (MYB95) | 1.931 | 1.942 | ||
| 255753_at | At1g18570 | Myb family transcription factor (MYB51) | 2.127 | 1.409 | ||
| 254652_at | At4g18170 | WRKY family transcription factor (WRKY28) | 3.564 | 2.709 | ||
| 261648_at | At1g27730 | Zinc finger (C2H2 type) family protein (ZAT10) | 3.665 | 3.663 | 4.978 | 5.127 |
Values are log2 fold changes.
Genes with an enhanced response in 35S:ABF3 plants
There are 407 genes that demonstrated an enhanced response in 35S:ABF3 plants. Of these, 332 are uniquely regulated in 35S:ABF3 plants with 9 differentially expressed at both time points, 87 differentially expressed only at 2 h and 236 differentially expressed at 24 h. In addition, 47 genes are differentially expressed in the control line at only 24 h and 27 genes are differentially expressed in the control line at only 2 h while in the 35S:ABF3 line they are differentially expressed at both time points. Finally, there is a transposable element gene that is differentially expressed in 35S:ABF3 plants at 2 h but is only differentially expressed in control plants at 24 h.
A number of genes in this category could contribute to the enhanced drought tolerance of 35S:ABF3 plants (Table 5). Several genes involved in the biosynthesis of the osmolyte trehalose show enhanced upregulation while two genes involved in osmolyte catabolism show enhanced downregulation. Two genes involved in detoxification show enhanced upregulation in 35S:ABF3 as do three LEA proteins. A number of transporters also show enhanced regulation in 35S:ABF3 lines.
Table 5.
Genes showing an enhanced response in 35S:ABF3 plants that may contribute to drought-tolerance
| 2 h | 24 h | |||||
|---|---|---|---|---|---|---|
| Probe Set ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| Osmolyte metabolism | ||||||
| 248404_at | At5g51460 | Trehalose-6-phosphate phosphatase (TPPA) | 1.402 | |||
| 247228_at | At5g65140 | Trehalose-6-phosphate phosphatase, putative | 1.479 | 1.587 | 2.224 | |
| 263452_at | At2g22190 | Trehalose-6-phosphate phosphatase, putative | 1.250 | |||
| 254806_at | At4g12430 | Trehalose-6-phosphate phosphatase, putative | 1.146 | |||
| 252983_at | At4g37980 | Mannitol dehydrogenase, putative/elicitor activated gene 3 (ELI3-1) | -1.061 | |||
| 251729_at | At3g56310 | Alpha-galactosidase, putative | -1.160 | |||
| Detoxification | ||||||
| 266299_at | At2g29450 | Glutathione S-transferase (class tau) 5 (AtGSTU5) | 2.268 | |||
| 258665_at | At3g08710 | Thioredoxin H-type 9 (ATH9) | 1.188 | |||
| LEA class proteins | ||||||
| 259516_at | At1g20450 | Dehydrin/early response to dehydration 10 (ERD10) | 1.584 | |||
| 252102_at | At3g50970 | dehydrin/XERO2/low-temperature-induced protein | 2.800 | |||
| 265211_at | At2g36640 | LEAprotein/embryonic cell protein 63 (AtECP63) | 1.652 | |||
| Transporters | ||||||
| 249063_at | At5g44110 | ABC transporter family protein | 1.017 | 1.540 | 1.826 | |
| 263918_at | At2g36590 | Proline transporter 3 (ProT3) | 1.907 | 1.845 | ||
| 245868_at | At1g58030 | Cationic amino acid transporter 2 (CAT2) | -1.406 | |||
| 260543_at | At2g43330 | Inositol transporter 1 (AtINT1) | 1.118 | 0.160 | ||
| 262756_at | At1g16370 | Organic cation/carnitine transporter 6 (AtOCT6)/carbohydrate transmembrane transporte | 1.992 | |||
| 245499_at | At4g16480 | Inositol transporter 4 (AtINT4) | -1.031 | |||
| 254291_at | At4g23010 | UDP-galactose transporter 2 (AtUTR2) | 1.081 | |||
| 267423_at | At2g35060 | K+ uptake permease 11 (KUP11)/potassium transporter family protein | -1.202 | |||
| 249298_at | At5g41330 | Potassium channel tetramerisation domain-containing protein | -1.157 | |||
| 256402_at | At3g06130 | Heavy-metal-associated domain-containing protein | -1.363 | |||
| 247128_at | At5g66110 | Heavy-metal-associated domain-containing protein | -1.716 | |||
| 261143_at | At1g19770 | Purine permease 14 (AtPUP14); purine transmembrane transporter | 1.222 | 1.571 | 1.569 | |
| 262649_at | At1g14040 | EXS family protein/ERD1/XPR1/SYG1 family protein | 1.103 | |||
| 250151_at | At5g14570 | High affinity nitrate transporter 2.7 (AtNRT2.7) | -1.317 | |||
| 251916_at | At3g53960 | Proton-dependent oligopeptide transport (POT) family protein | 1.278 | 1.548 | ||
| 252589_s_at | At3g45650/At3g45660 | Proton-dependent oligopeptide transport (POT) family protein | 1.164 | 0.740 | ||
| 254396_at | At4g21680 | Proton-dependent oligopeptide transport (POT) family protein | 2.426 | |||
Values are log2 fold changes.
Direct targets of ABF3 are likely found in this category of genes. In addition to showing enhanced regulation in 35S:ABF3 plants, it is expected that these genes possess at least one ABRE in their promoter. In addition, these genes are likely to be significantly differentially expressed at the 2 h time point. Amongst those genes showing an enhanced response in 35S:ABF3 plants that are significantly differentially expressed at the 2 h time point, 24 contain at least one ABRE according to the in silico analysis performed by Gómez-Porras et al [62] and are therefore identified as putative ABF3 targets (Table 6). Included in this group are four transcription factors, one choline kinase, one trehalose biosynthetic enzyme, one gene involved in ubiquitin-mediated protein degradation, and two transporters as well as seven other genes with undefined roles in the drought response and eight unknown genes.
Table 6.
