Abstract
Ribbon synapses of the vertebrate retina are specialized synapses that release neurotransmitter by synaptic vesicle exocytosis in a manner that is proportional to the level of depolarization of the cell. This release property is different from conventional neurons, in which the release of neurotransmitter occurs as a short-lived burst triggered by an action potential. Synaptic vesicle exocytosis is a calcium regulated process that is dependent on a set of interacting synaptic proteins that form the so-called SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptor) complex. Syntaxin 3B has been identified as a specialized SNARE molecule in ribbon synapses of the rodent retina. However, the best physiologically-characterized neuron that forms ribbon-style synapses is the rod-dominant or Mb1 bipolar cell of the goldfish retina. We report here the molecular characterization of syntaxin 3B from the goldfish retina. Using a combination of reverse transcription (RT) PCR and immunostaining with a specific antibody, we show that syntaxin 3B is highly enriched in the plasma membrane of bipolar cell synaptic terminals of the goldfish retina. Using membrane capacitance measurements we demonstrate that a peptide derived from goldfish syntaxin 3B inhibits synaptic vesicle exocytosis. These experiments demonstrate that syntaxin 3B is an important factor for synaptic vesicle exocytosis in ribbon synapses of the vertebrate retina.
Keywords: vesicle fusion, SNAP-25, Synaptobrevin, VAMP, splicing, isoform, SNARE
INTRODUCTION
Conventional and ribbon synapses are the two types of chemical synapses in the nervous system. Conventional synapses are found in many different types of neurons, while ribbon synapses have a more restricted distribution. Ribbon synapses are found in photoreceptors and bipolar cells of the retina, cochlear and vestibular hair cells and in pinealocytes. A defining feature of all ribbon synapses is a specialized structure called the synaptic ribbon, which is a sheet- or disc-like structure that lies perpendicular to the synaptic plasma membrane and to which synaptic vesicles are tethered. The tethered synaptic vesicles are believed to be fusion competent and comprise the readily releasable pool of vesicles (Heidelberger et al., 2002a; von Gersdorff H. et al., 1996). Both ribbon synapses and conventional synapses release neurotransmitter by Ca2+regulated synaptic vesicle exocytosis. However, ribbon-style synapses release neurotransmitter in manner that is graded with membrane potential. By contrast, neurons that form conventional synapses, release neurotransmitter in an all-or-none fashion in response to an action potential.
Why do conventional and ribbon synapses have different release properties? Synaptic vesicle exocytosis in conventional synapses is mediated by the interaction between the synaptic vesicle protein synaptobrevin2/VAMP2 and the plasma membrane proteins syntaxin 1 and SNAP-25 (reviewed in: Rizo and Rosenmund, 2008; Südhof and Rothman, 2009). These vesicular and plasma membrane proteins interact via the so called SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) domains and are thought to catalyze synaptic vesicle exocytosis. Ribbon synapses in the mammalian retina contain synaptobrevin2/VAMP2 and SNAP-25, but lack syntaxin 1. Instead, they contain the related isoform syntaxin 3B (Curtis et al., 2008; Sherry et al., 2006; Morgans et al. 1996,). Syntaxin 3B is one of four different isoforms (named syntaxin 3A, 3B, 3C and 3D) that are generated by differential splicing of the mouse syntaxin 3 gene (Curtis et al., 2008). Syntaxin 3A and 3B have an identical N-terminal region but differ in the SNARE domain and the trans-membrane domain. Syntaxin 3C and 3D lack a SNARE domain and a transmembrane domain. Syntaxin 3B is the only splice-variant that is expressed in the mouse retina (Curtis et al., 2008).
The rod-dominant or Mb1 bipolar neuron of the goldfish retina has been used extensively to study the physiological properties of ribbon synapses. The large diameter of the synaptic terminal (8–12 µm) of this neuron enables one to measure presynaptic calcium currents, intracellular, presynaptic calcium concentration, and synaptic vesicle exo-and endocytosis simultaneously (Heidelberger and Matthews, 1992, von Gersdorff and Matthews, 1994, Heidelberger, 1998;). Given these technical advantages, this neuron has served as a model system for ribbon synapses in general (Heidelberger and Matthews, 1991; Heidelberger and Matthews, 1992; von Gersdorff and Matthews, 1994; Heidelberger and Matthews, 1994; Lagnado et al., 1996; Zenisek et al., 2000). Although the Mb1 bipolar neuron is considered an ideal model cell in which to study neurotransmitter release from ribbon synapses, little is known about the proteins that mediate synaptic vesicle exocytosis from this cell. Most of the studies analyzing the molecular composition of synaptic proteins in retinal ribbon synapses have been performed in mammals, specifically mice and rats (Curtis et al., 2008; Reim et al., 2005; Sherry et al., 2006; Sherry et al., 2003; von Kriegstein and Schmitz, 2003; von Kriegstein et al., 1999; Wang et al., 2003). The few studies that have analyzed synaptic proteins in goldfish retinal ribbon synapses have used antibodies that were originally developed for mammalian proteins. The specificity of these antibodies in non-mammalian species is unclear and consequently the expression of synaptic proteins in these synapses is controversial. An example is the case of the Ca2+ sensor for synaptic vesicle exocytosis, synaptotagmin 1. Different studies have found different synaptotagmin isoforms present or absent in ribbon synapses of the goldfish (Berntson and Morgans, 2003; Heidelberger et al., 2003).
We show here using molecular biological approaches that syntaxin 3B is expressed in Mb1 bipolar neurons. We than generated a specific antibody against syntaxin 3 and show that syntaxin 3 is located at the pre-synaptic plasma membrane of Mb1 bipolar neurons of the goldfish. Finally we demonstrate that syntaxin 3B is an important factor for synaptic vesicle exocytosis in these neurons.
EXPERIMENTAL PROCEDURES
Animals
Goldfish (Carassius auratus) (7–14 cm length) were maintained on 12 hour light/dark cyclic lighting. Goldfish were killed by decapitation and pithing. Tissue for frozen sections for immunohistochemical studies were obtained from light adapted goldfish at the time of euthanasia. For all other experiments tissue was obtained from goldfish that were dark adapted for 20 minutes before euthanasia. All animal procedures conformed to National Institutes of Health (NIH) guidelines and were approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston.
Cloning
RNA was isolated from goldfish retina; cDNA was generated by reverse transcription as described (Curtis et al., 2008). Syntaxin 3B was amplified from the goldfish cDNA by PCR using the primers derived from the N and C-terminus of the zebrafish syntaxin 3B (Primers: CAGAAATATGAAGGACCGACTGGAACAACTAAA and AAACACAACAATCCCCAGAATCGCACAGCAAAACAT). The resulting fragment was subcloned and sequenced. Independent sequence was obtained by direct sequencing of fragments generated by separate PCR reactions. A bacterial expression construct coding for a fusion protein between glutathione sulfotransferase (GST) and the N-terminus of the zebrafish syntaxin 3B (amino acids 2–178) was generated by PCR and cloned into pGEX-KG. A syntaxin 3A standard for the real time PCR was generated by PCR from goldfish cDNA, subcloned and sequenced (primers: GATTCAGGGATTTCCAAACAAG and ACTGCTTTACCCACATTCACCT). Goldfish and zebrafish sequences have been submitted to GenBank (accession numbers: GU189280 and GU189281).
Bioinformatics
Database searches were performed using the BLAST program suite at the NCBI website using the default settings without filtering. The protein sequences were aligned using the ClustalW and Boxshade programs using default settings at the Biological Workbench (http://workbench.sdsc.edu/).
