Abstract
The Swi1-Swi3 replication fork protection complex and Mrc1 protein are required for stabilization of stalled replication forks in fission yeast. Hsk1 kinase also plays roles in checkpoint responses elicited by arrested replication forks. We show that both Swi1 and Swi3, the abundance of which are interdependent, are required for chromatin association of Mrc1. Co-immunoprecipitation experiments show the interactions of Swi1-Swi3, Mrc1 and Hsk1. Mrc1 interacts with Swi3 and Hsk1 proteins through its central segment (378-879) containing a SQ/TQ cluster, and this segment is sufficient for checkpoint reaction. The SQ/TQ cluster segment (536-673) is essential but not sufficient for the interactions and for resistance to replication inhibitor hydroxyurea. Mrc1 protein level is increased in hsk1-89 cells due to apparent stabilization, and we have identified a potential phosphodegron sequence. These results suggest that interactions of the Swi1-Swi3 complex and Hsk1 kinase with Mrc1 may play a role in cellular responses to stalled replication forks in fission yeast.
Introduction
Preservation of genome integrity is crucial for both the viability and vitality of all living organisms. Accurate and complete DNA replication is centrally important to this process. Indeed, the molecular machine at the replication fork (i.e. the replisome complex) not only needs to copy the genetic information with a minimum error rate, but it needs to do so in situations when replication fork progression is impeded by DNA lesions or by protein complexes bound to DNA. The ability of cells to detect arrested replication forks and to process them for repair and subsequent resumption of fork progression is crucial for cell survival (Boddy & Russell 2001; Kastan & Bartek 2004).
In recent years, progress has been made in identifying replication fork-associated proteins that are not essential for DNA replication but are required for preservation of genome integrity (Katou et al. 2003; Gambus et al. 2006). Some of these factors are thought to stabilize and maintain the structure of the stalled replication fork for subsequent activation of the replication checkpoint. Prominent amongst these proteins are Mrc1/Claspin, Tof1/Swi1/Tim1, and Csm3/Swi3/Tipin proteins. The latter two proteins form a complex that has been called “fork protection complex (FPC)” (Gotter 2003; Noguchi et al. 2003, 2004). They are required for activation of checkpoint kinases, Rad53 (budding yeast), Cds1 (fission yeast) or Chk1 (vertebrate) in response to replication fork arrest. Recently, others and we reported genetic and physical interactions between these factors and Cdc7 kinase, which has been known to play important roles in initiation of DNA replication (Masai & Arai 2002; Matsumoto et al. 2005; Sommariva et al. 2005). We have shown that Cdc7 is required for checkpoint kinase activation in fission yeast and human cells (Takeda et al. 2001; Kim et al. 2008), and presented evidence that Claspin may be the target of Cdc7 in this regulation (Kim et al. 2008). However, the precise modes of physical and functional interactions among these fork stabilization factors, Cdc7 kinase and checkpoint kinases are not known.
In this report, we examine interactions amongst these fork stabilization factors and Hsk1, the fission yeast orthologue of Cdc7. We show that chromatin association of Mrc1 requires Swi1 and Swi3 proteins and that Swi1, Swi3 and Hsk1 proteins interact with Mrc1. The domain of Mrc1 required for these interactions coincided with that required for resistance to HU. We also show that abundance of Swi1 and Swi3 are mutually dependent, and that Hsk1 affects the stability of Mrc1 but not that of Swi1 and Swi3. These results indicate physical and functional interactions between these factors regulate checkpoint responses to arrested replication forks.
Results
Physical and Genetic interactions between the Swi1-Swi3 fork protection complex and Hsk1-Dfp1/Him1 kinase
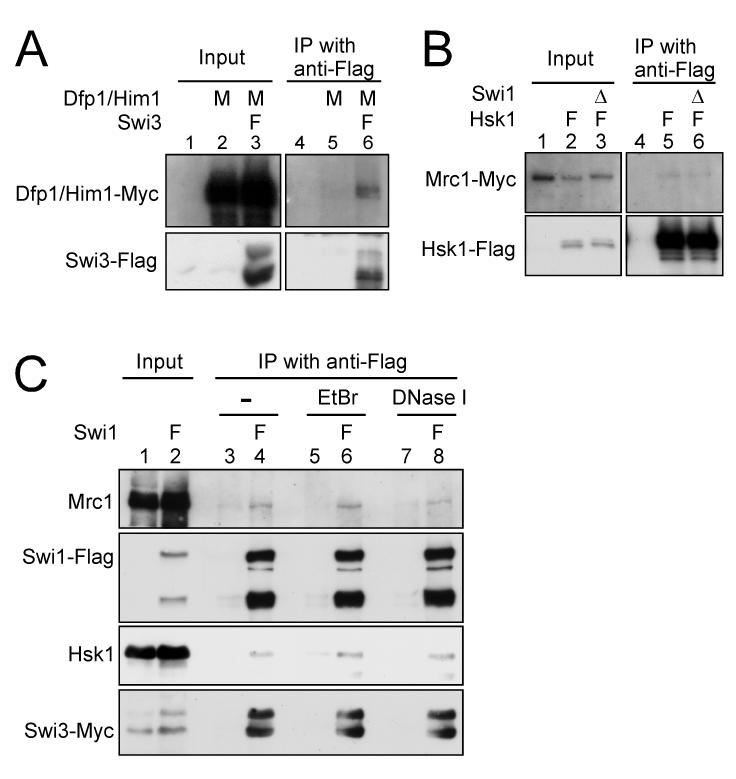
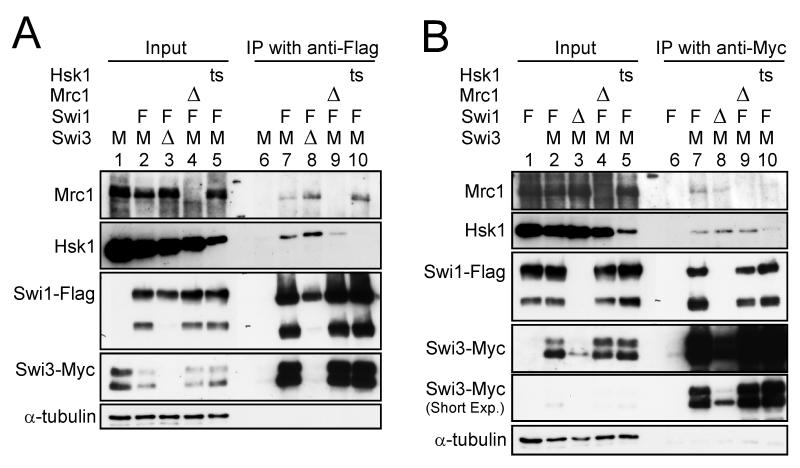
Previous studies have described physical and genetic interaction between Swi1 and Hsk1-Dfp1/Him1 (Matsumoto et al. 2005; Sommariva et al. 2005). We examined physical interactions between Swi1-Swi3, Mrc1 and Hsk1 by immunoprecipitation. Dfp1/Him1 protein was co-immunoprecipitated with Swi3 protein (Fig. 1A). Hsk1 was co-immunoprecipitated with Swi1 and Swi3 proteins (Fig. 2A and B, lane 7), and these interactions were observed also in swi3Δ and swi1Δ backgrounds, respectively (Fig. 2A and B, lane 8). Mrc1 was also co-immunoprecipitated with Hsk1 both in the presence and absence of Swi1 (Fig. 1B). These results suggest that Hsk1 can interact with Swi1, Swi3 and Mrc1 proteins individually or Hsk1 interacts with the large fork complex that includes these proteins. The interaction between Swi1 with Hsk1 was observed also after treating cell extracts with ethidium bromide or DNaseI (Fig. 1C), suggesting that the interaction is not dependent on DNA.