Potential targets of ABF3
| AGI | 2 h | 24 h | |||||
|---|---|---|---|---|---|---|---|
| Probe Set ID | Annotation | ABREs1 | Control | 35S:ABF3 | Control | 35S:ABF3 | |
| Osmolyte biosynthesis | |||||||
| 254806_at | At4g12430 | Trehalose-6-phosphate phosphatase, putative | 1 | 1.146 | |||
| Protein degradation | |||||||
| 262164_at | At1g78070 | WD-40 repeat family protein | 1 | 2.072 | 2.312 | 2.647 | |
| Signalling | |||||||
| 261506_at | At1g71697 | Choline kinase, putative (AtCK1) | 1 | 1.519 | 1.181 | 2.909 | |
| Transcription | |||||||
| 253259_at | At4g34410 | AP2 domain-containing transcription factor, putative | 2 | 3.865 | 5.126 | 4.940 | |
| 259432_at | At1g01520 | Myb family transcription factor | 3 | 2.367 | 2.762 | 3.247 | |
| 262098_at | At1g56170 | Nuclear factor Y, subunit C2 (NF-YC2) | 3 | 1.944 | |||
| 261892_at | At1g80840 | WRKY family transcription factor (WRKY40) | 2 | 3.762 | 4.724 | 5.138 | |
| Transporters | |||||||
| 260543_at | At2g43330 | Inositol transporter 1 (AtINT1) | 1 | 1.118 | |||
| 251916_at | At3g53960 | Proton-dependent oligopeptide transport (POT) family protein | 2 | 1.278 | 1.548 | ||
| Other | |||||||
| 251904_at | At3g54130 | Josephin family protein | 1 | 1.043 | 1.311 | ||
| 265634_at | At2g25530 | AFG1-like ATPase family protein | 2 | 1.399 | |||
| 252557_at | At3g45960 | Expansin family protein (EXPL3) | 1 | 1.467 | 1.850 | ||
| 253217_at | At4g34970 | Actin-depolymerizing factor, putative | 1 | -1.417 | -2.589 | -2.791 | |
| 262126_at | At1g59620 | Disease resistance protein (CC-NBS class), putative | 2 | 1.235 | |||
| 247246_at | At5g64620 | Invertase/pectin methylesterase inhibitor family protein | 1 | -1.125 | |||
| 246495_at | At5g16200 | 50S ribosomal protein-related | 1 | 1.193 | 2.080 | 2.162 | |
| Unknown | |||||||
| 261193_at | At1g32920 | Expressed protein | 1 | 1.541 | 1.793 | 2.457 | |
| 260227_at | At1g74450 | Expressed protein | 3 | 1.755 | 1.840 | 2.170 | |
| 253155_at | At4g35720 | Expressed protein | 3 | 2.671 | 2.010 | ||
| 261065_at | At1g07500 | Expressed protein | 2 | 1.201 | |||
| 256069_at | At1g13740 | Expressed protein | 5 | 2.090 | |||
| 260367_at | At1g69760 | Expressed protein | 1 | -1.278 | |||
| 260005_at | At1g67920 | Expressed protein | 1 | 2.251 | 2.051 | 2.825 | |
| 254356_at | At4g22190 | Expressed protein | 2 | -1.118 | -1.031 | -1.423 | |
Values are log2 fold changes.
1Number of ABA-responsive elements (ABREs) in the promoter region as determined by Gómez-Porras et al. [62]
Amongst those genes that show an enhanced response in 35S:ABF3 plants, there is an enrichment of genes that function in RNA processing pathways (Table 7). There are 8 genes involved in RNA processing that are similarly regulated in both control and 35S:ABF3 plants lines, 8 genes that show an attenuated response in 35S:ABF3, while 18 genes show an enhanced response. Many of these appear to be uniquely downregulated in 35S:ABF3 plants at 24 h. Eleven of these are predicted to function in RNA splicing or to be associated with the RNA splicing machinery while the others have roles in nucleocytoplasmic transport, deadenylation, or the function is not specifically known. Several genes that function in RNA processing have been found to function in ABA and abiotic stress signalling pathways [63-66]. This suggests that the differential regulation of RNA processing genes in 35S:ABF3 lines could contribute to its enhanced drought tolerance.
Table 7.
Genes showing enhanced regulation in 35S:ABF3 with predicted function in RNA processing
| 2 h | 24 h | |||||
|---|---|---|---|---|---|---|
| Probe Set ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| Splicing and splicing-related | ||||||
| 262110_at | At1g02840 | Pre-mRNA splicing factor SF2/SR1 protein | -1.731 | |||
| 263035_at | At1g23860 | Splicing factor RSZp21 (RSZP21) | -1.272 | |||
| 262295_at | At1g27650 | U2 snRNP auxiliary factor small subunit, putative (AtU2AF35A) | -1.129 | |||
| 262931_at | At1g65700 | Small nuclear ribonucleoprotein, putative | 1.386 | |||
| 267102_at | At2g41500 | LACHESIS (LIS)/related to yeast splicing factor PRP4 | -1.260 | |||
| 252182_at | At3g50670 | U1 small nuclear ribonucleoprotein 70 (U1-70k) | -1.525 | |||
| 251798_at | At3g55460 | SC35-like splicing factor, 30 kD (SCL30) | -1.048 | |||
| 253668_at | At4g30220 | Small nuclear ribonucleoprotein F (RUXF), putative | 1.057 | |||
| 249870_at | At5g23080 | SWAP domain-containing protein/TOUGH (TGH) | -1.476 | |||
| 246924_at | At5g25060 | RNA recognition motif (RRM)-containing protein | -1.043 | |||
| 248369_at | At5g52040 | Arginine/serine-rich splicing factor RSP41 (RSP41) | -1.009 | |||
| Nucleocytoplasmic transport | ||||||
| 257817_at | At3g25150 | Nuclear transport factor 2 (NTF2) family protein | -1.316 | |||
| Deadenylation | ||||||
| 252679_at | At3g44260 | CCR4-NOT transcription complex protein, putative | 2.108 | 3.479 | 3.354 | |
| Other | ||||||
| 262804_at | At1g20880 | RNA recognition motif (RRM)-containing protein | -1.335 | |||
| 261988_at | At1g33680 | KH domain-containing protein | -1.123 | |||
| 254355_at | At4g22380 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | 1.617 | |||
| 246088_at | At5g20600 | Expressed protein | -1.145 | |||
| 247004_at | At5g67570 | Pentatricopeptide (PPR) repeat-containing protein | -1.349 | |||
Values are log2 fold changes.
Interestingly, a member of the DREB family of transcription factors is also included in this group. DREB1D/CBF4 (At5g51990) was upregulated in 35S:ABF3 plants at both time points but was only upregulated in control plants at 24 h. DREB1D/CBF4 functions in both cold and drought stress signalling and, unlike the DREB2 subfamily, its expression is induced by ABA [38]. This suggests that CBF4 may act downstream of ABF3 in the ABA-dependent drought signalling pathway.
Genes with an attenuated response in 35S:ABF3 plants
There are 263 genes that show an attenuated response in 35S:ABF3 plants. Of these, 224 are uniquely regulated in control lines with 3 differentially expressed at both time points, 147 differentially expressed only at 2 h and 74 differentially expressed only at 24 h. In addition, 18 genes are differentially expressed in the 35S:ABF3 line at only 24 h and 5 genes are differentially expressed in the 35S:ABF3 line at only 2 h while in the control line they are differentially expressed at both time points. Finally, there are 16 genes that are differentially expressed in the control line at 2 h and in the 35S:ABF3 line at 24 h.
Surprisingly, a number of genes that typically function in conferring drought tolerance show an attenuated response in 35S:ABF3 plants. This includes several peroxidases and glutaredoxins involved in detoxification, two genes involved in biosynthesis of the osmolytes raffinose and spermine, a heat shock protein, and a number of transporters (Additional file 3). Similarly a number of transcription factors and other signalling components also show an attenuated response in 35S:ABF3 plants. The reason for the attenuated response of these genes in 35S:ABF3 plants is not clear but might reflect differences in the physiological state of 35S:ABF3 plants due to their enhanced drought resistance.