Antibodies
The syntaxin 3 polyclonal antibody was raised in rabbits by Cocalico Biologicals Inc. (Reamstown, PA) against a GST (glutathione S-transferase) fusion protein containing amino acids 2–178 of recombinant goldfish syntaxin 3B that had been expressed in E. coli. GST antibodies were removed by passing the serum over GST that had been chemically crosslinked to glutathione sepharose. The antibody was further purified by affinity chromatography on GST-syntaxin 3B (amino acids 2–178) crosslinked to GST-sepharose. The purified syntaxin 3 antibodies were used at dilution of 1:1000. The monoclonal antibody against SV2 was obtained from the Developmental Studies Hybridoma Bank (University of Iowa) and was used at a dilution of 1:2000. Secondary antibodies were from goat and were conjugated to either Alexa Fluor 568, Alexa Fluor 488 (Molecular Probes, Eugene, OR) or to peroxidase for western blots. Secondary fluorescent antibodies were used at a dilution of 1:400. Antibodies for immunostaining were diluted in 10% normal goat serum, 0.1% Triton X-100 in PBS (pH 7.4).
Immunofluorescent labeling of frozen sections
Eyecups were fixed in 4% formaldehyde in phosphate-buffered saline (PBS; pH 7.4). After fixation, eyecups were rinsed several times in PBS (pH 7.4), cryoprotected in 30% sucrose in PBS, embedded in OCT embedding medium, and fast frozen. Tissue was sectioned on a cryostat at a thickness of 30 µm. Immunolabeling was performed on frozen sections as previously described (Heidelberger et al., 2003).
Isolation and immunolabeling of goldfish bipolar cells
Bipolar neurons were isolated as previously described (Heidelberger et al., 2002a). Cells were plated onto glass coverslips, allowed to adhere for 15–30 minutes, and then fixed with 4% formaldehyde in PBS (pH 7.4) for 15–30 minutes. Cells were rinsed several times with PBS, and blocked as described above. Primary antibody was applied overnight at 4°C. Cells were rinsed in PBS, and a fluorescently labeled secondary antibody was applied for from 45 minutes to 1 hour at room temperature. Cells were washed with PBS, mounted in Vectashield (Vector Laboratories, Burlingame, CA), and examined by fluorescence microscopy. The characteristic soma size and shape, stout primary dendrite, long axon, and large synaptic terminals were used to identify Mb1 bipolar cells.
Confocal microscopy
Immunolabeled specimens were scanned with a 0.2-µm step size on a Zeiss Laser Scanning Microscope 510 Meta (Carl Zeiss, Inc., Thornwood, NY) with a Zeiss 40× 1.3 NA Plan-NEOFLUAR oil objective. Image scale was calibrated and image brightness and contrast were adjusted as necessary to highlight specific immunolabeling. Images from the two fluorescence channels in single optical sections (0.3–0.5 µm thickness) or stacks of optical sections were superimposed to assess colocalization of labeling. Bleed-through between channels was eliminated by adjusting laser intensity and detector sensitivity or by scanning channels sequentially.
Reverse-transcription PCR
Total RNA was isolated from goldfish tissue with TRI Reagent (Ambion) using the manufacturer’s protocol. cDNA was generated using the Transcriptor First Strand cDNA Synthesis Kit (Roche). One microgram of total RNA from each tissue was used in this reaction. The synthesized cDNA was then used for PCR. The following parameters were used for the PCR cycles: 94°C, 45 sec.; 58 °C, 45 sec.; 72 °C, 1 min.; 35 cycles. PCR products were separated on a 2.0% agarose gel and, after visualization, isolated from the gel, purified, and sequenced by Seqwright (Houston, TX). Primers used were as follows: syntaxin 3A: AAAGACATTGTGCGTCTGGAG and CACCTCAATGTTATCAACCATGT; syntaxin 3B: GATTCAGGGATTTCCAAACAAG and GACTGGTCC ATGTTGTTCTCAA; Beta-actin: AAGATCTGGCATCACACCTTCTA and ATCACCAGAGTCCATCACGATAC.
Real-Time PCR
The cDNA used for this PCR was the cDNA used in the Reverse-Transcription PCR. Expression of syntaxin 3A and 3B in goldfish retina was analyzed with a Smartcycler II (Cepheid) by the SYBR Green method. For the analytical samples, each reaction was composed of 24 µl of master mix (SYBR Green Jumpstart Taq Ready Mix (Sigma), 1.0 µM primers, and deionized water) and 1 ul of retina cDNA (1.0 g/l). For the standards, each reaction consisted of 24 µl of master mix (SYBR Green Jumpstart Taq Ready Mix (Sigma), 1.0 µM primers, and deionized water) and a defined amount of syntaxin 3A or 3B plasmid and 1ug of genomic E.coli DNA as carrier in a total volume of 25 µl. The cycling parameters for the reaction with syntaxin 3B were as follows: 94°C 2 min., 94°C 45 sec., 58 °C 1 min., 72 °C 1 min. 35 cycles. The cycling parameters for the reaction with syntaxin 3A were as follows: 94°C 2 min., 94°C 45 sec., 56 °C 1 min., 72 °C 1 min. 35 cycles. The primers for this PCR were the same ones that were used for the Reverse-Transcription PCR.
Single-cell reverse transcription PCR
Goldfish retina was dissociated as described (Heidelberger and Matthews, 1992). Single cells with the morphological characteristics of Mb1 bipolar neurons or horizontal cells were picked up with a pipette by applying gentle suction by either opening the pressure valve to atmospheric pressure or by gentle suction by mouth. The individual cells were then deposited into a PCR tube containing deionized water and frozen by dipping the tube into into liquid nitrogen. The cell was stored at −80 °C until needed. To generate cDNA, the cell was thawed and four microliters of the cell lysate was used for the reverse transcription reaction. The Transcriptor First Strand cDNA Synthesis Kit (Roche) was used to generate cDNA for PCR. Two rounds of PCR were performed. At the end of the first PCR, aliquots of the reactions were transferred to new tubes. Fresh PCR reagents were added to the tubes and a second PCR was performed. The following parameters were used for the PCR: 94°C 2 min., 94°C 45 sec., 58 °C 1 min., 72 °C 1 min., 72 °C 10 min. 40 cycles. The primers for this PCR were the same ones that were used for the Reverse-Transcription PCR.
Electrophysiology
Synaptic terminals of bipolar neurons were isolated as previously described (Heidelberger and Matthews, 1992) Briefly, 3–5” dark-adapted goldfish were decapitated and their eyeballs were enucleated. The eyeballs were dissected in oxygenated low calcium ringers solution containing (in mM), 120 NaCl, 2.6 KCl, 1.0 MgCl2, 0.5 CaCl2, 10 HEPES, 10 glucose (pH- 7.25–7.3 and 260–270 mosm). The retinas were removed cut into 8–10 pieces and incubated for 30 minutes at 20°C in a digestion solution containing (in mM), 115 NaCl, 2.5 KCl, 1.0 MgCl2, 0.5 CaCl2, 10 PIPES, 10 glucose, 2.7 cysteine, papain 30units/ml (pH- 7.25–7.3 and 260–270 mosm). After digestion the pieces were rinsed several times in the low calcium ringers solution and were stored at 10°C for up to 6–8 hours. Pieces were then triturated and plated onto recording dishes as needed. Electrophysiological recordings were made from isolated Mb1 type bipolar neuron synaptic terminals (Heidelberger and Matthews, 1992; von Gersdorff and Matthews, 1994; Heidelberger et al., 1994; Heidelberger, 2001). All experiments were performed at room temperature (20–21°C).
The extracellular recording solution was similar to the low calcium ringers solution except that the calcium concentration was increased to 2.5mM. The standard internal solution contained: (in mM) 100 CsGluconate, 10 TEA, 6.0 MgCl2, 5 EGTA, 2.5 CaCl2, 35 HEPES, 5.0 Na2 ATP, 0.5 GTP, pH 7.25–7.3 and 260–270 mosm. This solution was calculated to buffer the intracellular calcium concentration to ~ 150 nM (Maxchelator; http://maxchelator.stanford.edu) and verified experimentally (see also (Heidelberger et al., 2002a; Heidelberger et al., 2002b)). 0.5 mM of the syntaxin 3B SNARE peptide (Sequence: RHKDIMRLESSIKELHDMFVDVA) or the scrambled control peptide (Sequence: RIALKDDVIHMRESVDHKSFMEL) was dissolved in the standard internal solution. The final concentration of the peptide was reduced to 0.25mM by diluting the peptide containing internal solution with equal volume of standard internal solution. To minimize effects of osmolarity changes on synaptic vesicle dynamics (Heidelberger et al., 2002b), the osmolarity of internal solutions containing the peptides was readjusted to 260–270 mosm following addition of peptide. 7µl of the internal solution was loaded into each pipette.