Figure 1. Interaction of Hsk1-Dfp1/Him1 with Swi3 and Mrc1.
(A) Co-immunoprecipitation of Dfp1/Him1 with Swi3. (B) Co-immunoprecipitation of Mrc1 with Hsk1. The extracts were prepared from the strains indicated, and immunoprecipitation with anti-Flag antibody was performed as described in “Experimental procedures” (lanes 4-6 in (A) and lanes 4-6 in (B)). Input (lanes 1-3 in (A) and lanes 1-3 in (B)) represents 2.5 % of the extracts used for the immunoprecipitation. Western blotting analyses were conducted using the antibodies against the tag as indicated. (A) lanes 1 and 4, YM71; lanes 2 and 5, EN3404; lanes 3 and 6, SH1007. (B) lanes 1 and 4, KT2791; lanes 2 and 5, MS404; lanes 3 and 6, MS405. (C) Co-immunoprecipitation of Hsk1 with Swi1 from EtBr or DNaseI treated extracts. The extracts from the strains indicated were treated with either EtBr (50 μg/ml) or DNase I (0.35 U/μl) for 15 min at 0°C, followed by immunoprecipitation with anti-Flag antibody (lanes 3-8). Input (lanes 1 and 2) represents 1.7 % of the extracts used for the immunoprecipitation. Western blotting analyses were conducted using the antibodies indicated. Lanes 1, 3, 5 and 7, SH0987; lanes 2, 4, 6 and 8, SH1232. Genotypes are indicated above each lane with the abbreviation as follows. M, Myc-tagged; F, Flag-tagged; Δ, deletion.
Figure 2. Interactions among fork stabilization factors and Hsk1.
(A) Co-immunoprecipitation with Swi1-Flag protein. (B) Co-immunoprecipitation with Swi3-Myc protein. The extracts were prepared from the strains indicated and immunoprecipitated with anti-Flag (A) or anti-Myc (B) antibody as described in “Experimental procedures” (lanes 6-10 in (A) and (B)). Inputs (lanes 1-5 in (A) and (B)) represent 2 % of the extracts used for the immunoprecipitation. Western blotting analyses were conducted using the antibodies indicated. (A) lanes 1 and 6, SH0987; lanes 2 and 7, SH1232; lanes 3 and 8, SH1914; lanes 4 and 9, SH2303; lanes 5 and 10, SH1302. (B) lanes 1 and 6, EN3381; lanes 2 and 7, SH1232; lanes 3 and 8, SH3309; lanes 4 and 9, SH2303; lanes 5 and 10, SH1302. The degradation products of Swi1 (lower bands) are missing in swi3Δ, since they are further degraded under this background. Abbreviations of the genotypes are the same as those in Figure 1. ts, hsk1-89.
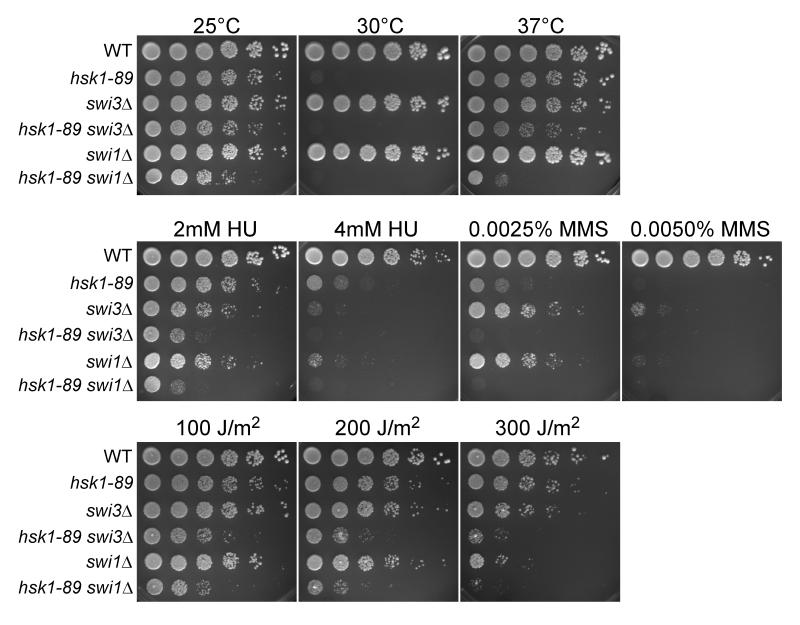
We then examined genetic interaction between Swi3 and Hsk1 by constructing a double mutant of swi3Δ and hsk1-89. hsk1-89 mutant cells show a growth defect at 30°C due to impaired kinase activity and instability of the mutant protein (data not shown; see Fig. 6B, lanes 8-14). However, this mutant can form colonies at 37°C, although it accumulates cells with abnormal nuclear structures (Takeda et al. 2001). The hsk1-89 swi3Δ double mutant cells exhibit growth only slightly reduced compared to that of hsk1-89 at all the temperatures, while hsk1-89 swi1Δ exhibits significantly reduced growth at 37°C compared to hsk1-89 (Matsumoto et al. 2005). This result may suggest that Swi1 and Swi3 have some distinct functions. Both hsk1-89 swi3Δ and hsk1-89 swi1Δ cells are significantly more sensitive to HU, MMS, and UV than either single mutant (Fig. 3). Enhanced sensitivity of hsk1-89 swi1Δ to these genotoxic agents was also reported (Matsumoto et al. 2005). These results indicate that Hsk1 and Swi1/Swi3 mediate resistance to HU in separate pathways.