A number of genes encoded by the chloroplast and mitochondrial genomes show an attenuated response in 35S:ABF3 plants (Table 8). Some of these genes encode proteins with electron transport activity or NADH dehydrogenase activity while others are predicted to function in transcription and translation processes. Most of these genes are upregulated in control lines only at the 2 h time point. The reason for the exclusive upregulation of these genes in the control line is not clear. The chloroplast NADH dehydrogenase (NDH) complex is predicted to be involved in cyclic electron transport around photosystem I, thereby dissipating energy and maintaining ATP supply under conditions of low CO2 availability following stomatal closure in response to stress [67]. The NDH complex may also be involved in detoxification in the chloroplast [68]. The upregulation of genes encoding NADH dehydrogenases may therefore reflect the greater sensitivity of the control plant line to stress. Rice plants overexpressing ABF3 were able to maintain higher photochemical efficiency during drought stress [11]. This suggests that one aspect of the drought tolerance conferred by ABF3 overexpression may be a minimization of the negative impact of drought stress on photosynthesis, which is reflected by differences in gene expression in the chloroplast. Similar effects may also occur in the mitochondria.
Table 8.
Genes encoded by the chloroplast and mitochondrial genomes showing attenuated regulation in 35S:ABF3 plants
| Probe Set | 2 h | 24 h | ||||
|---|---|---|---|---|---|---|
| ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| Electron transport activity | ||||||
| 266045_s_at | At2g07727/AtMg00220 | Cytochrome b (MTCYB) (COB) (CYTB)/apocytochrome b (cob) † | 2.060 | |||
| 244903_at | AtMg00660 | Hypothetical protein (orf149)† | 1.645 | |||
| 244977_at | AtCg00730 | Subunit IV of cytochrome b6/f complex (petD)* | 2.993 | 3.118 | ||
| NADH dehydrogenases | ||||||
| 244943_at | AtMg00070 | NADH dehydrogenase subunit 9 (nad9)† | 1.338 | 1.416 | ||
| 257337_at | AtMg00060 | NADH dehydrogenase subunit 5 (nad5) (Transplice part 3 of 3)† | 1.413 | |||
| 244933_at | AtCg01070 | NADH dehydrogenase ND4L (ndhE)* | 1.733 | |||
| 244994_at | AtCg01010 | Chloroplast encoded NADH dehydrogenase unit (ndhF)* | 3.240 | |||
| 244934_at | AtCg01080 | NADH dehydrogenase ND6 (ndhG)* | 2.465 | |||
| 244991_s_at | AtCg00890/AtCg01250 | NADH dehydrogenase ND2 (ndhB)* | 3.528 | |||
| RNA processing | ||||||
| 244999_at | AtCg00190 | RNA polymerase subunit beta (rpoB)* | 2.792 | |||
| Translation | ||||||
| 245005_at | AtCg00330 | Chloroplast ribosomal protein s14 (rps14)* | 3.796 | 3.644 | ||
| 244970_at | AtCg00660 | Ribosomal protein L20 (rpl20)* | 2.379 | |||
| 244939_at | AtCg00065 | Ribosomal protein s12 (Transplice part 1 of 2) (rps12.1)* | 1.653 | |||
| ATPase subunits | ||||||
| 244995_at | AtCg00150 | Subunit of ATPase complex CF0 (atpI)* | 2.049 | |||
| 266012_s_at | AtMg00410/AtMg01170/At2g07699/At2g07741 | ATPase subunit 6† | 1.924 | |||
| Other | ||||||
| 257319_at | AtMg01100 | Hypothetical protein (orf105a)† | 1.181 | |||
| 244989_s_at | AtCg00860/AtCg01280 | Expressed protein (ycf2)* | 3.493 | |||
| 245008_at | AtCg00360 | Protein required for photosystem I assembly and stability (ycf3)* | 2.371 | |||
| 244990_s_at | AtCg00870/AtCg01270 | Hypothetical protein (ycf15/orf77)* | 3.607 | 2.841 | ||
| 245016_at | AtCg00500 | Acetyl-CoA carboxylase carboxyl transferase subunit beta (accD)* | 1.452 | |||
| 266014_s_at | At2g07722/AtMg00170/AtMg00620 | Hypothetical protein† | 1.987 | |||
Values are log2 fold changes.
* indicates gene is encoded by the chloroplast genome.
† indicates gene is encoded by the mitochondrial genome.
Four transposable element genes were also included in the attenuated response category (Table 9). Only one other transposable element gene was significantly differentially expressed and it was included in the enhanced regulation category. Many transposable elements are transcriptionally activated in response to stress conditions [69,70]. The attenuated regulation of transposable element genes in 35S:ABF3 plants might suggest that they are experiencing a lower level of stress that is insufficient to activate the transposable element genes, consistent with the drought tolerance of these plants.
Table 9.
Transposable element genes showing attenuated regulation in 35S:ABF3 plants
| Probe Set | 2 h | 24 h | ||||
|---|---|---|---|---|---|---|
| ID | AGI | Annotation | Control | 35S:ABF3 | Control | 35S:ABF3 |
| Transposable element genes | ||||||
| 265709_at | At2g03540 | Transposable element gene | 1.305 | |||
| 263769_at | At2g06390 | Transposable element gene | 1.511 | |||
| 257777_x_at | At3g29210 | Transposable element gene | 1.117 | |||
| 257345_s_at | At3g33066 | Transposable element gene; gypsy-like retrotransposon family (Athila) | 2.379 | |||
Values are log2 fold changes.
Interestingly, DREB1A/CBF3 showed an attenuated response in 35S:ABF3 plants. DREB1A/CBF3 was upregulated in control plant lines at both time points but was only upregulated in 35S:ABF3 plants at 2 h. This may again reflect differences in the tolerance of 35S:ABF3 and control plants to the drought stress, with control plants requiring a stronger or longer activation of DREB1A/CBF3 expression in response to the increased stress.
Discussion
The Impact of Cre recombinase on the transcriptome is minimal
In order to eliminate position effects and focus on unintended pleiotropic effects of transcription factor overexpression, the Cre/lox recombination system was employed to create a series of control plant lines that contain the selectable marker at the site of transgene insertion but from which the ABF3 transgene was excised. The use of the Cre/lox recombination system also allowed us to determine the impact of Cre recombinase on the transcriptome.
In tomato, petunia, tobacco, and to a lesser extent Arabidopsis, expression of Cre recombinase has resulted in abnormal phenotypes, including leaf chlorosis, stunted growth, and sterility [49,50]. Similarly, expression of Cre recombinase resulted in reduced proliferation and chromosomal abnormalities in cultured embryonic mouse cells [55,57], toxicity in dividing cells of Drosophila melanogaster [54], and chromosomal rearrangements in mouse spermatids leading to male sterility [56]. These abnormal effects of Cre recombinase are suspected to result from Cre-mediated recombination using cryptic lox sites that may be found in eukaryotic genomes. Cryptic lox sites that can be recognized by Cre recombinase have been identified in the genomes of yeast and humans as well as the chloroplast genome of tobacco [52,53,71,72].