For whole-terminal capacitance recordings, patch pipettes of 5–7 mΩ were made from unfilamented borosilicate glass and coated with sylgard. An EPC-9 patch clamp amplifier controlled by Pulse software (ver 8.53, HEKA Electronik, Lambrecht, Germany) was utilized. For capacitance measurements, a sine wave voltage command (805 Hz, 15 mV peak amplitude) was applied about a holding potential of −60 mV. The Lindau-Neher technique (Gillis KD, 1995; Lindau and Neher, 1988) was used to calculate values for the membrane capacitance (Cm), series conductance (Gs) and membrane conductance (Gm). A train of four, 1 s depolarizing pulses (−60mV to 0mV), with an inter-pulse interval of 60s, was given. ΔCm recorded after each pulse was normalized to that of the first pulse. The average leak current between terminals dialyzed with the syntaxin 3B SNARE peptide and the scrambled control peptide were not statistically different. (33 ± 9 pA and 24 ± 5 pA respectively, n=5 for both groups). The series conductance was also not statistically different between the two groups. (73.7 ± 1.25 nS for terminals dialyzed with SNARE peptide, and 64.8 ± .6 nS for terminals dialyzed with the scrambled control peptide, n = 5 for both groups.)
Fluorescence measurements
To establish that fluorescein-tagged peptides entered the synaptic terminal from the internal recording solution, a computer-controlled photometry system (ASI/TILL Photonics.) was used to record fluorescein fluorescence from each patched synaptic terminal. Intra-terminal fluorescence started to increase immediately after break-in and a plateau in fluorescence was typically seen within 2–3 mins.
Data analysis
Data files from the electrophysiological recordings were exported to Igor Pro software (Wave Metrics) for analysis. All data are expressed in mean ± s.e.m. Comparisons between two groups were analyzed using unpaired t-test. A p value < 0.05 was considered significant.
RESULTS
Molecular Characterization of syntaxin 3B in the goldfish
As a first step towards assessing the functional roles of SNARE proteins at a retinal ribbon synapse, we have characterized the sequences of syntaxin 3B from zebrafish (Danio rerio) and goldfish (Carassius auratus). The EST database from zebrafish was screened using the program tblastn (NCBI) with the mouse syntaxin 3B sequence. One EST clone derived from a retina cDNA library which contained a sequence homologous to syntaxin 3B (IMAGE clone: 4786252) was identified and completely sequenced.
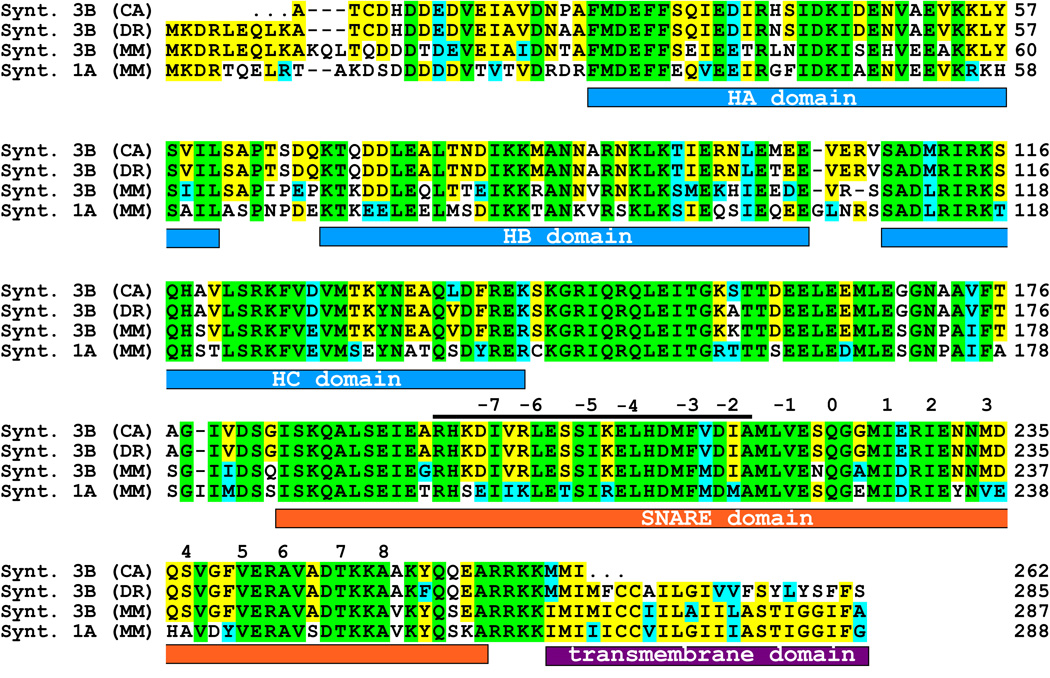
This clone contained a reading frame for the majority of the protein, but was lacking part of the n-terminus. In order to complement the sequence we screened the genomic zebrafish database and compared the sequence of our partial clone with the corresponding syntaxin 3 gene (Ensembl gene ID: ENSDARG00000001880 (WWW.ENSEMBL.ORG)). The exon/intron structure of the zebrafish syntaxin 3 gene is depicted in supplementary figure S1 and is very similar to the mouse gene (Curtis et al., 2008). The two main forms, syntaxin 3A and 3B, are generated by differential splicing of exons 8, 9 and 10, similar to the mouse (figure S1). The genomic sequence of the zebrafish was used to complement the region coding for the N-terminus of the predicted zebrafish syntaxin 3B protein. Reverse transcription (RT)-PCR using primers derived from the syntaxin 3B zebrafish sequence was then used to amplify the goldfish syntaxin 3B cDNA from mRNA isolated from goldfish retina. The obtained PCR fragments were sequenced and used to predict the goldfish syntaxin 3B protein sequence. We aligned the amino acid sequence of the predicted goldfish syntaxin 3B with mouse syntaxin 3B, mouse syntaxin 1A (the syntaxin isoform found in conventional synapses), and the predicted zebrafish syntaxin 3B sequence (Figure 1). The full-length protein sequences of the syntaxin 3B proteins were 75% identical between mouse and zebrafish, and the SNARE domains were 90% identical. Thus, syntaxin 3B is strongly conserved between mammals and fish. The sequence of the goldfish syntaxin 3B protein is highly homologous to the zebrafish syntaxin 3B sequence (98 % identity).
Fig. 1.
Sequence alignment of the different syntaxin isoforms. The protein sequences of the goldfish (CA) syntaxin 3B, zebrafish (DR) syntaxin 3B and the mouse (MM) syntaxin 3B and syntaxin 1A have been aligned for maximal homology using CLUSTALW. Sequences are identified on the left and residues numbered on the right. Residues that are conserved in all four proteins are labeled with green background. Residues conserved between three of the syntaxin isoforms are labeled with yellow background. Conservative exchanged residues are labeled with blue background. The positions of the conserved domains are marked below the sequence. The positions of the hydrophobic interacting layers are numbered in relation to the glutamine (Q) of the central 0 layer. The position of the peptide sequence used for the electrophysiological experiments is marked with a bar above the sequence.
Syntaxin 3B is expressed in bipolar neurons of the the goldfish retina
To evaluate the expression pattern of the syntaxin 3 gene in the fish, we searched the EST database from zebrafish using the syntaxin 3B cDNA sequence. We identified 20 EST-clones that could clearly be identified as syntaxin 3 transcripts. Most EST sequences corresponded to the 5’-end of the syntaxin 3B transcript, which is identical between syntaxin 3A or 3B; therefore, they could correspond to clones of syntaxin 3A or 3B. In contrast, no zebrafish EST clones corresponding to syntaxin 3C or 3D were found in the search. This result indicates that these spliceforms are either expressed at very low levels or not at all. Of the 20 identified EST clones, the majority of these EST clones were derived from mRNA isolated from embryonic or adult whole animals (9 clones). Among the clones generated from mRNA of defined tissues, 4 clones were from eye/retina, 3 from kidney, and the rest were from a variety of other tissues (brain, gut, olfactory ephitelium and ovary (one clone each)). This distribution pattern mirrors the expression pattern found for the syntaxin 3 gene in the mouse, with the highest level of expression of the syntaxin 3 gene in the retina and the kidney.