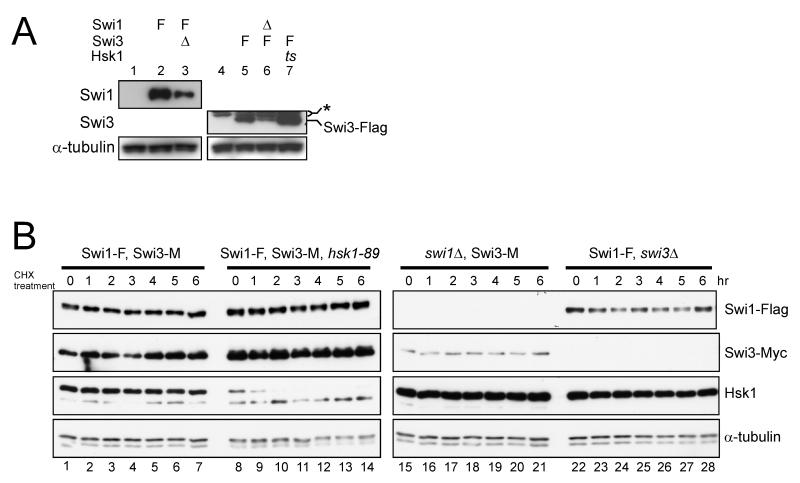
Figure 6. Stability of Swi1 and Swi3 proteins is mutually dependent.
(A) The whole cell extracts were analyzed by western blotting using the antibodies against the proteins indicated. * indicates a non-specific band reacting with the anti-Flag antibody. The arrow indicates the Flag-tagged Swi3 protein. Abbreviations of the genotypes are the same as those in Figures 1 and 2. (B) Cycloheximide was added to the asynchronously growing cultures, and cells were harvested at the times indicated. The whole cell extracts were analyzed by western blotting using the antibodies against the proteins indicated. (A) lane 1,YM71; lane 2, SH2219; lane 3, SH2504; lane 4, YM71; lane 5, SH1401; lane 6, SH2611; lane 7, SH1505. (B) lanes 1-7, SH1232; lanes 8-14,SH1302; lanes 15-21, SH3309; lanes 22-28, SH1914.
Figure 3. swi3Δ causes hypersensitivity to HU, UV and MMS in hsk1-89 cells.
Five-fold serial dilutions of exponentially growing S. pombe cells of the indicated genotypes were plated on YES agar medium and incubated at 25, 30, and 37°C for 5 days (upper panels). Identical five-fold serial dilutions of the indicated cultures were plated on YES plates supplemented with the indicated amounts of HU or MMS (middle panels) or on YES plates followed by UV irradiation as shown (lower panels), and incubated at 25°C for 5 days.
Chromatin association of Mrc1 is facilitated by Swi1 or Swi3
Mrc1 and Swi1-Swi3 are conserved proteins implicated in regulation of replication forks (Alcasabas et al. 2001; Tanaka & Russell 2001; Katou et al. 2003; Gambus et al. 2006). In budding yeast, deletion of mrc1 or tof1 (swi1 homolog) causes uncoupling of DNA synthesis and fork unwinding, resulting in destabilization of replication forks (Katou et al. 2003).
We fractionated fission yeast cells into Triton-soluble and -insoluble (chromatin-enriched) fractions. The amount of Mrc1 protein significantly increased in response to HU, as was reported previously (Tanaka & Russell 2001), and it was more enriched in Triton-insoluble fractions (Fig. 4A, lanes 1- 6). In contrast, Swi1 and Swi3 protein levels did not change but chromatin association of these factors slightly increased after replication stress (Fig. 4A, lanes 1-6), consistent with the previous observations using the GFP-fused proteins (Noguchi et al. 2003, 2004).
Figure 4. Chromatin association of Mrc1 is stimulated by HU treatment, and is reduced in swi1Δ or swi3Δ cells.
The total cell extracts (TCE), chromatin-free (Triton-soluble) and chromatin-enriched (Triton-insoluble) fractions were prepared from the yeast strains indicated and were analyzed by western blotting using the antibodies as indicated. (A) lanes 1-6, SH1232; lanes 7-12, SH1302. (B) lanes 1-6, SH1914; lanes 7-12, SH3309. HU: treated with 12 mM HU for three hrs. The lower bands in Swi1-Flag blots are major degradation products of Swi1 protein. In B, the blots with Swi1-Flag and Swi3-Myc were exposed for a longer time in order to visualize the proteins expressed at a lower level. Note that Mrc1 in the soluble fraction is highly unstable and is degraded during the extract preparation. Chromatin-bound Mrc1 is stable in wild-type or swi1Δ cells (see Figure S1). This is why the sum of soluble and insoluble Mrc1 is less than the TCE level in some lanes. Similarly, Swi3 in the absence of Swi1 is unstable in the soluble fraction and is degraded (B. lanes 8 and 11). (C) In situ chromatin binding assay of Mrc1-GFP in cells arrested at early S-phase by treatment with 15 mM HU for 3 h at 30°C. Spheroplasts of KT2884 (wild-type) (a), MS369 (swi1Δ) (b) and MS384 (swi3Δ) (c) were untreated (-Triton X-100) or preextracted with Triton X-100 to remove soluble nuclear proteins (+Triton X-100) and then fixed for microscopic analysis, as described in “Experimental procedures”. Fluorescence of Mrc1-GFP and DAPI are shown. (d) Quantification of Mrc1-GFP localization in nuclei. The GFP-positive cells were counted and the fraction (%) of GFP-positive cells in DAPI-positive nuclei are presented.
The amount of Mrc1 in asynchronously growing cells in the Triton-insoluble fraction decreased in swi1Δ or swi3Δ cells (Fig. 4B, lanes 3 and 9). The chromatin accumulation of Mrc1 in the presence of HU was also significantly reduced in both swi1Δ and swi3Δ cells (Fig. 4B, lanes 6 and 12), suggesting that the Swi1-Swi3 fork protection complex may play an important role in recruiting Mrc1 onto chromatin in the normal course of DNA replication as well as after replication stress. This conclusion was also supported by the localization of GFP-tagged Mrc1 protein (Fig. 4C). The Mrc1 signal in nuclei significantly reduced in swi1Δ or swi3Δ cells. The reduction in swi1Δ or swi3Δ cells was observed both with or without Triton treatment, suggesting that the Swi1 and Swi3 may be required for efficient nuclear localization of Mrc1 and, once localized in nuclei, Mrc1 could bind to the chromatin, at least with this assay system. The chromatin association of Mrc1 was not affected by the hsk1-89 mutation (Fig. 4A, lanes 9 and 12).
Interaction of Swi1 and Swi3 with Mrc1
We next examined interactions of Swi1 or Swi3 with Mrc1. Immunoprecipitation analyses revealed that endogenous Mrc1 was coimmunoprecipitated with Swi1-Flag or Swi3-Myc protein expressed from its own promoter (Fig. 2A, lane 7 and Fig. 2B, lane 7). These interactions were also confirmed by reciprocal immunoprecipitation in which Swi1 or Swi3 was coimmunoprecipitated with Mrc1-Myc (data not shown). Interaction between Swi1 and Mrc1 was detected in the presence of ethidium bromide or DNaseI (Fig. 1C), indicating that the interaction does not depend on DNA.
