Abstract
Several quassinoids were identified in a high-throughput screening assay as inhibitors of the transcription factor AP-1. Further biological characterization revealed that while their effect was not specific to AP-1, protein synthesis inhibition and cell growth assays were inconsistent with a mechanism of simple protein synthesis inhibition. Numerous plant extracts from the plant family Simaroubaceae were also identified in the same screen; bioassay-guided fractionation of one extract (Ailanthus triphylla) yielded two known quassinoids, ailanthinone (3) and glaucarubinone (4), which were also identified in the pure compound screening procedure.
The quassinoids are a group of several hundred complex nortriterpenoids originally isolated from plants in the family Simaroubaceae on the basis of their bitter taste.1 A number of quassinoids have been found to have antileukemic and other anticancer activity,2–5 most notably bruceantin, which advanced to phase II clinical trials in breast cancer and melanoma before failing due to toxicity (hypotension, nausea, vomiting and fever) and poor efficacy.6,7 Although the first pharmacologic studies of quassinoids identified inhibition of protein synthesis as a probable mechanism of action, recent workers have proposed a number of other mechanisms to explain the cell growth inhibition caused by these compounds.8 Among these are inhibition of plasma membrane NADH oxidase activity, induced differentiation of leukemia and lymphoma cells via c-myc down-regulation and G1 arrest,8,9 and mitochondrial membrane depolarization with caspase-3 activation.9,10 Quassinoids have also shown activity in antimalarial assays, and this activity has been correlated with inhibition of protein synthesis in the parasite.11 Quassinoids comprise a relatively homogeneous group of structures with a common tetracyclic skeleton, which can be categorized by the presence or absence of a bridging ether oxygen from carbon 17, and whether that bridge is connected to carbon 11 or 13.
We have conducted a high-throughput screening assay for inhibitors of the transcription factor activating protein-1 (AP-1).12 Among the confirmed hits in the screen were two purified quassinoids, as well as numerous extracts and their fractions from the Simaroubaceae. This contribution reports the evaluation of a set of quassinoids for specificity against the AP-1 target, the structure-activity requirements for such activity, as well as their patterns of cell growth inhibition in the NCI 60-cell screen and their effect on protein synthesis.
The two pure compounds initially identified in the AP-1 screen were 6α-tigloyloxyglaucarubol (1, NSC#374763)13 and 15-methylcarbamoyl-bruceolide (2, NSC#377097),14 however, samples of these compounds were not available for further testing due to limited supply. Other quassinoid samples were obtained from the NCI repository, and were checked for purity and identity by NMR spectroscopy. Of these, the ten compounds in Table 1 yielded spectra that matched literature values,15 and were judged to be of adequate purity for further biological characterization.
Table 1.
Activity of Pure Quassinoids in AP-1 Assay, and on Protein Synthesis Inhibition.
| NSC | name | AP-1 IC50 (µM) |
NCI-60 mean GI50 (µM) |
in vitro protein synthesis inhibition19 (µM) |
IC50, cellular inhibition of protein synthesis19 (µM) |
|
|---|---|---|---|---|---|---|
| 3 | 367114 | ailanthinone24 | 0.032 | 31 | 0.42±0.06 | 0.0034±0.0007 |
| 4 | 132791 | glaucarubinone25 | 0.040 | 1.8 | 0.29±0.04 | 0.27±0.05 |
| 5 | 341651 | 6α-senecionylchaparrin29 | 0.071 | 0.26 | 0.91±0.16 | 0.67±0.12 |
| 6 | 368671 | SUN 023730 | toxic | 0.024 | < 1031 | >231 |
| 7 | 165563 | bruceantin5 | toxic | 0.005 | 0.41±0.05 | 0.051±0.015 |
| 8 | 126765 | holacanthone32 | toxic | 0.03 | n.d. | n.d. |
| 9 | 344025 | sergiolide33 | toxic | 0.0038 | 0.07±0.0219 | ≪119 |
| 10 | 14974 | glaucarubol34 | inactive | 41 | >3019 | >219 |
| 11 | 36342 | quassin35 | inactive | not tested | >3019 | ≫219 |
| 12 | 172924 | brusatol36 | inactive | 0.26 | 0.2319 | ≪119 |
Each of the ten quassinoids was tested in dose response format using the screening assay.12 Compounds 3–5 inhibited β-lactamase under the control of the AP-1 promoter at nanomolar concentrations, while inhibiting cell growth less than 25 percent at 1 µM. Quassinoids 6–9 were toxic to cells at micromolar concentrations. Compounds 10–12 had no effect in these assays, suggesting that the apparent AP-1 inhibitory activity was actually due to cell killing.