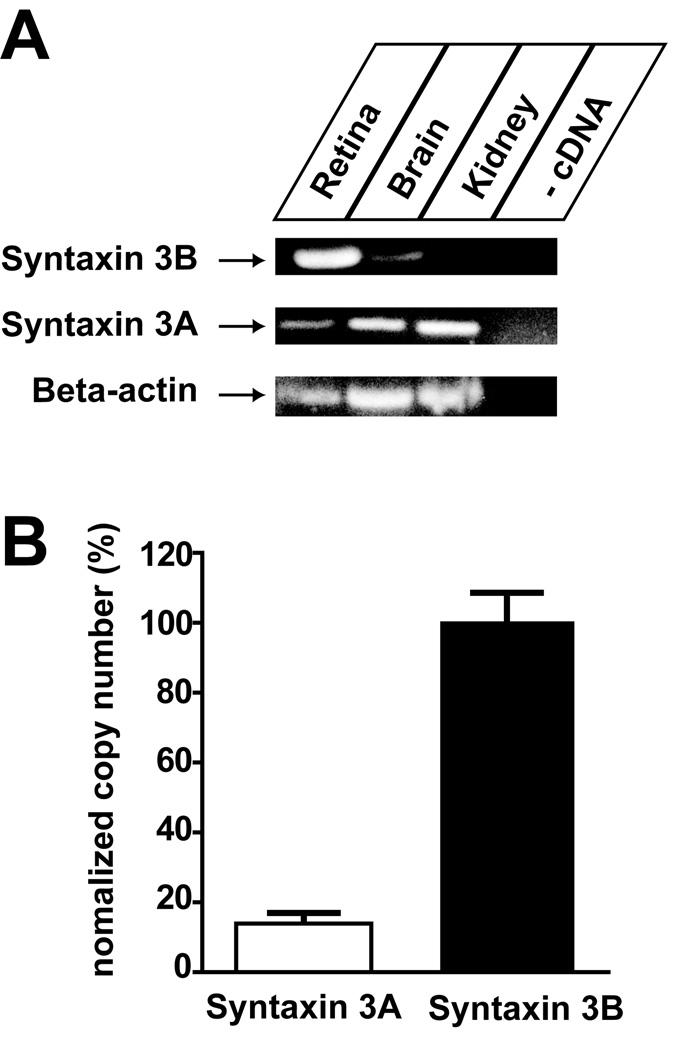
Next, the tissue distribution of syntaxin 3A and 3B in the goldfish was investigated (Figure 2). RT-PCR analysis of mRNA isolated from retina, brain and kidney was performed using primers specific for syntaxin 3A and 3B. The PCR reaction with the syntaxin 3B primers produced a strong signal in the goldfish retina and a weak signal in the brain sample. No syntaxin 3B could be detected in the kidney samples. In contrast, PCR reactions using the syntaxin 3A primers generated strong signals in the kidney and brain, and a weak signal in the retina sample. To compare the expression level of syntaxin 3A to syntaxin 3B in the retina, we used quantitative real-time PCR to measure the copy number of each of the two different syntaxin 3 transcripts (Figure 2B). The results of the experiment demonstrate that syntaxin 3B mRNA is about 7 times more abundant than syntaxin 3A mRNA. This demonstrates that syntaxin 3B is the major syntaxin 3 form expressed in the goldfish retina.
Fig. 2.
Expression of Syntaxin 3B in the Goldfish. A. Reverse-transcription PCR was performed to analyze the expression of Syntaxin 3A and 3B and beta actin in retina, brain and kidney. B. Real time PCR analysis of expression of syntaxin 3 A and 3B in goldfish retina. Data have been normalized to the value of syntaxin 3B. (Syntaxin 3A: 14.0 % ± 2.2 % (n=6); Syntaxin 3B: 100 % ± 9.5 % (n=6) (mean +/− s.e.m.)).
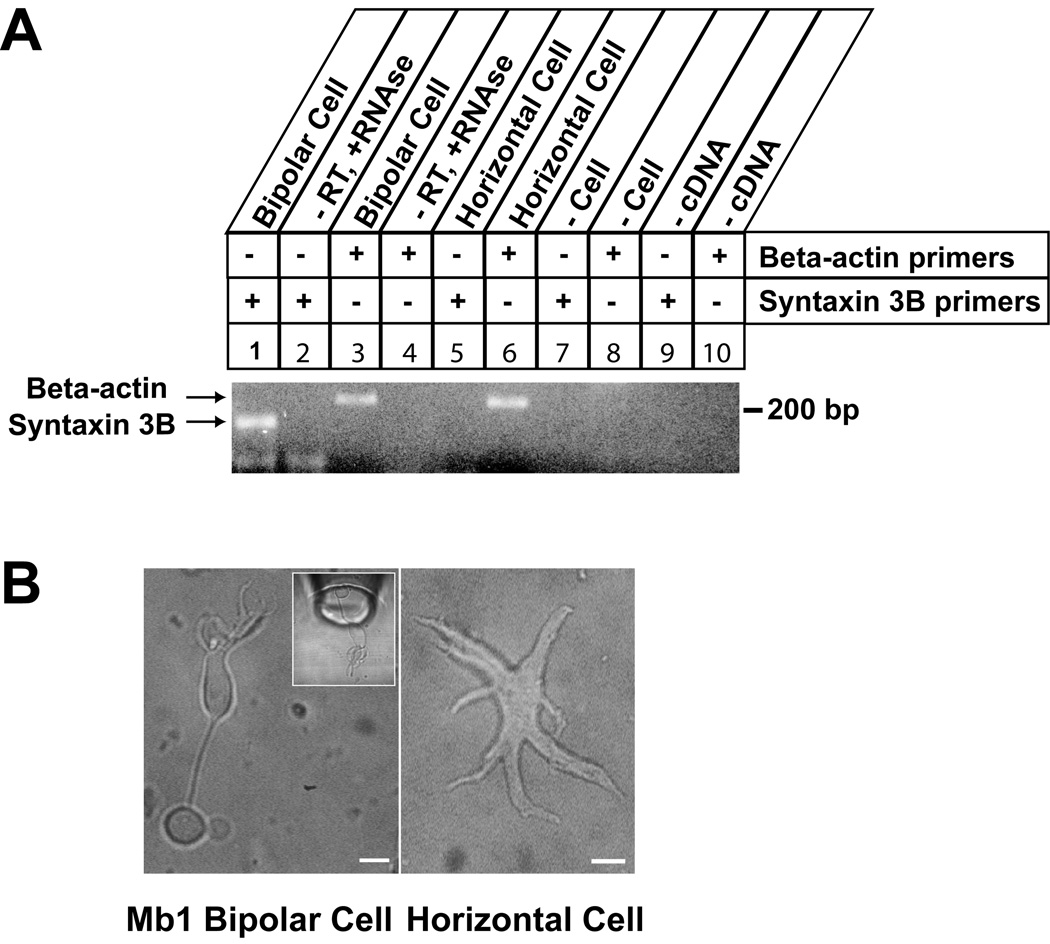
As a first step towards identifying which cell types in the goldfish retina express syntaxin 3B, RT-PCR analysis was performed using mRNA from single cells isolated from dissociated retina. Cells were classified as Mb1 goldfish bipolar neurons or as horizontal cells based on morphological criteria (Heidelberger and Matthews, 1992; Dowling, 1987). The analysis showed that the Mb1 bipolar neurons express syntaxin 3B mRNA. In contrast, syntaxin 3B mRNA could not be detected in the horizontal cells under the same conditions (Figure 3).
Fig. 3.