Interaction of Mrc1 with Swi1 or Swi3 was detected also in the absence of Swi3 or Swi1 protein (Fig. 2A and B, lanes 8). These interactions were not affected by hsk1-89 (Fig. 2A and B, lanes 10). Thus, these results indicate that Mrc1 interacts with Swi1 or Swi3 individually, or it associates with the replication fork complex which may contain the Swi1-Swi3 complex or either factor alone. Hsk1 kinase does not appear to significantly affect this interaction. The reason for slight increase of Mrc1 in the Swi1-immunoprecipitate in swi3Δ and hsk1-89 cells (Fig. 2A) is not clear
Mrc1 segments required for resistance to HU and protein interactions
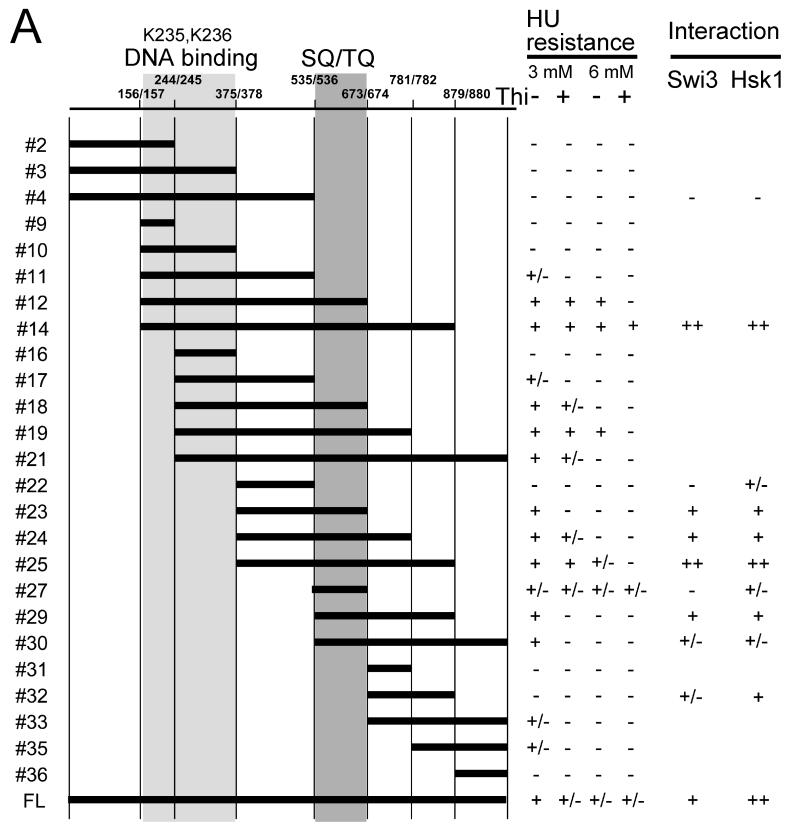
In order to identify the domain of Mrc1 required for interaction with Hsk1 and Swi1 proteins, we have generated a series of deletion derivatives of Mrc1 and expressed them in fission yeast cells. The fragments were cloned into a pREP41-based vector and a Flag-tag was attached at the C-terminus. The plasmids were introduced into mrc1Δ cells, and HU sensitivity was examined (Fig. 5A). The segment 536-673 containing the SQ/TQ cluster (#27) is essential for exhibition of HU resistance (Zhao et al. 2003). The minimum segment showing resistance to 3 mM HU was 376-879 (#25), and resistance to 6 mM HU required the additional segment 157-375 containing DNA binding domain (Zhao & Russell 2004) (#14). 376-673 (#23) or 536-879 (#29) showed 3 mM HU resistance only after the overproduction, suggesting the requirement of both N-terminal and C-terminal sides of the SQ/TQ cluster for efficient checkpoint function.
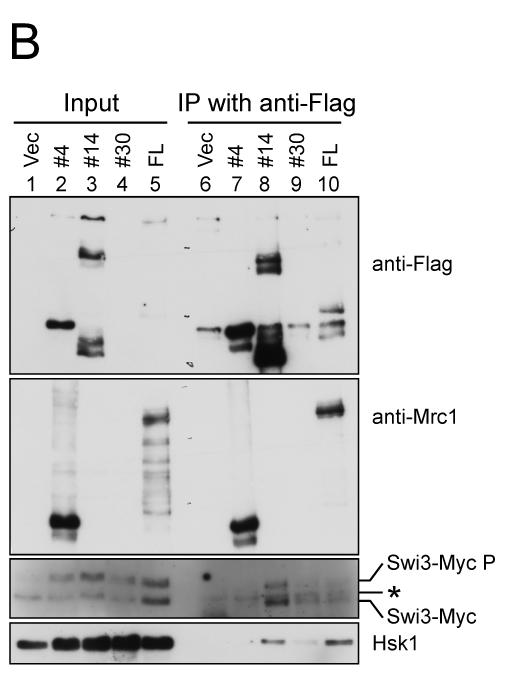
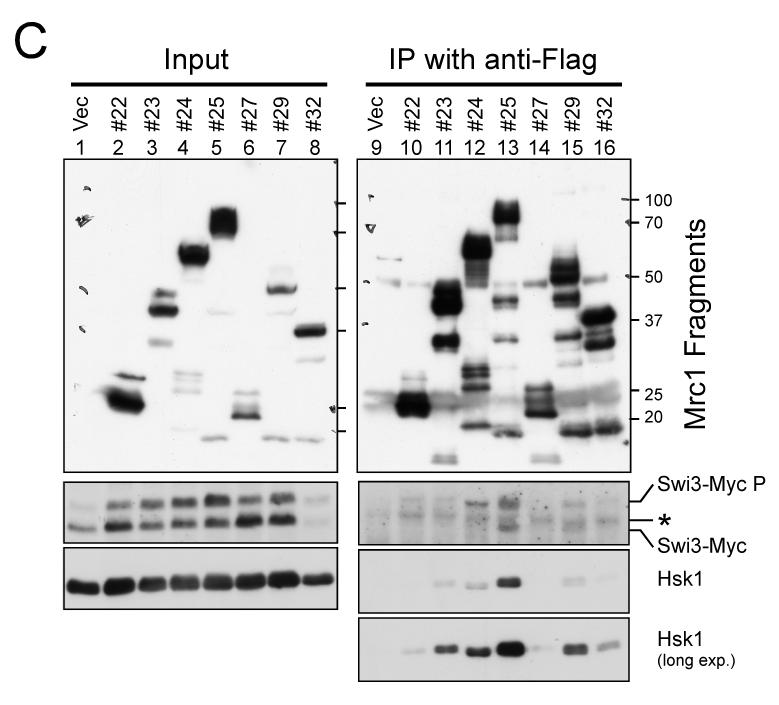
Figure 5. Mrc1 segments required for resistance to HU and protein interactions.