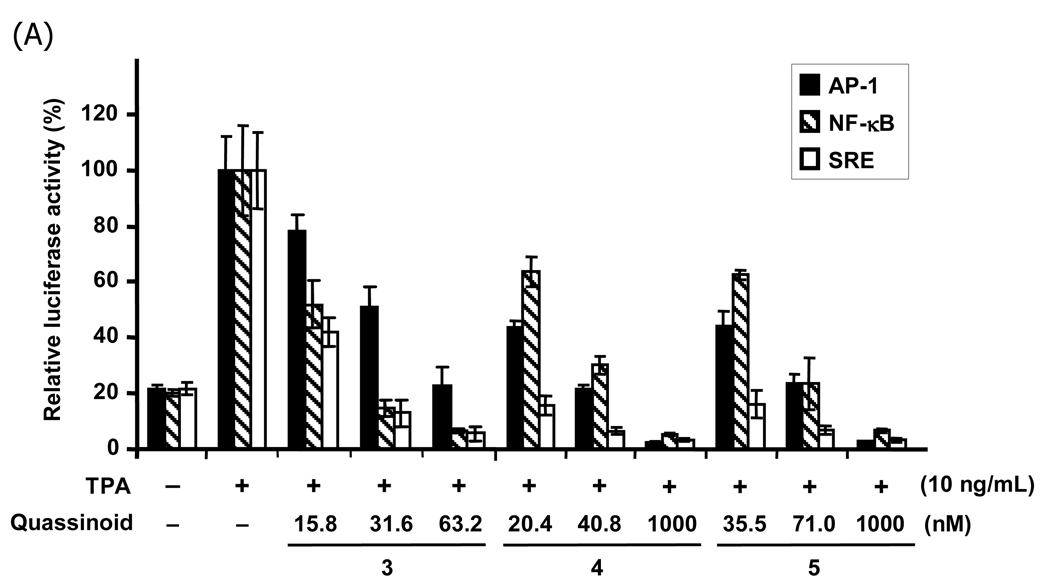
In order to confirm the specificity of the putative AP-1 inhibitors (3–5), they were tested further for their ability to inhibit TPA-induced nuclear factor kappa B (NF-κB). Both AP-1 and NF-κB transcription factors are important regulators in carcinogenesis,16,17 and are also inhibited by dominant negative c-Jun,18 while serum response element (SRE) is a target unaffected by the dominant negative c-Jun. Test concentrations were based on the apparent IC50s for AP-1 inhibition in the screening assay. The three AP-1 active non-toxic quassinoids 3–5 also affected the other targets dose-dependently (Figure 1A), while inactive quassinoids 11–12 did not show inhibition activity (Figure 1B). In addition, similar inhibition potencies for inhibitor of NF-κB (IκB)- α phosphorylation were seen with non-toxic quassinoids 3–5 (Figure 2). Thus, they are likely to be nonspecific transcription or translation inhibitors. However, phosphorylation at Tyr705 of Stat3, a well-known tyrosine kinase target, did not change in response to quassinoids 3–5 (Figure 2). Moreover, a significant differential in sensitivity was shown in the transcription reporters to quassinoids 3–5 (Figure 1A). Therefore, it may be possible that quassinoids 3–5 are selective inhibitors for one or a limited number of transcription factors by targeting transduction pathways, not as general inhibitors of all transcriptional or translational activity, in which case they would be expected to show severe toxicity.
Figure 1.
Inhibition of tetradecanoylphorbol acetate (TPA)-induced AP-1, NF-κB or SRE-dependent transcription reporter activity by quassinoids. Inhibition activity of quassinoids 3–5 (panel A) and non-inhibition with quassinoids 11–12 (panel B). HEK293 cells were transfected with AP-1, NF-κB, or SRE promoted luciferase reporter. The transfected cells were pre-treated with varied concentrations of quassinoids or DMSO for 1 h before 18 h stimulation by TPA (10 ng/mL) or DMSO as vehicle. Luciferase activities with TPA induction alone were set at 100% and the relative activities with TPA in the presence of quassinoids are shown. Assays were performed in quadruplicate.
Figure 2.
Quassinoids 3–5 inhibit phosphorylation of IκB-α but not Stat3. HEK293 cells were treated with various concentrations of quassinoids or DMSO for 1 h followed by induction with TPA (10 ng/mL) for 18 h. Endogenous protein expression was measured with whole cell extract by immunoblot analysis. Immunoblots were probed with antibodies for phospho-IκB-α and phospho-Stat3. Tubulin was used as a loading control.