Expression of syntaxin 3B in isolated retina bipolar cells. A. Single-cell Reverse-Transcription PCR was performed to confirm the presence of Syntaxin 3B in the Mb1 bipolar cell. Primers used for RT-PCR are marked on the right. Arrows mark the position of the specific PCR products. Input is labeled on top. Controls lane 2 and 4 show PCR reactions performed on mRNA from bipolar cells without reverse transcriptase and with added RNase A. Controls lanes 7 and 8 show PCR reactions performed on bath solution without cells. B. Representative pictures of an Mb 1 bipolar cell (left) and a horizontal cell (right) collected for single cell PCR. Inset is taken from an actual single cell PCR experiment and shows a bipolar cell being taken up into a collection pipette. Scale bars are 10µm.
Syntaxin 3 protein is located in ribbon synapses in the goldfish retina
In order to investigate the distribution of syntaxin 3 protein in the goldfish retina, we generated a specific antibody against a recombinant protein that consisted of the N-terminus of syntaxin 3 fused to GST. This syntaxin 3 epitope is identical in syntaxin 3A and 3B; therefore, the antibody is predicted to recognize both isoforms of syntaxin 3. However, the real time PCR results indicate that syntaxin 3B is the major form of syntaxin 3 expressed in the goldfish retina. Thus, the majority of the protein recognized by the antibody should correspond to syntaxin 3B.
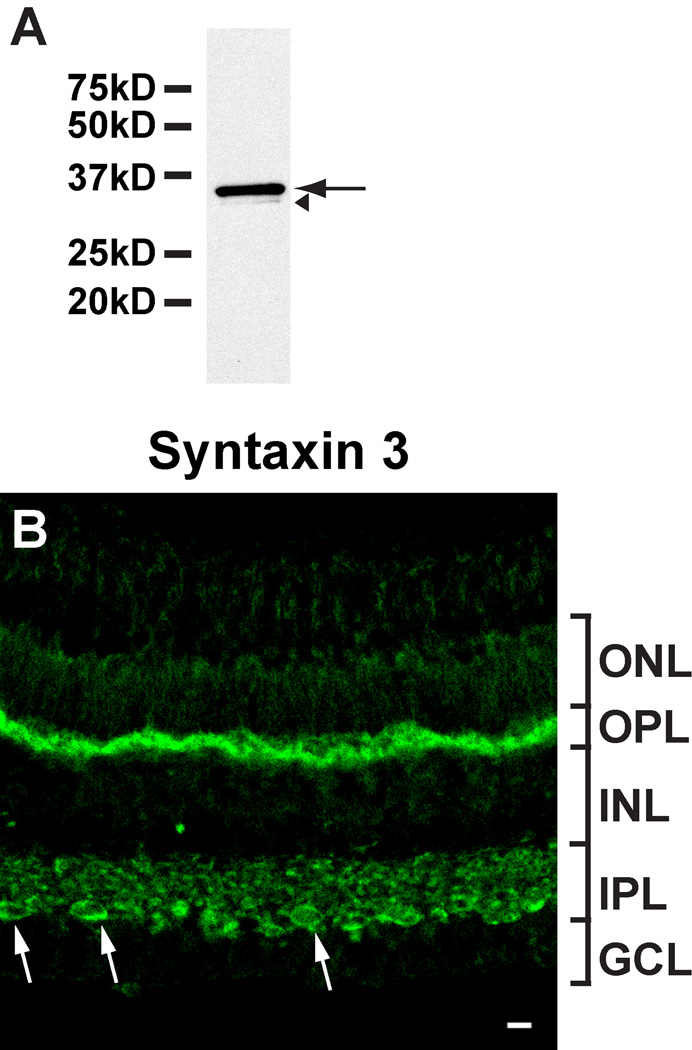
The antibody was affinity purified by using the antigen and tested by western blot analysis of goldfish retina extract and recombinant GST-syntaxin fusion proteins. The purified antibody strongly reacted with a protein of about 32 kD (the predicted size of syntaxin 3B) in goldfish retina extract (Figure 4A). The antibody did react strongly with goldfish GST-syntaxin 3B and weakly with mouse GST-syntaxin 3B. In contrast, the antibody did not crossreact with mouse GST-syntaxin 1A indicating that it is specific for syntaxin 3 (supplementary figure S2). The purified antibody was then used to investigate the distribution of syntaxin 3 in the goldfish retina. The syntaxin 3 antibody strongly labeled the inner and outer plexiform layers in the goldfish retina (Fig.4B). Strongly labeled puncta were visible throughout the inner plexiform layer (IPL). In the innermost IPL, strongly labeled terminals showed the distinctive size, morphology, and location of Mb1 bipolar cell terminals. Labeling in the OPL, which houses the ribbon synapses of the photoreceptors, was also strong. The staining pattern indicates that syntaxin 3 in the goldfish retina is found in ribbon synapse-containing synaptic layers. This is similar to distribution that has been observed in the mouse (Curtis et al., 2008).
Fig. 4.
Syntaxin 3 is located in ribbon synapses in the goldfish retina. A. The purified syntaxin 3 antibody strongly reacted with a protein of the predicted size of syntaxin 3B in goldfish retina extract (arrow). A very weak, slower migrating band (labeled by an arrowhead) probably corresponds to a breakdown product. B. The syntaxin 3 antibody labels synaptic layers (inner and outer plexiform layers (IPL, OPL)) in a section of goldfish retina. Potential Mb1 bipolar terminals are marked by arrows. (ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer). Scale bar = 10 µm for all panels.
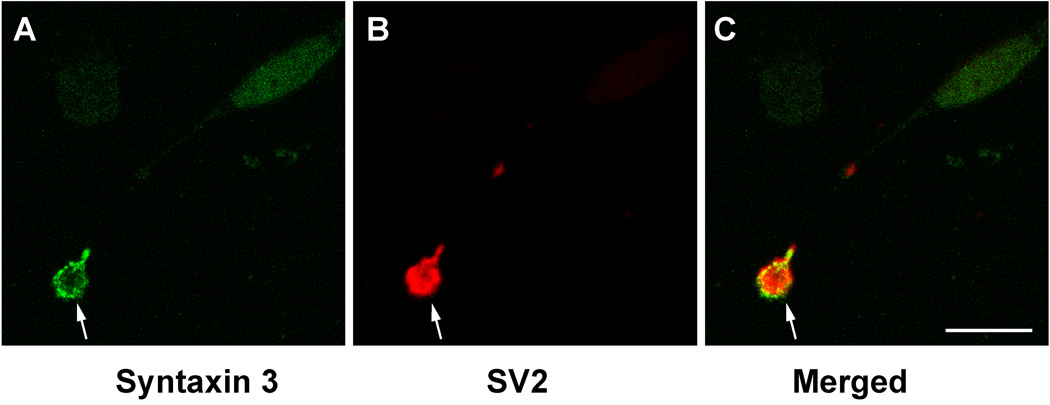
To confirm that the large terminals labeled in the innermost IPL correspond to Mb1 bipolar terminals, we enzymatically dissociated goldfish retinas and immunolabeled the dissociated cells for syntaxin 3 and the synaptic vesicle marker protein SV2. Mb1 bipolar neurons were then identified by their characteristic morphology, and analyzed by confocal microscopy (Fig. 5). The Mb1 bipolar cell terminals showed strong syntaxin 3 labeling of the plasma membrane (Fig. 5 A,C) and strong labeling for SV2 proteins within the terminals (Fig. 5 B,C). The labeling of syntaxin 3 on the plasma membrane was not uniform but rather in patches indicating the presence of clusters of syntaxin 3 on the synaptic plasma membrane. Very little overlap between the syntaxin 3 signal inside the terminal and the SV2 signal could be detected, indicating that the majority of the syntaxin 3 is located at the synaptic plasma membrane.
Fig. 5.
Syntaxin 3 is located at the plasma-membrane of Mb1 bipolar terminals. Confocal image of a bipolar terminal labeled with antibodies against syntaxin 3 (A,C) and the synaptic vesicle marker SV2 (B,C). An optical section of 0.5 µm is depicted. The terminals of isolated Mb1 bipolar cells show strong labeling for Syntaxin 3B on the plasma-membrane (A,C). Arrow points to the synaptic terminal. Scale bar = 10 µm for all panels.