(A) A series of deletion derivatives of Mrc1 indicated were cloned into a pREP41X-3Flag which permits addition of 3xFlag-tag at the C terminus of the cloned fragment, and were expressed in mrc1Δ cells (MS252). HU resistance of each transformants were examined on EMM plates containing 3 mM or 6 mM HU. The extent of growth is indicated in the right. +. +/− and – indicate full growth, partially impaired growth and no growth, respectively. The interaction of selected deletion derivatives with Swi3 and Hsk1 is also shown. ++, +, +/− and – represents relative strength of interaction. Light gray and gray segments indicate those involved in DNA binding and containing SQ/TQ clusters, respectively. (B) Mrc1 segments 1-535 (#4), 157-879 (#14), 536-1019 (#30) and a full length of Mrc1 (FL) expressed in mrc1Δ cells (SH2309) were immunoprecipitated with anti-Flag antibody. Western blotting analyses were conducted using the antibodies indicated. (C) Mrc1 segments 376-535 (#22), 376-673 (#23), 376-781 (#24), 376-879 (#25), 536-673 (#27), 536-879 (#29) and 674-879 (#32) expressed in mrc1Δ cells (SH2309) were immunoprecipitated with anti-Flag antibody. Western blotting analyses were conducted using the antibodies indicated. * indicates a non-specific band detected in anti-Flag immunoprecipitates. Swi3-MycP indicates the mobility-shifted form of Swi3-Myc due to phosphorylation.
We then examined which segment of Mrc1 is required for interaction with Swi3 or Hsk1 protein. Efficient interaction with both Swi3 and Hsk1 was observed with the segment 157-879 (#14), the minimum Mrc1 sufficient for its full checkpoint function, while the N-terminal segment (1-535; #4) did not show interaction with either Hsk1 or Swi3. On the other hand, the C-terminal segment (536-1019; #30) exhibited weak interaction with both proteins (Fig. 5B). Further analyses with smaller segments of Mrc1 indicated strong interaction of the segment 376-879 (#25) with both proteins. Weaker interaction with Hsk1 was observed also with 376-673 (#23), 376-781 (#24) or 536-879 (#29). 376-781 (#24) and 536-879 (#29) also showed weak interaction with Swi3. The segments lacking the SQ/TQ cluster (376-535 [#22] or 674-879 [#32]) showed very reduced or no interaction with Swi3 or Hsk1 (Fig. 5C). Although the SQ/TQ cluster is essential for the interactions, this segment (#27; 536-673) alone did not interact with either protein (Fig. 5C). Thus, these results suggest the requirement of the SQ/TQ cluster for the interaction, and the stimulatory role of the adjacent region for the interaction. General correlation of HU resistance and ability to bind to Swi1/3 or Hsk1 (Fig. 5A) supports the importance of these interactions in cellular responses to stalled replication forks .
The stability of Swi1 and Swi3 proteins is interdependent
The total protein level of Swi1 or Swi3 is affected in swi3Δ or swi1Δ mutant, respectively (Fig. 6A, lanes 3 and 6), as was noted previously (Noguchi et al. 2004), suggesting that the complex formation stabilizes both proteins. While the amount of Swi3 protein dramatically decreased in swi1Δ cells, the Swi1 protein level was only mildly reduced in swi3Δ, consistent with the previous report (Noguchi et al. 2004; Fig. 2A and B, lanes 3 and Fig. 6, lanes 3 and 6). Mutual dependency of the levels of Tim and Tipin proteins, mammalian orthologs of Swi1 and Swi3, respectively, was reported previously (Chou & Elledge 2006; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata & Masai 2007). The protein levels of Swi1 and Swi3 were not significantly affected by mrc1Δ (Fig. 2A and B, lanes 4).
We then examined the stability of Swi1 and Swi3 in the wild type and mutant cells. In the wild type background, both Swi1 and Swi3 proteins are stable with no sign of degradation for six hrs in the presence of cycloheximide (Fig. 6B). In contrast, Mrc1 protein was unstable under the same condition with a half-life of less than 1 hr (data not shown; see Fig. 7B and C). In swi1Δ or swi3Δ cells, the stability of Swi3 and Swi1, respectively, was not significantly affected, although their steady-state levels before addition of cycloheximide were lower than those in the wild-type cells (Fig. 6B). These results indicate that the presence of both proteins is required for efficient expression and/or accumulation of Swi1 and Swi3.
Figure 7. Mrc1 is stabilized in hsk1-89.
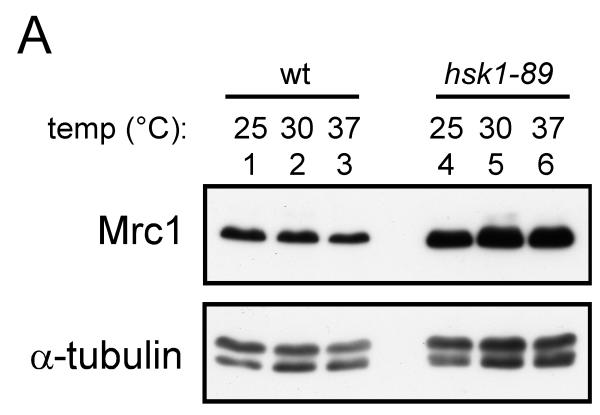
(A) Whole cell extracts were prepared from hsk1+ (YM71; lanes 1-3) or hsk1-89 (KO147; lanes 4-6) cells grown at 25°C (lanes 1 and 4), 30°C (lanes 2 and 5) and 37°C (lanes 3 and 6) for 6 hrs, and were analyzed by western blotting. (B) Cycloheximide (0.1 mg/ml) was added to the asynchronously growing cells (at 25°C) and the cells were harvested at the times indicated. The whole cell extracts were prepared and analyzed by western blotting. Lanes 1-5, KT2791; lanes 6-10, MS346; lanes 11-15, SH0603. (C) The band intensities of (B) were scanned, and the intensities of the Mrc1 bands were normalized to those of tubulin bands, and the normalized values are presented as relative Mrc1 protein level. For each strain, the values at time 0 were taken as 100. In (A) and (B), anti-Mrc1, anti-Myc and anti-tubulin antibodies were used for western blotting. (D) A putative phosphodegron sequence in Mrc1 protein. The mutated residues are indicated. (E) The whole cell extracts from cells carrying the wild-type or a phosphodegron mutant (SSAA) grown at 30°C were analyzed by western blotting using anti-Flag antibody. Lanes 1-4 wild-type; lanes 5-8, mutant; lanes 3-8; tagged with 3-Flag at the C-terminus. Lanes 1-2, YM71, lanes 3-4, MS424, lanes 5-6, MS164, lanes 7-8, MS165.