To explore this further, compounds 3–5 were tested in cell-free and cellular assays of protein synthesis inhibition.19 All three compounds were inhibitory in both assays, but the potency in the cellular assay varied widely, from 3.4 nM for 3 to 670 nM for 5 (Table 1). Combined with previous results on quassinoids19 in the same assays, it is apparent that the AP-1 activity observed and inhibition of protein synthesis are not well correlated.
The quassinoid sergiolide (9)20 was previously observed to be active in the NCI 60-cell screen. Since that time, approximately 30 other quassinoids have been tested in the 60-cell screen, with mean GI50 values ranging from 1 nM to over 50 µM (Table S1, Supporting Information). We used the COMPARE algorithm21 to match the patterns for quassinoids 3–10 and 12 at the GI50 level. While compounds 3–8 formed a single cluster with mutual Pearson correlations between compounds greater than 0.45, compounds 9, 10 and 12 were not part of the cluster (Table S4, Supporting Information). It is apparent that pattern of cell growth inhibition as measured by the NCI 60-cell assay is not correlated to AP-1 inhibition, at least for this set of compounds.
A total of nine crude extracts prepared from plants in the family Simaroubaceae were found to be active in the AP-1 assay (Table S2, Supporting Information). Five genera were represented; all have been previously found to contain quassinoids (Ailanthus,3 Cedronia,20 Quassia,13 Simaba,22 and Simarouba23). Bioassay-guided fractionation of the organic extract of Ailanthus triphysa (Dennst.) Alston (Simaroubaceae) yielded ailanthinone (3) and glaucarubinone (4), which were identified by comparison of spectroscopic values to literature data.24,25 This species has not previously been noted to contain quassinoids. While the family Simaroubaceae has been shown to be polyphyletic in recent molecular analyses,26 the subfamily Simarouboideae, which contains all of the quassinoid-containing genera, remains intact as a monphyletic group within the family sensu lato.
In addition to the crude extracts, we also screened a library of prefractionated extracts which included samples from the Simaroubaceae. These are tabulated in Table S3, Supplemental Information. In addition to fractions from the crude extracts noted to be active above, samples from three other genera were found active. These also have been previously found to contain quassinoids (Brucea,5 Hannoa,13 Samadera4). The active samples were from organic extracts, with three exceptions, and the fractions were largely of intermediate polarity (e.g., B and C are the second and third fractions of five, in an elution of increasing polarity), which is consistent with the activity originating from quassinoids.
Using a high-throughput screen for samples that inhibit the function of the transcription factor AP-1, we have identified pure quassinoids, crude extracts, and fractions of crude extracts with activity in the assay. While the observed activity of the pure quassinoids was not considered to be specific for AP-1, it was also not well correlated with cell growth inhibition or with protein synthesis inhibition. Thus, a subset of quassinoids may possess the ability to modulate transcription factor activity independent of their inhibition of protein synthesis.
Inspection of the active structures leads to tentative structure-activity generalizations. The active compounds 1, 3, 4 and 5 all contain an ether bridge from C-11 to C-17. However, 2 possesses a C-17 to C-13 bridge. Thus, the AP-1 activity is mostly associated with C-17 ether bridging. The direction of ether bridge does not appear to affect cell growth inhibition or protein synthesis, as compounds with either connection inhibit both cell growth and protein synthesis. The lack of an ether bridge, as well as lack of ester substitution, lead to inactivity in all three assays, as was previously noted.19
The results for plant extracts and prefractionated samples indicate that prefractionation is a useful strategy for natural products screening. Screening the prefractionated samples identified plants that were not found by screening the crude extracts, presumably by either concentrating the quassinoid-containing material, or by removing cytotoxic materials that would mask activity in this assay.
Experimental Section
General Experimental Procedures
Optical rotations ([α]D) were obtained on a Perkin-Elmer 241 polarimeter in a 100 by 2 mm cell (units 10−1 deg cm2 g−1), while ultraviolet (UV) absorption spectra were obtained using a Varian Cary 50 Bio UV-Visible spectrophotometer. NMR experiments were performed on a Varian INOVA 500 MHz NMR spectrometer. 1H and 13C spectra were referenced to the deuterated solvent peaks. High-performance liquid chromatography (HPLC) was performed using a Varian ProStar 210/215 solvent delivery module equipped with a Varian ProStar 325 UV-Vis detector, operating under Star 6.41 chromatography workstation software.