If cryptic lox sites exist in plant genomes, expression of Cre recombinase could induce deletions or inversions of genome segments or even chromosome translocations that could adversely impact the plant. These deletions or inversions may alter the expression of genes found within this segment, which would be detectable by microarray. In three independent plants lines in which Cre recombinase was employed to excise the ABF3 transgene from the T-DNA insertion, only a small number of genes were found to be differentially expressed (Table 1). Of these, two genes were found to be differentially expressed in two out of the three control lines. The two genes are the chloroplast encoded rps7 (AtCg00900 and AtCg01240) and rps12.1 (AtCg00065) genes. In tobacco, one of the cryptic lox sites identified in the chloroplast genome is found just downstream of the start site of the rps12.2/rps7 operon [52,53]. The altered expression of rps7 might suggest that this cryptic lox site is conserved in Arabidopsis and may have undergone Cre-mediated recombination. Cre recombinase may therefore also be able to act on cryptic lox sites in the Arabidopsis chloroplast genome, resulting in a change in expression of the affected genes. It is, however, unclear how Cre recombinase is targeted to the chloroplast since it is only predicted to be targeted to the nucleus. Cre-mediated chloroplast genome deletions are not likely to be of great concern since chloroplast genomes containing deletions in essential genes are typically rapidly lost due to selection pressures [73-75], especially once the Cre recombinase has been removed. The impact of Cre recombinase on the nuclear transcriptome was negligible, which demonstrates that in Arabidopsis this technology does not produce unintended effects.
The activity of ABF3 is strictly controlled
Microarray analysis of Arabidopsis plants overexpressing the transcription factor ABF3 suggests that alterations to the transcriptome are minimal when position effects are eliminated as a source of variation. In the absence of stress, a small number of genes were differentially expressed in three 35S:ABF3 plant lines (Table 2), but no genes were differentially expressed in more than one independent line.
Members of the ABF/AREB family of transcription factors bind to ABREs found in the promoters of ABA-responsive genes [29,31]. If the genes identified by microarray analysis are actual downstream targets of ABF3, they would be expected to contain at least one ABRE. An in silico analysis of the Arabidopsis nuclear genome has identified 3829 genes containing one or more ABREs [62]. None of the nuclear genes identified by microarray analysis of 35S:ABF3 transgenic plants are predicted to contain an ABRE. Other members of the ABF/AREB subfamily of transcription factors localize to the nucleus [76,77], and it is likely that ABF3 similarly functions in the nucleus. Therefore, it is unlikely that the chloroplast genes identified by microarray analysis are functional targets of ABF3. This suggests that overexpression of ABF3 alone is not sufficient to alter the transcriptome.
This result was unexpected as previous work identified a number of genes with altered expression in Arabidopsis [7] and rice [11] overexpressing ABF3. Similarly, overexpression studies of many other transcription factors have revealed alterations in gene expression and this approach is typically used to identify the gene network controlled by that particular transcription factor [39,42,58]. The absence of differentially expressed genes in 35S:ABF3 transgenic plants suggests that an additional signal is required to activate ABF3 that is not present in unstressed plants.
There is accumulating evidence that members of the ABF/AREB family of transcription factors are regulated by phosphorylation. ABF2/AREB1 transactivation of a reporter gene in the presence of ABA was inhibited by the addition of the protein kinase inhibitor staurosporine [31] and ABI5 is phosphorylated following ABA treatment [78]. Several studies have suggested a role for members of the SnRK2 family of protein kinases in the phosphorylation of ABF/AREB transcription factors [79-84]. ABF3 and ABF4/AREB2 interact with the calcium-dependent protein kinase AtCPK32 and evidence suggests that it phosphorylates a highly conserved serine residue in ABF4/AREB2 that is necessary for activity [85]. The protein kinases CPK4 and CPK11 are also likely to phosphorylate ABF1 and ABF4/AREB2 and their activity is enhanced by ABA [86]. It is possible that in the absence of stress, ABF3 is not phosphorylated and therefore cannot activate gene expression.
Furthermore, other factors necessary for the activity of ABF3 may not be expressed in the absence of abiotic stress. Members of the ABF/AREB family have been shown to interact with diverse proteins that are predicted to modulate their transcriptional activity. ABF2/AREB1 interacts with an arm-repeat protein that is predicted to positively regulate its activity [87]. ABI5, ABF1, ABF3, and ABF4/AREB2 can interact with the transcription factor ABI3 [88,89]. Furthermore, the rd29a promoter contains both an ABRE as well as a dehydration-responsive elements that is bound by members of the DREB/CBF family of transcription factors and the two elements function interdependently to activate expression of rd29a [90]. Members of the ABF/AREB can also heterodimerize [91], suggesting that other members of this family may need to be expressed in order for ABF3 to be functional. Therefore, it is possible that other components of the stress response pathway are necessary in order for ABF3 to be active, preventing ABF3 from altering gene expression in the absence of stress.
While the 35S:ABF3 plants did not show any changes in transcription in the absence of drought stress, there were some phenotypic differences compared to control plants. Most notably, the 35S:ABF3 plants were smaller in size than control plants of the same age, with the difference becoming more pronounced with increased age (Figure 2B, C). This is a common observation for plants overexpressing transcription factors [7,33,39,60] and is often overcome by using tissue-specific or inducible promoters [32,60]. At least some of the growth retardation may be attributable to reduced transpiration rates of 35S:ABF3 plants compared to control plants (Figure 2A), which is consistent with the observation that Arabidopsis plants overexpressing ABF3 typically have stomata with smaller openings than do wild-type plants [7]. This would suggest that ABF3 may govern gene networks involved in stomatal closure. Consistent with this, ABF3 is expressed in guard cells and its expression is further induced in these cells in response to ABA [7,92]. Since our analysis was performed on whole plants, it is likely that changes in the transcriptional network of guard cells would not be readily detectable.
Overexpression of ABF3 results in transcriptional reprogramming of the drought response
Overexpression of ABF3 confers drought tolerance to Arabidopsis plants and since ABF3 is a transcription factor, it can be predicted that this will occur through changes to the transcriptional network of the plants. Consistent with this, the expression profile of Arabidopsis plants overexpressing ABF3 differed from that of control plants. As might be expected, there were a number of genes with expression patterns that appeared to be enhanced in 35S:ABF3 plants compared to control plants. Whether this occurred through alterations in the timing or strength of expression could not be established with the two time points considered in this study. Many of the genes with enhanced expression are known to function in mitigating drought stress, suggesting that they could contribute incrementally to the enhanced drought tolerance of 35S:ABF3 plants (Table 5). Those genes showing enhanced regulation in 35S:ABF3 plants likely include some direct targets of ABF3. In particular, those genes that are differentially expressed at 2 h and contain at least one ABRE are the most likely targets of ABF3 (Table 6).