We also examined effect of hsk1-89 mutation on Swi1 and Swi3. The levels of Swi1 and Swi3 proteins were not affected by hsk1-89 mutation (Fig. 4A, lane 7; Fig. 6B, lanes 8-14), and co-immuoprecipitation of Swi1 and Swi3 was not affected by hsk1-89 mutation (Fig. 2A and B, lanes 10). The abundance of Swi1 and Swi3 and their interaction were not affected by mrc1Δ mutation, either (data not shown; Fig. 2A and B, lane 9). These results indicate that the stability and complex formation of Swi1-Swi3 is independent of Hsk1 and Mrc1.
Mrc1 protein is stabilized in hsk1-89 cells
We have noticed that the level of Mrc1 protein is increased at 25°C in hsk1-89 cells. Therefore, we examined the level of Mrc1 protein at various temperatures. In the wild-type cells, the Mrc1 protein levels were similar at 25°C and 30°C and slightly decreased at 37°C. In hsk1-89 cells, the Mrc1 protein level was higher than in the wild-type cells at all the temperatures (Fig. 7A). This suggests that Hsk1 may regulate the stability of Mrc1. Therefore, we have examined the stability of Mrc1 in hsk1+, hsk1-89, and cds1Δ cells at 25°C. At this temperature, all the cells exhibit DNA contents of typical asynchronously growing cells (peak at 2C, post-replication state; data not shown), precluding the potential cell cycle effect on Mrc1 stability. Mrc1 protein is rapidly degraded in the wild-type cells (Fig. 7B, lanes 1-5); its level became less than 20% in 2 hrs in the presence of cycloheximide (Fig. 7C). The level of Mrc1 decreased rapidly also in cds1Δ cells (Fig. 7B, lanes 11-15). In contrast, the amount of Mrc1 protein did not significantly change in hsk1-89 cells under the same condition (Fig. 7B, lanes 6-10; Fig. 7C). This indicates that Mrc1 protein is stabilized in hsk1-89 cells, suggesting that degradation of Mrc1 may be facilitated by Hsk1-dependent phosphorylation events.
A putative phosphodegron sequence, DSGVS, was found at 859-864 on Mrc1 (Fig. 7D). We therefore replaced the two serine resides at 860 and 864, the putative phosphorylation sites, with alanine. The resulting SSAA mutant grows normally and did not show any increased sensitivity to HU (data not shown). The level of the SSAA mutant both in untreated or HU-treated cells remarkably increased compared to the wild-type cells (Fig. 7E). This result may be consistent with the possibility that Hsk1 destabilizes Mrc1 protein through phosphorylation of the putative phosphodegron sequence, although this needs to be experimentally examined.
Discussion
The process of DNA replication is strictly monitored to ensure that the entire genome is replicated in coordination with other cell cycle events. It is now being recognized that the normal course of DNA replication is perturbed more frequently than anticipated. The identification of conserved replication fork factors, Tof1/Swi1/Tim1, Csm3/Swi3/Tipin, and Mrc1/Claspin, whose main functions are to protect and stabilize the replication forks, underscores the physiological importance of the strict monitoring system of the replication forks (Katou et al. 2003; Gambus et al. 2006).
Cdc7 kinase was first identified in budding yeast, and has been regarded as a factor regulating the initiation of DNA replication (Masai et al. 2000, 2006). Although the functions for initiation are conserved throughout evolution (Masai et al. 1995; Sato et al. 1997; Kim et al. 2002; Masai & Arai 2002), diverse roles of Cdc7 kinase in other chromosome transactions have been suggested (Takeda et al. 2001; Bailis et al. 2003; Ogino et al. 2006). Among them, the roles in DNA replication checkpoint have been reported in various organisms (Brown & Kelly 1999; Weinreich & Stillman 1999; Snaith et al. 2000; Duncker et al. 2002; Costanzo et al. 2003). We reported the role of Cdc7 in checkpoint kinase activation, and more recently its potential roles in stabilization of arrested replication forks in conjunction with Swi1 protein (Takeda et al. 2001; Matsumoto et al. 2005; Sommariva et al. 2005).
Interactions between fork stabilization factors and Hsk1 kinase
In this manuscript, we first explored physical and genetic interactions of Hsk1, the fission yeast homologue of Cdc7, and the replication fork stabilizing factors. We have presented evidence for physical interactions between Hsk1, Swi1, Swi3 and Mrc1. The interactions of Hsk1 with Swi1 and Swi3 occur in either swi3Δ or swi1Δ cells, respectively, as well as in mrc1Δ cells (Fig. 1 and 5). Similarly, Hsk1 also interacts with Mrc1 protein in the presence or absence of Swi1 protein (Fig. 1). The interactions do not involve DNA (Fig. 1). Synergistic genetic interaction between hsk1-89, a temperature-sensitive mutation of hsk1+, and swi1Δ or swi3Δ (Fig. 3) also supports our conclusions that Cdc7 plays an important role in maintaining the replication fork integrity in conjunction with these fork protection complex factors (Matsumoto et al. 2005). It should be noted that synthetic growth defect in the absence of a fork stress is observed in swi1Δ, but not in swi3Δ, although hsk1-89 swi3Δ double mutant is more sensitive to HU, MMS and UV than the single hsk1-89 mutant (Fig. 3). This may indicate that Swi1 and Swi3 play distinct roles in interaction with Hsk1. These results suggest that Hsk1 may interact with these individual proteins, or alternatively it may interact with a large replication fork complex containing helicase components as well as other regulatory factors (Nedelcheva et al. 2005; Gambus et al. 2006).
We have also shown that the chromatin binding of Mrc1 depends on Swi1 and Swi3 proteins (Fig. 4). Similar observation was previously reported in mammalian cells (Yoshizawa-Sugata & Masai 2007). Thus, Swi1-Swi3 may serve as a landing pad for chromatin binding of Mrc1 protein. Since chromatin localization of Mrc1 in the absence of replication stress also depends on Swi1 and Swi3, Mrc1 may be loaded onto chromatin in a manner dependent on Swi1-Swi3 even during the normal course of DNA replication. The increased chromatin binding of Mrc1 upon replication stress may be due to accumulation of S phase population or to recruitment of Mrc1 at the stalled replication forks. Chromatin binding of Swi1 and Swi3 only slightly increased after replication stress in the current experimental conditions (Fig. 4). In fact GFP-fused Swi1 or Swi3 was readily detected in the Triton-extracted nuclei after HU treatment (Noguchi et al. 2003, 2004). Thus, interaction of Mrc1 with Swi1 and Swi3 is likely to play a role in chromatin binding of Mrc1. However, the stable interaction of Mrc1 with chromatin may require interaction of Mrc1 with other fork proteins or stable replication fork structures (Wang et al. 2006).