Isolation of glaucorubinone (3) and ailanthinone (4) from Ailanthus triphysa
804 g of dried stem bark of A. triphysa (U44Z1548) was extracted to yield 20.19 g of organic extract. A portion (3.09 g) of the organic extract was taken through a solvent/solvent partition scheme as previously described.27 The EtOAc-soluble materials (251 mg) were further separated by VLC using C18 reversed-phase material eluting with a stepwise gradient (70% MeOH (aq), 80% MeOH (aq), 90% MeOH (aq), MeOH and CH2Cl2) to yield six fractions. The materials that eluted with 70% MeOH (aq) were then separated by reversed-phase HPLC (gradient 50–66% MeOH(aq) over 24 min.; ramp to 100% MeOH) to yield 3 (9.1 mg, 0.007%) and 4 (10 mg; 0.008%).
Transfection and Luciferase Reporter Assay
All expression plasmids were transiently transfected into HEK293 cells in a 96-well plate (0.1 µg/well) using Effectene transfection reagent (Qiagen), according to the manufacturer's instructions. HEK293 cells were maintained in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and incubated at 37 °C in a 5% CO2 incubator. After 24 h incubation, the medium was changed to DMEM with 0.2% FBS, and the transfectants were exposed to various concentrations of quassinoids and TPA (10 ng/mL). Cells were lysed with passive lysis buffer and the luciferase assay was performed using the firefly-luciferase reporter assay system (Promega) and an MLX luminometer (Dynex technology).
Plasmids
Three different luciferase reporter genes, AP-1 reporter containing four copies of the TRE consensus sequence originated from GCN4, and SRE reporter purchased from Stratagene, respectively were as described previously.28 The NF-κB reporter is driven by five copies of the NF-κB responsive element inserted into Cis-reporter backbone (Stratagene).
Immunoblot Assay
To measure endogenous level of several proteins in response to TPA induction and quassinoids, treated HEK293 cells were lysed with RIPA buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.1% sodium dodecyl sulfate, 0.1% deoxycholate, and protease inhibitor). Whole cell extracts were subjected to immunoblot analysis with phospho-IκB-α and phospho-Stat3 (Tyr 705) purchased from Cell Signaling.
Supplementary Material
Figure 3.
Acknowledgment
We would like to thank Djaja Doel Soejarto for identification of the Ailanthus specimen, Tom McCloud for plant extract preparation, David Newman for plant collection contracting, David Covell for COMPARE analyses, and the Drug Synthesis & Chemistry Branch, DTP, NCI, for repository samples of quassinoids. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funded in part by the Division of Cancer Treatment and Diagnosis, National Cancer Institute. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Dedicated to Dr. David G.I. Kingston of Virginia Polytechnic Institute and State University for his pioneering work on bioactive natural products.
Supporting Information Available. Tables of data for crude extracts and prefractionated samples, experimental details of translation assays, 60-cell summary and correlations for known quassinoids are available online at www.pubs.acs.org.
References and Notes
- 1.Polonsky J. Fortschr. Chem. Org. Naturst. 1973;30:101–150. doi: 10.1007/978-3-7091-7102-8_3. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh PC, Larrahondo JE, LeQuesne PW, Raffauf RF. Lloydia. 1977;40:364–369. [PubMed] [Google Scholar]
- 3.Ogura M, Cordell GA, Kinghorn AD, Farnsworth NR. Lloydia. 1977;40:579–584. [PubMed] [Google Scholar]
- 4.Wani MC, Taylor HL, Wall ME, McPhail AT, Onan KD. J. Chem. Soc., Chem. Commun. 1977:295–296. [Google Scholar]
- 5.Kupchan SM, Britton RW, Ziegler MF, Sigel CW. J. Org. Chem. 1973;38:178–179. doi: 10.1021/jo00941a049. [DOI] [PubMed] [Google Scholar]
- 6.Arseneau JC, Wolter JM, Kuperminc M, Ruckdeschel JC. Invest. New Drugs. 1983;1:239–242. doi: 10.1007/BF00208896. [DOI] [PubMed] [Google Scholar]