Syntaxin 3B is essential for synaptic vesicle exocytosis in goldfish bipolar cells
In order to study the role of syntaxin 3B in neurotransmitter release in a ribbon synapse, we designed a peptide derived from goldfish syntaxin 3B (figure 1). The peptide contained two conservative exchanges to the goldfish sequence (peptide sequences listed in the methods). The peptide covered the N-terminal half of the SNARE domain and was coupled to fluorescein to enable us to monitor the loading of the peptide into the synaptic terminal. We refer to it as the syntaxin 3B SNARE peptide. A scrambled peptide served as a control.
One minute after gaining whole-cell access to the synaptic terminal, a sequence of four depolarizing pulses (−60 mV to 0 mV, each 1 s duration) were given. Each depolarization was sufficient to deplete the releasable pool of synaptic vesicles (Heidelberger, 1998; von Gersdorff H. and Matthews, 1997). Refilling of the releasable pool following a depleting stimulus takes about 20 s under standard conditions (τ ≈ 7 s.,e.g. Heidelberger et al., 2002b; von Gersdorff H. and Matthews, 1997). To allow sufficient time for refilling, the interpulse interval was set to 60 s.
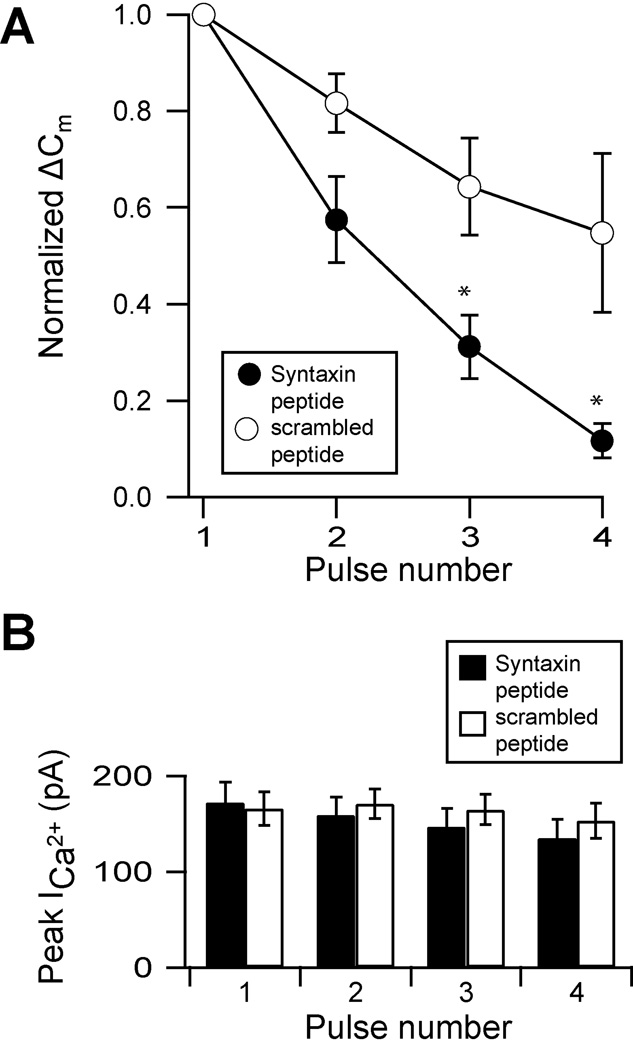
The amplitude of the exocytotic response evoked by the first depolarization was not significantly different between cells dialysed with the syntaxin 3B SNARE peptide (ΔCm = 138 ± 27.4 fF, n= 5) and the scrambled peptide (ΔCm = 162 ± 22.2 fF, n=5), This amplitude was also consistent with the reported magnitude of the releasable pool of synaptic vesicles (Heidelberger et al., 2005), indicating that neither peptide initially altered the exocytotic response. uent stimulation, terminals dialyzed with the scrambled peptide displayed a mild rundown in exocytosis (see also Augustine and Neher, 1992; von Gersdorff and Matthews, 1994). However, terminals dialyzed with the syntaxin 3B SNARE peptide showed a pronounced, progressive decline in the exocytotic response (figure 6A). By the fourth stimulus, exocytosis was reduced by ≈ 89% in terminals with the syntaxin 3B SNARE peptide (figure 6A; black circles), whereas it was reduced by only ≈ 45% with the scrambled peptide (figure 6A, white circles). This difference was significant (p < 0.05).
Fig. 6.
A peptide derived from the SNARE domain of syntaxin 3B inhibits functional refilling in bipolar neurons. A. Isolated terminals were dialyzed with internal solution containing either the syntaxin 3B SNARE peptide (filled circles, n = 5) or a scrambled control peptide (open circles, n = 5.) Four 1 s depolarizing pulses (−60 to 0 mV) were given with an interpulse interval of 60 seconds. The change in membrane capacitance (ΔCm) measured for each pulse was normalized to the magnitude of the response to the first pulse. Data are expressed in mean ± s.e.m. p-values < 0.05 are marked with asterisks. B. The successive decrease in exocytosis is not due to decreased calcium influx. There was no significant difference in the mean peak amplitudes of the calcium current between cells dialyzed with the syntaxin 3B SNARE peptide (black bars, n = 5) and those dialyzed with the scrambled control (white bars, n = 5.)
To address whether the effect of the SNARE peptide on exocytosis was mediated by changes in calcium entry, we analyzed the calcium currents evoked by each pulse in the train. The data demonstrate that there was no difference in the peak current amplitude in terminals dialyzed with scrambled peptide versus the SNARE peptide (figure 6B). Thus, it is unlikely that the syntaxin 3B SNARE peptide disrupted exocytosis via an effect on calcium entry.
DISCUSSION
Syntaxin 3 is an evolutionary conserved gene
The analysis of the syntaxin 3 gene in goldfish revealed the same intron/exon structure as in the mouse. In mouse and goldfish, two major forms, syntaxin 3A and 3B are generated by differential splicing. The expression pattern is also comparable between fish and mice, with syntaxin 3B being the major form expressed in the retina and syntaxin 3A being mainly expressed in non-neuronal tissues such as kidney. In the mammalian system, syntaxin 3A has been shown to be involved in the trafficking of vesicles from the trans-golgi to the apical membrane of epithelial cells (Low et al., 1998). The high expression level of syntaxin 3A in the goldfish kidney suggests that syntaxin 3A is likely involved in intracellular trafficking in fish epithelial cells.
Goldfish are thought to be tetraploid, in contrast to the diploid zebrafish (Ciudad et al., 2002). Several cases of duplications of genes in the goldfish have been described. In the case of the t-SNARE protein SNAP-25, one gene has been identified in mammals; two very similar genes coding for two closely related forms of SNAP-25) named SNAP-25A and B have been found in the zebrafish and three genes have been described in the goldfish (Risinger and Larhammar, 1993; Risinger et al., 1998). We have analyzed the most recent release of the zebrafish genome database for syntaxin 3 homologous genes. Besides the gene coding for the syntaxin 3B described above, another predicted gene was discovered that could encode a protein with 71% identity to the mouse syntaxin 3B, but only a 58% identity to the mouse syntaxin 1A (VEGA gene ID: OTTDARG00000027188 (http://vega.sanger.ac.uk/Danio_rerio/index.html). Is this syntaxin3-related gene also expressed in the retina? Searches of zebrafish EST databases with the predicted transcript sequence of the syntaxin 3 related gene detected only two homologous EST clones derived from embryonic tissue and ovaries. This pattern indicates, in comparison to the large number of EST clones found for syntaxin 3, that the syntaxin 3-related gene is only expressed at low levels and is probably not expressed at biologically relevant levels in the retina.