Interestingly, there seemed to be a number of genes involved in RNA processing that showed enhanced expression in 35S:ABF3 plant lines. Most of these were downregulated at the 24 h time point. A number of RNA processing mutants impaired in ABA response have demonstrated the importance of RNA processing to ABA signalling [93]. Loss-of-function mutations in two genes encoding subunits of a nuclear cap-binding complex cause ABA hypersensitivity [63,66] as do mutations in a pre-mRNA splicing factor that is important for both mRNA splicing and turnover [94], a poly(A)-specific ribonuclease that is predicted to function in mRNA degradation [65], and a protein with homology to the Sm-like small nuclear ribonucleoproteins (snRNPs) that function in mRNA splicing, export, and degradation [95]. Two phosphatases that belong to the family of proteins that dephosphorylate the C-terminal domain of RNA polymerase II negatively regulate stress-responsive gene transcription [96] and a mutation in one of these results in decreased ABA sensitivity [97]. The enrichment of genes involved in RNA processing that show enhanced expression in the 35S:ABF3 line might suggest that ABF3 plays an important role in the regulation of ABA-responsive RNA processing events. The downregulation of many of the RNA processing genes is consistent with the negative regulatory role observed for several of the RNA processing proteins previously identified in the ABA-signalling pathways.
In addition to those genes showing enhanced regulation in 35S:ABF3 plants, there were also a number of genes with attenuated expression. Many of these are known to encode proteins with roles in minimizing drought stress and their attenuated expression is not consistent with the drought tolerance of the 35S:ABF3 plants. These genes may be reflective of a greater transcriptional reprogramming in 35S:ABF3 plants than merely enhancing the rate or level of expression of a subset of genes. The strong activation of the ABF3 pathway may result in co-ordinated feedback that modulates other drought responsive pathways, resulting in attenuated gene expression in some cases. This might reflect cooperativity between some of the drought signalling pathways. In many cases, drought-responsive transcription factors have been shown to function in concert to activate gene expression [39,40,43,90]. Furthermore, it has been observed that downregulation of the phosphoinositide pathway in Arabidopsis results in an upregulation of the DREB2A gene as well as several DREB2A-regulated genes, suggesting a negative interaction between two drought signalling pathways [98].
The degree of stress experienced by the control plants should be greater than that experienced by the 35S:ABF3 plants and this may ultimately have secondary consequences on the transcriptional network that is reflected by the genes showing an attenuated transcriptional response. This is consistent with the observation that several transposable element genes show attenuated expression in 35S:ABF3 plants (Table 9). Many transposons are activated in response to stress [69,70] and their delayed activation in 35S:ABF3 plants could be a result of the drought tolerance of these plants.
Similarly, the enrichment of genes encoded by the chloroplast and mitochondrial genomes in the attenuated category (Table 8) may also reflect the greater drought sensitivity of the control plant lines. Drought has a significant and complex impact on the activities of both the mitochondria and chloroplasts and this is reflected in changes in gene expression [99,100]. Several of the chloroplast and mitochondria-encoded genes showing attenuated expression encode NADH dehydrogenases. The chloroplast NDH complex is predicted to function in cyclic electron transport around photosystem I to help dissipate excess energy during abiotic stresses such as drought and thereby to alleviate oxidative stress [67]. The elevated expression of chloroplast NDH complex subunits and other chloroplast and mitochondria-encoded genes in control plants may therefore be indicative of the increased stress of these plants.
Drought stress can change the composition of a plant, altering levels of oils, proteins and other constituents, which can affect the commercial and nutritional value of the plant [101-107]. In some cases, drought can also initiate the accumulation of higher levels of dangerous toxins and anti-nutrients [102,105,108]. The altered expression of transposable element genes and genes that are encoded by the chloroplast and mitochondrial genomes in control plants suggests that these plants are experiencing a higher level of drought stress than 35S:ABF3 plants. It is therefore possible that control plants exposed to drought will exhibit more compositional changes compared to unstressed plants than 35S:ABF3 plants. This is an important consideration because compositional analysis is one of the parameters used to determine the substantial equivalence of transgenic plants to their non-transgenic comparators during the risk assessment process. These results might suggest that 35S:ABF3 plants are better able to maintain compositional standards under drought stress than their non-transgenic counterparts.
Overexpression of ABF3 does not activate unintended gene networks during the drought response
While differences were observed in the patterns of gene expression of 35S:ABF3 and control plant lines, at the functional level the response was very similar. Functional categorization of the significantly differentially expressed genes demonstrated that the percentage of genes in each category was relatively similar between the two plant lines (Additional file 2). This suggests that overexpression of ABF3 does not activate new gene networks but simply functions to modify existing gene networks that function in drought response. Although there were genes that were uniquely regulated in each of the two plant lines, many of these genes did observe a change in expression in the other plant line that was not of a high enough magnitude to meet our criteria for differential expression (Additional file 3). This suggests that these genes are showing stronger and/or earlier differential regulation as opposed to being uniquely regulated.
Among the genes that were differentially regulated between the 35S:ABF3 and control plant lines, there was no indication that overexpression of ABF3 activated any unintended gene networks. Several closely related members of the ABF/AREB transcription factor family function in seed germination and seed and early seedling developmental pathways, including ABI5 whose role in these pathways has been best characterized [109-111]. Although ABF3 is primarily associated with abiotic stress signalling in vegetative tissues, there is some evidence that it may also function in seed and early seedling developmental processes, although its role may be relatively minor. Microarray data from GENEVESTIGATOR [112] indicates that ABF3 is expressed during seed development, although at low levels compared to ABI5. Also, levels of ABF3 are enhanced by dehydration and salinity stresses or by ABA treatment in germinating embryos [113] and an alternative splice form of the ABF3 gene was identified from a cDNA library prepared from immature seed [91]. Double mutant analysis has also revealed several redundancies between ABF3 and ABI5, including sensitivity to ABA during germination, stress sensitivity of root growth, resistance to glucose, and regulation of the ABA-induced vegetative expression of RAB18 and RD29B [88]. ABI5 and ABF3 also appear to antagonistically cross-regulate each other [88,114].
If ABF3 does function in seed developmental pathways, it is possible that altering its pattern of expression could ectopically activate gene networks involved in those pathways. Another member of the ABF/AREB family of transcription factors, ABF2/AREB1, which functions in abiotic stress signalling and glucose response, is also expressed in embryonic axes in dry siliques [33,77]. Overexpression of a phosphorylated active form of ABF2/AREB1 led to the activation of several seed storage protein genes in vegetative tissues, many of which also have binding sites for ABI3, another transcription factor involved in seed development [81]. However, none of the seed storage genes activated by the phosphorylated active form of ABF2/AREB1 were differentially expressed in the 35S:ABF3 plants, nor were AtEm1 and AtEm6, two LEA-class genes that are ABI5 targets expressed during seed maturation (data not shown). This further demonstrates that overexpression of ABF3, while modifying patterns of gene expression, did not activate unintended pathways in response to drought stress in Arabidopsis. This suggests that overexpression of a transcription factor to confer an abiotic stress tolerance trait may not necessarily produce unintended pleiotropic effects.