Chromatin binding of Mrc1 as well as interaction of Mrc1 with Swi1-Swi3 was not significantly affected by hsk1 mutation (Fig. 4 and 5). In contrast, chromatin accumulation of Claspin in response to replication stress (HU) is severely impaired by Cdc7 depletion in human cells (Kim et al. 2008).
Regulation of stability of fork stabilization factors
Swi1 and Swi3 are relatively stable proteins and no significant degradation was observed for 6 hrs in the presence of cycloheximide (Fig. 6). In contrast, Mrc1 is unstable and more than 80% was degraded within one hr after addition of cycloheximide (Fig. 7). The abundance of Swi1 and Swi3 is interdependent and loss of either protein leads to a significant decrease of the other protein (Fig. 6), similar to what was previously reported in mammalian cells (Chou & Elledge 2006; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata & Masai 2007). As reported previously (Noguchi et al. 2004), Swi3 protein level dramatically decreased in swi1Δ, while the effect of swi3Δ on the Swi1 protein level was milder. Overall stability of Swi1 and Swi3 in the presence of cycloheximide was not significantly affected by the loss of their partner proteins. However, we noticed that the level of Swi1 or Swi3 in the Triton-soluble fraction is significantly decreased and degradation product could be detected in some cases (Fig. 4B, lanes 2, 5, 8, and 11). On the other hand, the protein levels in the Triton-insoluble fractions were less affected by the deletion. Thus, loss of either Swi1 or Swi3 causes selective destabilization of the partner protein in the chromatin-free fraction, while those bound to chromatin appears to be stable.
Abundance of Mrc1 significantly increased in hsk1-89 mutant. This appears to be due to the stabilization of the protein. Stability of Claspin/ Mrc1 is known to be regulated by cell cycle-dependent proteolysis (Yoo et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006). Although, we do not see significant cell cycle effect of hsk1-89 mutation, we cannot completely rule out the possibility that the hsk1 mutation indirectly affects the stability of Mrc1. There is a putative phosphodegron-like amino acid sequences, DSGVGS (859-864), on Mrc1. Mutation of the serine residues in the putative phosphodegron sequence resulted in stabilization of Mrc1 (Fig. 7E), suggesting that these serine residues could be the targets of phosphorylation. It remains to be determined whether Hsk1 targets these residues for degradation of Mrc1 The synthetic growth defect between mrc1-degSSAA and hsk1-89 (Matsumoto et al. unpublished data) may suggest that there may be other target sequences of Hsk1 on Mrc1 for destabilization. At this moment, we cannot entirely rule out the possibility that Mrc1 is destabilized in hsk1-89 due to some secondary effect on cell cycle. Further studies are required to understand precise roles of the possible Hsk1-mediated Mrc1 degradation during normal cell cycle progression and/ or in the replication checkpoint. It has been reported that checkpoint signaling may be eventually attenuated to resume cell cycle through a process called adaptation (Yoo et al. 2004; Nedelcheva et al. 2005). Hsk1 might play a role in shutting off the checkpoint signaling by promoting the degradation of Mrc1 in the late stage of checkpoint reaction (Fig. 7C). Role of Claspin degradation in shut-off of checkpoint responses have been proposed in other organisms (Yoo et al. 2004; Nedelcheva et al. 2005).
Interaction of Mrc1 with Swi1-Swi3 and Hsk1
The Mrc1 protein is required for replication stress checkpoint regulation and serves as an adapter in signal transduction from the Rad3 kinase to the Cds1 kinase. Mrc1 contains a segment (536-673) containing the clusters of SQ and TQ sites, putative targets of the Rad3 kinase. The Mrc1-6A mutant lacks the putative Rad3 target sequences and is deficient in checkpoint responses (Zhao et al. 2003). Examination of deletion derivatives of Mrc1 confirmed that the SQ/TQ clusters are essential for the checkpoint functions. The shortest form of checkpoint-proficient Mrc1 derivative contained the amino acids from 157-879. The segment 376-879 was partially checkpoint-proficient, supporting resistance to 3 mM HU but not to 6 mM. This segment also showed efficient binding to both Swi1 and Hsk1. As for the checkpoint functions, the segment 536-673 containing the SQ/TQ cluster is required but not sufficient for the binding. The presence of an adjacent segment at N-terminus or C-terminus of this segment resulted in weak binding, but the presence of the both segments was required for full-level binding. Similarly, weak HU resistance (growth on 3 mM HU only after overproduction) was observed with the segment 376-673 or 536-879, but higher resistance required the presence of the both segments surrounding the SQ/TQ cluster. Thus, these results suggest that Swi1-Swi3 and Hsk1 may interact with Mrc1 through the SQ/TQ cluster and the adjacent segments (either directly or indirectly), and that this interaction may be important for checkpoint signaling. We have observed that hyperphosphorylation of Mrc1 in response to HU requires Hsk1 (data not shown). It would be necessary to identify the phosphorylation events on Mrc1 during the course of replication fork stress checkpoint signaling.
Experimental procedures
General techniques of fission yeast
Methods for genetic and biochemical analyses of fission yeast have been described previously (Alfa et al. 1993).
Fission yeast strains
The following strains were used for this study: YM71 (h−), NI358 (h+ hsk1-89:ura4+), KO147 (h− hsk1-89:ura4+), EN3366 (h− swi3::kan), MS381 (h− hsk1-89:ura4+ swi3::kan), EN3182 (h− swi1::kan), MS307 (h− hsk1-89:ura4+ swi1::kan), SH0987 (h+ swi3-13myc:kan), MS404 (h− hsk1-3FLAG:kan mrc1-13myc:kan), MS405 (h− hsk1-3FLAG:kan mrc1-13myc:kan swi1::kan),KT2791 (h− mrc1-13myc:kan), EN3381 (h− swi1-3FLAG:kan), EN3404 (h− dfp1-13myc:kan), SH1007 (h+ dfp1-13myc:kan swi3-3FLAG:kan), SH1232 (h+ swi1-3FLAG:kan swi3-13myc:kan), SH1302 (h− hsk1-89:ura4+ swi1-3FLAG:kan swi3-13myc:kan), SH1914 (h− swi1-3FLAG:kan swi3::kan), SH3309 (h− swi1::kan swi3-13myc:kan), SH2303 (h+ swi1-3FLAG:kan swi3-13myc:kan mrc1::kan), SH2309 (h− swi3-13myc:kan mrc1::kan), SH2219 (h− swi1-3FLAG:kan mrc1-13myc:kan), SH2504 (h− swi1-3FLAG:kan mrc1-13myc:kan swi3::kan), SH1401 (h+ swi3-3FLAG:kan mrc1-13myc:kan), SH2611 (h+ swi1::kan swi3-3FLAG:kan mrc1-13myc:kan), SH1505 (h+ hsk1-89:ura4+ swi3-3FLAG:kan mrc1-13myc:kan), MS346 (h− mrc1-13myc:kan hsk1-89:ura4+), SH0603 (h− mrc1-13myc:kan cds1::ura4+), MS252 (h− mrc1::kan), KT2884 (h− mrc1-GFP:kan), MS369 (h− swi1::kan mrc1-GFP:kan), MS384 (h− swi3::kan mrc1-GFP:kan), MS424 (h− mrc1-3FLAG:kan), MS164 (h− mrc1-degSSAA-3FLAG:kan), MS165 (h− mrc1-degSSAA-3FLAG:kan). All the strains are leu1-32 and ura4-D18.