It is likely that a similar gene exists in the goldfish and it is possible that other copies of related genes are also present in the goldfish genome. However, based on the sequence of the syntaxin 3 related gene from the zebrafish, our PCR primers should have amplified the syntaxin 3 related gene from the goldfish. However, we have detected only one type of mRNA in our RT-PCR experiments when we amplified syntaxin 3B from goldfish mRNA, indicating that syntaxin 3-related genes are not expressed at relevant levels in the goldfish retina.
Syntaxin 3B is expressed at high levels in bipolar cell synaptic terminals of the goldfish retina
Using different approaches we have demonstrated that syntaxin 3B is expressed in goldfish bipolar neurons. The RT-PCR analysis has shown that syntaxin 3B is expressed at high levels in the retina compared to brain and kidney. In contrast, syntaxin 3A is expressed at high levels in the kidney, but only expressed at low levels in retina or brain. This expression pattern is the same as the one found in the mouse, showing that the tissue-specific transcription and splicing of the syntaxin 3 gene is evolutionary conserved between fish and mammals. Using a combination of single cell RT-PCR, quantitative real time RT-PCR and immunostaining, we could show that syntaxin 3B is found at high levels in the ribbon synapse-containing synaptic terminals of bipolar neurons in the goldfish retina. High levels of of syntaxin 3 staining, that should mainly reflect the presence of syntaxin3B, were also detected in the ribbon synapse-containing OPL. Based on the expression pattern observed, it is likely that syntaxin 3B is, as has been shown in the mouse, is mainly found in ribbon synapses of the goldfish retina. Syntaxin 3B in the bipolar neuron is located at the presynaptic plasma membrane. This distribution pattern is consistent with a role of syntaxin 3B as a t-SNARE for synaptic vesicle fusion in bipolar neurons.
Synaptic vesicle fusion in bipolar neurons is thought to occur mainly at the base of synaptic ribbons but also to a lesser degree at fusion sites that are located away from the synaptic ribbons. We found that syntaxin 3B is distributed in patches at the plasma membrane. It is likely that these patches represent synaptic vesicle fusion sites at the plasma membrane. Studies have shown that ribbon synapses show multi-vesicular fusion (Matthews and Sterling, 2008). A possible mechanism that has been proposed for this form of vesicle fusion is that synaptic vesicles fuse with vesicles that have already fused with the synaptic plasma membrane in a process called “compound exocytosis”. Such type of inter-vesicular fusion would require the presence of t-SNARE molecules on synaptic vesicles. However, the majority of the syntaxin 3 in the Mb1 bipolar cells did not co-localize with the synaptic vesicle marker SV2, indicating that most of the synaptic vesicles lack syntaxin 3. Still, it is possible that a small subset of synaptic vesicles in the Mb1 bipolar cells contain syntaxin 3. Alternatively, unidentified t-SNARE proteins could be responsible for compound exocytosis in ribbon synapses.
The role of syntaxin3B in synaptic vesicle exocytosis at ribbon synapses
The t-SNARE protein syntaxin 1 has been shown to be essential for synaptic vesicle exocytosis in conventional synapses (Broadie et al., 1995; Mishima et al., 2002; Mochida et al., 1995; O'Connor et al., 1997). Ribbon synapses in the mouse retina contain syntaxin 3B instead of syntaxin 1 (Morgans et al., 1996; Sherry et al., 2006; Curtis et al., 2008). Here we show that ribbon synapse-containing synaptic terminals of the goldfish Mb1 bipolar cells also contain syntaxin 3B. However the role of syntaxin 3B in ribbon synapses has not been investigated until now. Here we used a short peptide based on the first half of the N-terminus of the syntaxin 3B protein to probe its functional role in ribbon synapses.
The initial exocytotic response in the presence of either peptide was similar in magnitude to the depletion of the entire releasable pool of synaptic vesicles. Subsequently, there was a progressive and significant decrease in the magnitude of the exocytotic response in terminals dialyzed with the syntaxin 3B SNARE peptide (Fig 6A). This decrease in exocytosis was not due to a decrease in calcium entry (Fig 6B). It also could not be attributed to a slowing of the rate endocytosis, as the time course of membrane retrieval in the presence of the syntaxin 3B SNARE peptide was not prolonged relative to terminals dialyzed with scrambled peptide and was in keeping with literature values for these terminals (data not shown). Thus, we propose that it was due to a progressive decrease in the extent to which the releasable pool of vesicles was functionally-refilled.
How might the syntaxin 3B SNARE peptide progressively impede excitation-secretion coupling? Given that we dialyzed the peptides into the terminal and the four-pulse stimulation train began at one minute after break-in, contributing factors to the progressive decline include: 1) the time required for the peptide to dialyze into the terminals relative to the timing of the stimuli, 2) competition with endogenous protein for binding partners, 3) the turnover of vesicles. Mechanistically, the impairment in functional refilling of the releasable pool could arise from a decrease in the number of vesicles physically present at the active zone or in the number of vesicles at the active zone that are in a fusion-competent state. Syntaxin proteins play a critical role in SNARE mediated vesicle fusion and syntaxin 3B has been shown to function as a t-SNARE in a liposomal fusion assay (Curtis et al., 2008), Therefore, we postulate that the syntaxin 3B SNARE peptide inhibits functional refilling of the releasable pool of synaptic vesicles in synaptic terminals of retinal bipolar neurons by interfering with the formation of SNARE complexes, thereby reducing the number of fusion-competent vesicles. Thus, our results suggest that the syntaxin 3B isoform, present in ribbon style synapses of the retina, is important for exocytosis of synaptic vesicles.
Why do ribbon synapses in the retina use a different syntaxin isoform to catalyze synaptic vesicle exocytosis than conventional synapses? As is apparent in figure 1 the sequences of syntaxin 1A and syntaxin 3B are quite homologous with some clusters of less conserved regions in the N-terminus of the protein. Despite the high degree of homology it has been shown that mouse syntaxin 3B is less efficient in binding SNAP-25 and catalyzing vesicle fusion than syntaxin 1A (Curtis et al., 2008). Syntaxin 1A has been shown to exist in an open and closed conformation with only the open form capable of binding SNAP-25 and forming a SNARE complex (Dulubova et al., 1999). It is possible that syntaxin 3B exists mostly in the closed conformation and is therefore less efficient in binding SNAP-25. This could indicate that specific factors are required to convert syntaxin 3B to a more open form. Such factors could potentially modulate the efficiency of synaptic vesicle exocytosis in retinal ribbon synapses. Future experiments will ascertain the extent to which differences in release properties between conventional and ribbon-style synapses are due to the molecular differences in the exocytotic machinery.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH EY016452 (R.J), by NIH EY012128 (RH) and by core and equipment grants NIH EY010608 and NIH RR022531. Leigh Curtis was supported by NIH training grants T32 NS007467 and T32 EY007024. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Augustine GJ, Neher E. Calcium requirements for secretion in bovine chromaffin cells. J Physiol. 1992;450:247–271. doi: 10.1113/jphysiol.1992.sp019126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntson AK, Morgans CW. Distribution of the presynaptic calcium sensors, synaptotagmin I/II and synaptotagmin III, in the goldfish and rodent retinas. J Vis. 2003;3:274–280. doi: 10.1167/3.4.3. [DOI] [PubMed] [Google Scholar]