Conclusions
The Cre/lox recombination system allowed us to create paired plant lines with identical T-DNA insertion sites either with or without the ABF3 transgene in order to eliminate position effects in our analysis of unintended effects. This approach also allowed us to examine unintended effects resulting from the expression of Cre recombinase. We found that Cre recombinase had a minimal impact on the transcriptome, which suggests that it produces few unintended effects in Arabidopsis. Microarray analysis of Arabidopsis plants overexpressing ABF3 demonstrated that the impact on the transcriptome is minimal. In the absence of drought stress, there were no differentially expressed genes. In response to drought stress, a reprogramming of the drought response was observed, suggestive of changes in the timing or strength of expression of some genes in 35S:ABF3 plants. Some of these changes may be directly related to the action of ABF3 while others may reflect an altered physiological state as a result of the enhanced drought tolerance of 35S:ABF3 plants. Amongst the differentially expressed genes, no unintended pathways appeared to be activated as a result of ABF3 overexpression. These results are significant because they demonstrate that plant responses to abiotic stresses such as drought may be strictly coordinated at multiple regulatory steps and this limits the extent of unintended pleiotropic effects. This demonstrates that engineering stress tolerance through manipulation of endogenous plant pathways may not necessarily produce unintended pleiotropic effects despite the complexity of such traits. This is an important finding for establishing the safety of such traits as they begin to enter the market in the near future.
Methods
Generation of transgenic plants
To generate 35S:ABF3 plant lines, Arabidopsis thaliana Col-0 plants were transformed by the floral dip method [115] using Agrobacterium tumefaciens strain GV3101 harbouring the pCAMBIA3300 vector containing the Cauliflower mosaic virus 35S promoter, ABF3 coding region, and nopaline synthase transcriptional terminator, bordered on either side loxP sites. Homozygous single insert transformants were identified by Southern blot and segregation analysis and three lines with high levels of ABF3 expression, as determined by RT-PCR analysis, were selected for further analysis.
To generate plant lines expressing Cre recombinase, Arabidopsis thaliana Col-0 plants were transformed as above with a pCAMBIA1200 vector containing the Cauliflower Mosaic Virus 35S promoter, Cre recombinase coding region, and nopaline synthase transcriptional terminator.
To generate control plant lines, 35S:ABF3 plants were crossed with plants expressing Cre recombinase in order to excise the 35S-ABF3-nos transgene. The F1 plants were PCR genotyped to identify those that underwent successful excision, using the primers ABF-F (5'-ATGGGGTCTAGATTAAAC-3') and Nos-R (5'-CCGATCTAGTAACATAGATG-3'), which are specific for the ABF3 gene and the nopaline synthase transcriptional terminator, respectively. One plant from each cross, Control-48; Cre 1.1.2, Control-57; Cre 1.2.2 and Control-59; Cre 2.2.1, was selected from which loss of the 35S-ABF3-nos transgene was confirmed by PCR (Additional File 1). F2 progeny from the selected F1 plants were then backcrossed to wild-type plants to eliminate Cre recombinase. The F1 plants from this cross were again PCR genotyped to identify those from which the Cre gene was lost, using primers CRE.F (5'-CCAGGCGTTTTCTGAGCATACCTG-3') and CRE.R (5'-CTCTGACCAGAGTCATCCTTAGCG-3'), which are both specific for the Cre recombinase gene. One plant from each cross, Control-48 1.1.2.5, Control-57 1.2.2.5 and Control-59 2.2.1.4, was selected from which loss of the Cre recombinase gene was confirmed by PCR (Additional File 1). The selected F1 plants represent the control plant lines and the F2 progeny of these plants were used in subsequent experiments.
Growth of Arabidopsis plants
Seeds were surface sterilized by soaking for 20 min in 25% (v/v) commercial Clorox (final concentration of 1.3% sodium hypochlorite) and 0.05% (v/v) Triton X-100 (Fisher Scientific, Hampton, NH, USA), and then rinsing four times with distilled water. Seeds were germinated on MS media and grown at 22°C with a 16 h photoperiod. To determine the growth rate of plants, three-day-old seedlings were transplanted onto fresh MS plates and after one week or four weeks of growth the fresh weight of the seedlings was measured. To measure transpiration rate, leaves of a similar developmental stage were excised from four-week-old plants and the loss of weight over a 24 h-period was measured. For microarray analysis, seeds were germinated on MS media and one-week-old seedlings were collected. For the drought stress, seedlings were transferred onto paper towels and harvested after 2 and 24 h.
Microarray Analysis
For each sample, three biological replicates were prepared. RNA was extracted from seedlings using the RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA), following the manufacturer's protocol. A 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA, USA) was used to determine the quality of the RNA. Microarray analysis was performed using the GeneChip® Arabidopsis ATH1 Genome Array (Affymetrix Inc., Santa Clara, CA, USA). Standard RNA processing, hybridization, and scanning protocols were followed as recommended by the GeneChip® Expression Analysis Technical Manual (Affymetrix Inc., Santa Clara, CA, USA). Hybridization, and scanning were performed at Agriculture and Agri-Food Canada (Winnipeg, MB, Canada) by Mark Jordan. RMA procedure [116] was performed to normalize data using the AffylmGUI R software package from Bioconductor (http://www.bioconductor.org/)[117]. Analysis of differential expression was done using a moderated t-test with empirical Bayes smoothing [118]. Microarray data from this study have been deposited at ArrayExpress (accession number E-MEXP-2435).
Reverse transcription-PCR (RT-PCR)
DNaseI-treated RNA was used for first strand cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo (dT)18 primers according to the manufacturer's protocol. Between 24 and 30 cycles of PCR amplification was performed using gene-specific primers. As an internal control, elongation factor 1-α (EF-1) was amplified. Sequences of all primers used in RT-PCR analysis can be found in Additional file 4.
List of Abbreviations
ABF: ABA-responsive element binding factor; AREB: ABA-responsive element binding protein; ABA: abscisic acid; ABRE: ABA-responsive element; DREB: dehydration responsive element binding protein; CBF: C-repeat binding factor; NAC: NAM, ATAF1,2, CUC2; ZFHD: zinc finger homeodomain; CaMV 35S promoter: Cauliflower Mosaic Virus 35S promoter; RT-PCR: reverse transcription polymerase chain reaction; LEA proteins: late embryogenesis abundant proteins; NDH complex: NADH dehydrogenase complex; ABI: ABA-insensitive.
Authors' contributions
AA participated in the design of the study and carried out the vector construction, plant transformation and molecular analysis, microarray analysis, and contributed to the drafting of the manuscript. JS participated in the final microarray analysis, carried out the RT-PCR work, and drafted the manuscript. BM conceived of the study and participated in its design and coordination, and participated in the drafting of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
PCR genotyping of control plant lines. (A) Control plant lines, derived from 35S:ABF3 plants crossed with plants expressing Cre recombinase, were PCR genotyped to identify plants from which the 35S-ABF3-nos transgene was excised. (B) Control plant lines following a backcross with Col-0 plants were genotyped to identify plants that lost the Cre gene by genetic segregation. Plant lines with the desired genotype that were selected following each cross are indicated with asterisks.
Functional categorization of drought responsive genes in 35S:ABF3 and control plants. Distribution of genes differentially expressed in 35S:ABF3 and control plant lines at 2 h and 24 h following drought stress into functional categories.