Preparation of extracts from yeast cells for immunoprecipitation
2-7 × 108 cells from 50-100 ml culture were harvested and washed once with PBS. The cells were then resuspended in 2 ml of SP buffer (1.2 M sorbitol, 0.1M potassium phosphate buffer [pH6.5], and 2 mM PMSF) and digested with Zymolyase. Spheroplasts were washed with SP buffer twice and resuspended in 1.4 ml of IP buffer (20 mM Hepes•KOH [pH7.6], 50 mM potassium acetate, 5 mM magnesium acetate, 0.1 M sorbitol, 0.1 % TritonX-100, 2 mM DTT, 20 mM NaOVa, 50 mM β-glycerophosphate, protease inhibitors). Cell suspensions were sonicated, and high speed supernatants were used for immunoprecipitation. Whole cell extracts from approximately 107 cells were made by the “boiling method”, as described previously (Takeda et al. 1999), for immunoblotting.
Immunoprecipitation
The primary antibody was bound to Protein G Dynabeads (Invitrogen, Inc.), as recommended by the supplier. Three hundred μl of the extracts were mixed with 5 μl of the antibody-bound Protein G Dynabeads and incubated at 4°C for 1 hr with continuous rotation. Beads were washed extensively with IP wash buffer (0.2 M NaCl, 125 mM Tris•HCl [pH7.4], 1 mM EDTA and 0.5% TritonX-100), and finally washed with 100 μl of 25mM Tris•HCl (pH 7.6). Proteins associated with the beads were analyzed by immunoblotting
Fractionation of cells into chromatin-soluble and chromatin-enriched fractions
Exponentially growing S. pombe cells in 100 ml culture were treated with 0.1% NaN3 and harvested. Cells were washed with Sorbitol Buffer (1.2 M sorbitol, 1 mM DTT, 10 mM Hepes•KOH [pH7.6], 40 mM potassium glutamate, 1 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 0.5× protease inhibitor cocktail, and 1 mM PMSF) and suspended in 4 ml of Sorbitol solution. Cells were treated with Zymolyase for 20 min at 30°C, and resulting spheroplasts were washed three times with Sorbitol Buffer. To the washed spheroplast pellet was added 2× volume of NE buffer (10 mM Hepes•KOH [pH7.6], 40 mM potassium glutamate, 1 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail, 0.1 M AEBSF (4-[s-aminoethyl]-benzenesulfonyl fluoride hydrochloride), 0.1 mM Na3VO4, and 50 mM NaF) containing 1% Triton-X100, and the suspension was kept on ice for 20 min. Unlysed spheroplasts were removed by low-speed centrifugation, and the supernatant was recovered as the total cell extracts, which were further centrifuged at a high speed to yield the supernatant as Triton-soluble fraction (chromatin-free). The pellets were washed once with NE buffer and the final pellet was recovered as Triton-insoluble fraction (chromatin-enriched). In situ chromatin binding assays were carried out as previously described (Kearsey et al. 2000).
Antibodies
Mouse anti-FLAG M2 monoclonal antibody (Sigma), mouse anti-cMyc (A-14) monoclonal antibody (Santa Cruz), mouse anti-α tubulin monoclonal antibody (B-5-1-2, Sigma), and rabbit anti-Hsk1 antibody (Masai et al. 1995) were used. Rabbit anti-Mrc1 antibody was developed against GST-tagged recombinant N-terminal (1-180) Mrc1 protein expressed in E. coli.
Construction of expression vectors for Mrc1 polypeptides
Various segments of the Mrc1 coding frame were amplified by PCR and subcloned into the pREP41X-3FLAG vector (Matsumoto et al. unpublished) which permits the addition of 3×-FLAG tag at the C-terminus of the insert. The polypeptides were expressed in various yeast strains and growth at different temperatures or in the presence of various concentrations of HU was determined on EMM plates.
Supplementary Material
Acknowledgements
We thank all the laboratory members for useful discussion. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.M.). E.N. is supported by the NIH grant GM077604. P.R. is supported by the NIH grant GM59447.
References
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 1993. [Google Scholar]
- Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 2003;5:1111–1116. doi: 10.1038/ncb1069. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Russell P. DNA replication checkpoint. Curr. Biol. 2001;11:R953–R956. doi: 10.1016/s0960-9822(01)00572-3. [DOI] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc. Natl. Acad. Sci. USA. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc. Natl. Acad. Sci. USA. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Duncker BP, Shimada K, Tsai-Pflugfelder M, Pasero P, Gasser SM. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA. 2002;99:16087–16092. doi: 10.1073/pnas.252093999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gotter AL. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with Timeless and disrupts its self-association. J. Mol. Biol. 2003;331:167–176. doi: 10.1016/s0022-2836(03)00633-8. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kakusho N, Yamada M, Kanoh Y, Takemoto N, Masai H. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2008;27:3475–3482. doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- Kim JM, Nakao K, Nakamura K, Saito I, Katsuki M, Arai K, Masai H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002;21:2168–2179. doi: 10.1093/emboj/21.9.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, Medema RH, Freire R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 2006;16:1950–1955. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex: In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J. Biol. Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, Tamai K, Ohtani K, Arai K. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- Nedelcheva MN, Roguev A, Dolapchiev LB, Shevchenko A, Taskov HB, Shevchenko A, Stewart AF, Stoynov SS. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J. Mol. Biol. 2005;347:509–521. doi: 10.1016/j.jmb.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR, III, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta A, Arai K, Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. USA. 2006;103:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFβTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Brown GW, Forsburg SL. Schizosaccharomyces pombe Hsk1p is a potential Cds1p target required for genome integrity. Mol. Cell. Biol. 2000;20:7922–7932. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 Are Components of a Novel S-Phase Response Pathway to Alkylation Damage. Mol. Cell. Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1+/dfp1+ encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Tatebayashi K, Ikeda H, Arai K, Masai H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, Elledge SJ, Li L. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004;117:575–588. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J. Biol. Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- Zhao H, Russell P. DNA binding domain in the replication checkpoint protein Mrc1 of Schizosaccharomyces pombe. J. Biol. Chem. 2004;279:53023–53027. doi: 10.1074/jbc.M410449200. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tanaka K, Noguchi E, Noguchi C, Russell P. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol. Cell. Biol. 2003;23:8395–8403. doi: 10.1128/MCB.23.22.8395-8403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.