- Broadie K, Prokop A, Bellen HJ, O'Kane CJ, Schulze KL, Sweeney ST. Syntaxin and synaptobrevin function downstream of vesicle docking in Drosophila. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- Ciudad J, Cid E, Velasco A, Lara JM, Aijon J, Orfao A. Flow cytometry measurement of the DNA contents of G0/G1 diploid cells from three different teleost fish species. Cytometry. 2002;48:20–25. doi: 10.1002/cyto.10100. [DOI] [PubMed] [Google Scholar]
- Curtis LB, Doneske B, Liu X, Thaller C, McNew JA, Janz R. Syntaxin 3b is a t-SNARE specific for ribbon synapses of the retina. J Comp Neurol. 2008;510:550–559. doi: 10.1002/cne.21806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JE. The Retina: An Approachable Part of the Brain. Cambridge: Belknap Press of Harvard University Press; 1987. [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Südhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis KD. Techniques for membrane capacitance measurements. In: Neher E, Sakmann B, editors. Single Channel Recording. New York: Plenum Press; 1995. pp. 155–198. [Google Scholar]
- Heidelberger R. Adenosine triphosphate and the late steps in calcium-dependent exocytosis at a ribbon synapse. J Gen Physiol. 1998;111:225–241. doi: 10.1085/jgp.111.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R. ATP is required at an early step in compensatory endocytosis in synaptic terminals. J Neurosci. 2001;21:6467–6474. doi: 10.1523/JNEUROSCI.21-17-06467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R, Heinemann C, Neher E, Matthews G. Calcium dependence of the rate of exocytosis in a synaptic terminal. Nature. 1994;371:513–515. doi: 10.1038/371513a0. [DOI] [PubMed] [Google Scholar]
- Heidelberger R, Matthews G. Inhibition of calcium influx and calcium current by gamma-aminobutyric acid in single synaptic terminals. Proc Natl Acad Sci U S A. 1991;88:7135–7139. doi: 10.1073/pnas.88.16.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R, Matthews G. Calcium influx and calcium current in single synaptic terminals of goldfish retinal bipolar neurons. J Physiol (Lond) 1992;447:235–256. doi: 10.1113/jphysiol.1992.sp019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R, Matthews G. Dopamine enhances Ca2+ responses in synaptic terminals of retinal bipolar neurons. Neuroreport. 1994;5:729–732. doi: 10.1097/00001756-199402000-00018. [DOI] [PubMed] [Google Scholar]
- Heidelberger R, Sterling P, Matthews G. Roles of ATP in depletion and replenishment of the releasable pool of synaptic vesicles. J Neurophysiol. 2002a;88:98–106. doi: 10.1152/jn.2002.88.1.98. [DOI] [PubMed] [Google Scholar]
- Heidelberger R, Zhou ZY, Matthews G. Multiple components of membrane retrieval in synaptic terminals revealed by changes in hydrostatic pressure. J Neurophysiol. 2002b;88:2509–2517. doi: 10.1152/jn.00267.2002. [DOI] [PubMed] [Google Scholar]
- Heidelberger R, Wang MM, Sherry DM. Differential distribution of synaptotagmin immunoreactivity among synapses in the goldfish, salamander, and mouse retina. Vis Neurosci. 2003;20:37–49. doi: 10.1017/s095252380320105x. [DOI] [PubMed] [Google Scholar]
- Heidelberger R, Thoreson WB, Witkovsky P. Synaptic transmission at retinal ribbon synapses. Prog Retin Eye Res. 2005;24:682–720. doi: 10.1016/j.preteyeres.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagnado L, Gomis A, Job C. Continuous vesicle cycling in the synaptic terminal of retinal bipolar cells. Neuron. 1996;17:957–967. doi: 10.1016/s0896-6273(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Lindau M, Neher E. Patch-clamp techniques for time-resolved capacitance measurements in single cells. Pflugers Arch. 1988;411:137–146. doi: 10.1007/BF00582306. [DOI] [PubMed] [Google Scholar]
- Low SH, Chapin SJ, Wimmer C, Whiteheart SW, Kömüves LK, Mostov KE, Weimbs T. The SNARE machinery is involved in apical plasma membrane trafficking in MDCK cells. J. Cell Biol. 1998;141:1503–1513. doi: 10.1083/jcb.141.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews G, Sterling P. Evidence that vesicles undergo compound fusion on the synaptic ribbon. J Neurosci. 2008;28:5403–5411. doi: 10.1523/JNEUROSCI.0935-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Fujiwara T, Akagawa K. Reduction of neurotransmitter release by the exogenous H3 domain peptide of HPC-1/syntaxin 1A in cultured rat hippocampal neurons. Neurosci Lett. 2002;329:273–276. doi: 10.1016/s0304-3940(02)00662-6. [DOI] [PubMed] [Google Scholar]
- Mochida S, Saisu H, Kobayashi H, Abe T. Impairment of syntaxin by botulinum neurotoxin C1 or antibodies inhibits acetylcholine release but not Ca2+ channel activity. Neuroscience. 1995;65:905–915. doi: 10.1016/0306-4522(94)00508-3. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Brandstätter JH, Kellerman J, Betz H, Wässle H. A SNARE complex containing syntaxin 3 is present in ribbon synapses of the retina. J Neurosci. 1996;16:6713–6721. doi: 10.1523/JNEUROSCI.16-21-06713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor V, Heuss C, De Bello WM, Dresbach T, Charlton MP, Hunt JH, Pellegrini LL, Hodel A, Burger MM, Betz H, Augustine GJ, Schäfer T. Disruption of syntaxin-mediated protein interactions blocks neurotransmitter secretion. Proc Natl Acad Sci U S A. 1997;94:12186–12191. doi: 10.1073/pnas.94.22.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim K, Wegmeyer H, Brandstäatter JH, Xue M, Rosenmund C, Dresbach T, Hofmann K, Brose N. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol. 2005;169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger C, Larhammar D. Multiple loci for synapse protein SNAP-25 in the tetraploid goldfish. Proc Natl Acad Sci U S A. 1993;90:10598–10602. doi: 10.1073/pnas.90.22.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger C, Salaneck E, Soderberg C, Gates M, Postlethwait JH, Larhammar D. Cloning of two loci for synapse protein Snap25 in zebrafish: comparison of paralogous linkage groups suggests loss of one locus in the mammalian lineage. J Neurosci Res. 1998;54:563–573. doi: 10.1002/(SICI)1097-4547(19981201)54:5<563::AID-JNR1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DM, Mitchell R, Standifer KM, du Plessis B. Distribution of plasma membrane-associated syntaxins 1 through 4 indicates distinct trafficking functions in the synaptic layers of the mouse retina. BMC Neurosci. 2006;7:54. doi: 10.1186/1471-2202-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DM, Wang MM, Frishman LJ. Differential distribution of vesicle associated membrane protein isoforms in the mouse retina. Mol Vis. 2003;9:673–688. [PubMed] [Google Scholar]
- Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gersdorff H, Matthews G. Dynamics of synaptic vesicle fusion and membrane retrieval in synaptic terminals. Nature. 1994;367:735–739. doi: 10.1038/367735a0. [DOI] [PubMed] [Google Scholar]
- von Gersdorff H, Matthews G. Depletion and replenishment of vesicle pools at a ribbon-type synaptic terminal. J Neurosci. 1997;17:1919–1927. doi: 10.1523/JNEUROSCI.17-06-01919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gersdorff H, Vardi E, Matthews G, Sterling P. Evidence that vesicles on the synaptic ribbon of retinal bipolar neurons can be rapidly released. Neuron. 1996;16:1221–1227. doi: 10.1016/s0896-6273(00)80148-8. [DOI] [PubMed] [Google Scholar]
- von Gersdorff H, Matthews G. Dynamics of synaptic vesicle fusion and membrane retrieval in synaptic terminals. Nature. 1994;367:735–739. doi: 10.1038/367735a0. [DOI] [PubMed] [Google Scholar]
- von Kriegstein K, Schmitz F. The expression pattern and assembly profile of synaptic membrane proteins in ribbon synapses of the developing mouse retina. Cell Tissue Res. 2003;311:159–173. doi: 10.1007/s00441-002-0674-0. [DOI] [PubMed] [Google Scholar]
- von Kriegstein K, Schmitz F, Link E, Sudhof TC. Distribution of synaptic vesicle proteins in the mammalian retina identifies obligatory and facultative components of ribbon synapses. Eur J Neurosci. 1999;11:1335–1348. doi: 10.1046/j.1460-9568.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- Wang MM, Janz R, Belizaire R, Frishman LJ, Sherry DM. Differential distribution and developmental expression of synaptic vesicle protein 2 isoforms in the mouse retina. J Comp Neurol. 2003;460:106–122. doi: 10.1002/cne.10636. [DOI] [PubMed] [Google Scholar]
- Zenisek D, Steyer JA, Almers W. Transport, capture and exocytosis of single synaptic vesicles at active zones. Nature. 2000;406:849–854. doi: 10.1038/35022500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.