Genes differentially expressed in response to drought in 35S:ABF3 and control plants. This table includes a complete list of genes differentially expressed in both 35S:ABF3 and control plant lines in response to drought at 2 h and 24 h. Genes are divided into three groups including those that are similarly regulated in both 35S:ABF3 and control plants, those that show enhanced regulation in 35S:ABF3 plants, and those that show attenuated regulation in 35S:ABF3 plants.
Sequences of primers used in RT-PCR analysis. This table includes sequences of primers used to amplify selected genes in the RT-PCR analysis performed to confirm microarray results.
Contributor Information
Ashraf Abdeen, Email: ashraf.abdeen@mail.mcgill.ca.
Jaimie Schnell, Email: Jaimie.Schnell@inspection.gc.ca.
Brian Miki, Email: brian.miki@agr.gc.ca.
Acknowledgements
We would like to thank Hélène Labbé for her assistance in preparing RNA samples for microarray hybridization. Probe synthesis, hybridization, and scanning were performed by Mark Jordan at Agriculture and Agri-Food Canada (Winnipeg, MB, Canada). The gene encoding Cre recombinase was provided by Teresa Martin. This research was supported by funding from the Plant Biosafety Office, Feeds Section, and the Plant and Biotechnology Risk Assessment Unit of the Canadian Food Inspection Agency. Both AA and JS were supported by Visiting Fellowships in Canadian Government Laboratories granted by NSERC.
References
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Burke EJ, Brown SJ, Christidis N. Modeling the recent evolution of global drought and projections for the twenty-first century with the Hadley Centre climate model. J Hydrometeorology. 2006;7:1113–1125. doi: 10.1175/JHM544.1. [DOI] [Google Scholar]
- Meehl GA, Washington WM, Santer BD, Collins WD, Arblaster JM, Hu A, Lawrence DM, Teng H, Buja LE, Strand WG. Climate change projections for the twenty-first century and climate change commitment in the CCSM3. J Climate. 2006;19:2597–2616. doi: 10.1175/JCLI3746.1. [DOI] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008;147:20–29. doi: 10.1104/pp.108.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J, Herrera-Estrella LR. Plant biotechnology: Fast-forward genomics for improved crop production. Curr Opin Plant Biol. 2006;9:177–179. doi: 10.1016/j.pbi.2006.01.018. [DOI] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36:17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG. et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicov I, Bastola DR. Transgenic overexpression of the transcription factor alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol. 1999;120:473–480. doi: 10.1104/pp.120.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki B, Abdeen A, Manabe Y, MacDonald P. Selectable marker genes and unintended changes to the plant transcriptome. Plant Biotechnol J. 2009;7:211–218. doi: 10.1111/j.1467-7652.2009.00400.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Gerats T, Vandenbussche M. Evolutionary complexity of MADS complexes. Curr Opin Plant Biol. 2007;10:32–38. doi: 10.1016/j.pbi.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Abdeen A, Miki B. The pleiotropic effects of the bar gene and glufosinate on the Arabidopsis transcriptome. Plant Biotechnol J. 2009;7:266–282. doi: 10.1111/j.1467-7652.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, Beale MH. A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol J. 2006;4:381–392. doi: 10.1111/j.1467-7652.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Baudo MM, Lyons R, Powers S, Pastori GM, Edwards KJ, Holdsworth MJ, Shewry PR. Transgenesis has less impact on the transcriptome of wheat grain than conventional breeding. Plant Biotechnol J. 2006;4:369–380. doi: 10.1111/j.1467-7652.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Albo AG, Mila S, Digilio G, Motto M, Aime S, Corpillo D. Proteomic analysis of a genetically modified maize flour carrying CRY1AB gene and comparison to the corresponding wild-type. Maydica. 2007;52:443–455. [Google Scholar]
- Catchpole GS, Beckmann M, Enot DP, Mondhe M, Zywicki B, Taylor J, Hardy N, Smith A, King RD, Kell DB. et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc Natl Acad Sci USA. 2005;102:14458–14462. doi: 10.1073/pnas.0503955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Beaulieu J, Iquira E, Belzile FJ, Fortin MG, Strömvik MV. Effect of transgenes on global gene expression in soybean is within the natural range of variation of conventional cultivars. J Agric Food Chem. 2008;56:3057–3067. doi: 10.1021/jf073505i. [DOI] [PubMed] [Google Scholar]
- Corpillo D, Gardini G, Vaira AM, Basso M, Aime S, Accotto GP, Fasano M. Proteomics as a tool to improve investigation of substantial equivalence in genetically modified organisms: The case of a virus-resistant tomato. Proteomics. 2004;4:193–200. doi: 10.1002/pmic.200300540. [DOI] [PubMed] [Google Scholar]
- El Ouakfaoui S, Miki B. The stability of the Arabidopsis transcriptome in transgenic plants expressing the marker genes nptII and uidA. Plant J. 2005;41:791–800. doi: 10.1111/j.1365-313X.2005.02350.x. [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Brinch-Pedersen H, Holm PB. A microarray-based comparative analysis of gene expression profiles during grain development in transgenic and wild type wheat. Transgenic Res. 2005;14:887–905. doi: 10.1007/s11248-005-1526-y. [DOI] [PubMed] [Google Scholar]
- Lehesranta SJ, Davies HV, Shepherd LVT, Nunan N, McNicol JW, Auriola S, Koistinen KM, Suomalainen S, Kokko HI, Kärenlampi SO. Comparison of tuber proteomes of potato varieties, landraces, and genetically modified lines. Plant Physiol. 2005;138:1690–1699. doi: 10.1104/pp.105.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Kang JY, Kim SY. Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotechnol J. 2004;2:459–466. doi: 10.1111/j.1467-7652.2004.00090.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T. et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M. et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/A:1006321900483. [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2006;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- Gilbertson L. Cre-lox recombination: Cre-ative tools for plant biotechnology. Trends Biotechnol. 2003;21:550–555. doi: 10.1016/j.tibtech.2003.09.011. [DOI] [PubMed] [Google Scholar]
- De Buck S, Peck I, De Wilde C, Marjanac G, Nolf J, De Paepe A, Depicker A. Generation of single-copy T-DNA transformants in Arabidopsis by the CRE/loxP recombination-mediated resolution system. Plant Physiol. 2007;145:1171–1182. doi: 10.1104/pp.107.104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa TTC, Bong BB, Huq E, Hodges TK. Cre/lox site-specific recombination controls the excision of a transgene from the rice genome. Theor Appl Genet. 2002;104:518–525. doi: 10.1007/s001220100748. [DOI] [PubMed] [Google Scholar]
- Osborne BI, Wirtz U, Baker B. A system for insertional mutagenesis and chromosomal rearrangement using the Ds transposon and Cre-lox. Plant J. 1995;7:687–701. doi: 10.1046/j.1365-313X.1995.7040687.x. [DOI] [PubMed] [Google Scholar]
- Russell SH, Hoopes JL, Odell JT. Directed excision of a transgene from the plant genome. Mol Genet Genet. 1992;234:49–59. doi: 10.1007/BF00272344. [DOI] [PubMed] [Google Scholar]
- Coppoolse ER, de Vroomen MJ, Roelofs D, Smit J, van Gennip F, Hersmus BJM, Nijkamp HJJ, van Haaren MJJ. Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol. 2003;51:263–279. doi: 10.1023/A:1021174726070. [DOI] [PubMed] [Google Scholar]
- Que Q, Wang HY, Jorgensen RA. Distinct patterns of pigment suppression are produced by allelic sense and antisense chalcone synthase transgenes in petunia flowers. Plant J. 1998;13:401–409. doi: 10.1046/j.1365-313X.1998.00038.x. [DOI] [Google Scholar]