- 7.Wiseman CL, Yap HY, Bedikian AY, Bodey GP, Blumenschein GR. Am. J. Clin. Oncol. 1982;5:389–391. doi: 10.1097/00000421-198208000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Cuendet M, Christov K, Lantvit DD, Deng Y, Hedayat S, Helson L, McChesney JD, Pezzuto JM. Clin. Cancer Res. 2004;10:1170–1179. doi: 10.1158/1078-0432.ccr-0362-3. [DOI] [PubMed] [Google Scholar]
- 9.Morre DJ, Grieco PA, Morre DM. Life Sci. 1998;63:595–604. doi: 10.1016/s0024-3205(98)00310-5. [DOI] [PubMed] [Google Scholar]
- 10.Rosati A, Quaranta E, Ammirante M, Turco MC, Leone A, De Feo V. Cell Death Differ. 2004;11:S216–S218. doi: 10.1038/sj.cdd.4401534. [DOI] [PubMed] [Google Scholar]
- 11.Kirby GC, O'Neill MJ, Phillipson JD, Warhurst DC. Biochem. Pharmacol. 1989;38:4367–4374. doi: 10.1016/0006-2952(89)90644-8. [DOI] [PubMed] [Google Scholar]
- 12.Ruocco KM, Goncharova EI, Young MR, Colburn NH, McMahon JB, Henrich CJ. J. Biomol. Screen. 2007;12:133–139. doi: 10.1177/1087057106296686. [DOI] [PubMed] [Google Scholar]
- 13.Luyengi L, Vanhaelen M. Phytochemistry. 1984;23:2121–2123. [Google Scholar]
- 14.Lidert Z, Wing K, Polonsky J, Imakura Y, Masayoshi, Okano, Tani S, Lin YM, Kiyokawa H, Lee KH. J. Nat. Prod. 1987;50:442–448. [Google Scholar]
- 15.Hamburger MO, Cordell GA. Planta Med. 1988;54:352–355. doi: 10.1055/s-2006-962456. [DOI] [PubMed] [Google Scholar]
- 16.Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karin M, Cao Y, Greten FR, Li ZW. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 18.Young MR, Yang HS, Colburn NH. Trends Mol. Med. 2003;9:36–41. doi: 10.1016/s1471-4914(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 19.Fukamiya N, Lee KH, Muhammad I, Murakami C, Okano M, Harvey I, Pelletier J. Cancer Lett. 2005;220:37–48. doi: 10.1016/j.canlet.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Tischler M, Cardellina JH, II, Boyd MR, Cragg GM. J. Nat. Prod. 1992;55:667–671. doi: 10.1021/np50083a018. [DOI] [PubMed] [Google Scholar]
- 21.Monks A, Scudiero DA, Johnson GS, Paull KD, Sausville EA. Anticancer Drug Des. 1997;12:533–541. [PubMed] [Google Scholar]
- 22.Ozeki A, Hitotsuyanagi Y, Hashimoto E, Itokawa H, Takeya K, de Mello AS. J. Nat. Prod. 1998;61:776–780. doi: 10.1021/np980023f. [DOI] [PubMed] [Google Scholar]
- 23.Bhatnagar S, Polonsky J, Prange T, Pascard C. Tetrahedron Lett. 1984;25:299–302. [Google Scholar]
- 24.Kupchan SM, Lacadie JA. J. Org. Chem. 1975;40:654–656. doi: 10.1021/jo00893a024. [DOI] [PubMed] [Google Scholar]
- 25.Gaudemer A, Polonsky J. Phytochemistry. 1965;4:149–153. [Google Scholar]
- 26.Fernando ES, Gadek PA, Quinn C. J. Am. J. Bot. 1995;82:92–103. [Google Scholar]
- 27.Meragelman KM, McKee TC, Boyd MR. J. Nat. Prod. 1999;63:427–428. doi: 10.1021/np990570g. [DOI] [PubMed] [Google Scholar]
- 28.Suzukawa K, Weber TJ, Colburn NH. Environ. Health Perspect. 2002;110:865–870. doi: 10.1289/ehp.02110865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arisawa M, Kinghorn AD, Cordell GA, Farnsworth NR. J. Nat. Prod. 1983;46:218–221. doi: 10.1021/np50026a015. [DOI] [PubMed] [Google Scholar]
- 30.Honda T, Imao K, Nakatsuka N, Nakanishi T. U.S. Pat. 1987;4(665):201. [Google Scholar]
- 31.Chan J, Khan SN, Harvey I, Merrick W, Pelletier J. RNA. 2004;10:528–543. doi: 10.1261/rna.5200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viala B, Polonsky J. C. R. Acad. Sci. Paris, Ser. B. 1970;271:410–413. [Google Scholar]
- 33.Moretti C, Polonsky J, Vuilhorgne M, Prange T. Tetrahedron Lett. 1982;23:647–650. [Google Scholar]
- 34.Geissman TA, Ellestad GA. Tetrahedron Lett. 1962:1083–1088. [Google Scholar]
- 35.Valenta Z, Papadopoulos S, Podešva C. Tetrahedron. 1961;15:100–110. [Google Scholar]
- 36.Sim KY, Sims JJ, Geissman TA. J. Org. Chem. 1968;33:429–431. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.