- Corneille S, Lutz K, Svab Z, Maliga P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J. 2001;27:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- Corneille S, Lutz KA, Azhagiri AK, Maliga P. Identification of functional lox sites in the plastid genome. Plant J. 2003;35:753–762. doi: 10.1046/j.1365-313X.2003.01845.x. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Gilbertson L, Staub JM. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 2001;27:161–170. doi: 10.1046/j.1365-313x.2001.01067.x. [DOI] [PubMed] [Google Scholar]
- Heidmann D, Lehner CF. Reduction of Cre recombinase toxicity in proliferating Drosophila cells by estrogen-dependent activity regulation. Dev Genes Evol. 2001;211:458–465. doi: 10.1007/s004270100167. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/S1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, Mayer JE, Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/S0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2365. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005;44:972–984. doi: 10.1111/j.1365-313X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30:1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- Casano LM, Zapata JM, Martín M, Sabater B. Chlororespiration and poising of cyclic electron transport. Plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J Biol Chem. 2000;275:942–948. doi: 10.1074/jbc.275.2.942. [DOI] [PubMed] [Google Scholar]
- Grandbastien M-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. doi: 10.1016/S1360-1385(98)01232-1. [DOI] [Google Scholar]
- Madlung A, Comai L. The effect of stress on genome regulation and structure. Ann Bot. 2004;94:481–495. doi: 10.1093/aob/mch172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J Mol Biol. 1992;223:911–928. doi: 10.1016/0022-2836(92)90252-F. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/S0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:97–104. doi: 10.1046/j.1365-313x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- Legen J, Wanner G, Herrmann RG, Small I, Schmitz-Linneweber C. Plastid tRNA genes trnC-GCA and trnN-GUU are essential for plant cell development. Plant J. 2007;51:751–762. doi: 10.1111/j.1365-313X.2007.03177.x. [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Giraudat J, Parcy F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot. 2005;56:597–603. doi: 10.1093/jxb/eri050. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol. 2007;63:151–169. doi: 10.1007/s11103-006-9079-x. [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell. 2002;14:3177–3189. doi: 10.1105/tpc.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Choi H, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005;139:1750–1761. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ. et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi H, Ryu HJ, Park JH, Kim MD, Kim SY. ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol. 2004;136:3639–3648. doi: 10.1104/pp.104.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SSL, Lynch TJ, Thomas TL, Rock CD. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 2002;130:688–697. doi: 10.1104/pp.003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI. Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol. 2003;6:463–469. doi: 10.1016/S1369-5266(03)00084-0. [DOI] [PubMed] [Google Scholar]
- Lee B, Kapoor A, Zhu J, Zhu JK. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006;18:1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell. 2001;1:771–781. doi: 10.1016/S1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M. et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA. 2002;99:10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell. 2008;20:2876–2893. doi: 10.1105/tpc.108.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin OK, Macherel D. The crucial role of plant mitochondria in orchestrating drought tolerance. Ann Bot. 2009;103:581–597. doi: 10.1093/aob/mcn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho IS, Chaves MM, Pinto Ricardo C. Influence of Water Stress on the Chemical Composition of Seeds of Two Lupins (Lupinus albus and Lupinus mutabilis) J Agron Crop Sci. 2005;191:95–98. doi: 10.1111/j.1439-037X.2004.00128.x. [DOI] [Google Scholar]
- Champolivier L, Merrien A. Effects of water stress applied at different growth stages to Brassica napus L. var. oleifera on yield, yield components and seed quality. Eur J Agron. 1996;5:153–160. doi: 10.1016/S1161-0301(96)02004-7. [DOI] [Google Scholar]
- Dornbos DL Jr, Mullen RE. Soybean seed protein and oil contents and fatty acid composition adjustments by drought and temperature. J Am Oil Chem Soc. 1992;69:228–231. doi: 10.1007/BF02635891. [DOI] [Google Scholar]
- Gigon A, Matos AR, Laffray D, Zuily-Fodil Y, Pham-Thi AT. Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia) Ann Bot. 2004;94:345–351. doi: 10.1093/aob/mch150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CR, Mogensen VO, Mortensen G, Fieldsend JK, Milford GFJ, Andersen MN, Thage JH. Seed glucosinolate, oil and protein contents of field-grown rape (Brassica napus L.) affected by soil drying and evaporative demand. Field Crops Res. 1996;47:93–105. doi: 10.1016/0378-4290(96)00026-3. [DOI] [Google Scholar]
- Rotundo JL, Westgate ME. Meta-analysis of environmental effects on soybean seed composition. Field Crops Res. 2009;110:147–156. doi: 10.1016/j.fcr.2008.07.012. [DOI] [Google Scholar]
- Dakhma WS, Zarrouk M, Cherif A. Effects of drought-stress on lipids in rape leaves. Phytochemistry. 1995;40:1383–1386. doi: 10.1016/0031-9422(95)00459-K. [DOI] [Google Scholar]
- Bejarano L, Mignolet E, Devaux A, Espinola N, Carrasco E, Larondelle Y. Glycoalkaloids in potato tubers: the effect of variety and drought stress on the α-solanine and α-chaconine contents of potatoes. J Sci Food Agric. 2000;80:2096–2100. doi: 10.1002/1097-0010(200011)80:14<2096::AID-JSFA757>3.0.CO;2-6. [DOI] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–610. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313X.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. Regulation of Arabidopsis thaliana Em genes: role of ABI5. The Plant Journal. 2002;30:373–383. doi: 10.1046/j.1365-313X.2002.01295.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002;129:1533–1543. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR genotyping of control plant lines. (A) Control plant lines, derived from 35S:ABF3 plants crossed with plants expressing Cre recombinase, were PCR genotyped to identify plants from which the 35S-ABF3-nos transgene was excised. (B) Control plant lines following a backcross with Col-0 plants were genotyped to identify plants that lost the Cre gene by genetic segregation. Plant lines with the desired genotype that were selected following each cross are indicated with asterisks.
Functional categorization of drought responsive genes in 35S:ABF3 and control plants. Distribution of genes differentially expressed in 35S:ABF3 and control plant lines at 2 h and 24 h following drought stress into functional categories.
Genes differentially expressed in response to drought in 35S:ABF3 and control plants. This table includes a complete list of genes differentially expressed in both 35S:ABF3 and control plant lines in response to drought at 2 h and 24 h. Genes are divided into three groups including those that are similarly regulated in both 35S:ABF3 and control plants, those that show enhanced regulation in 35S:ABF3 plants, and those that show attenuated regulation in 35S:ABF3 plants.
Sequences of primers used in RT-PCR analysis. This table includes sequences of primers used to amplify selected genes in the RT-PCR analysis performed to confirm microarray results.