Abstract
Integrase is an essential retroviral enzyme that binds both termini of linear viral DNA and inserts them into a host cell chromosome. The structure of full-length retroviral integrase, either separately or in complex with DNA, has been lacking. Furthermore, although clinically useful inhibitors of HIV integrase have been developed, their mechanism of action remains speculative. Herein we report a crystal structure of full-length integrase from the prototype foamy virus in complex with its cognate DNA. The structure reveals the organization of the retroviral intasome comprising an integrase tetramer tightly associated with a pair of viral DNA ends. All three canonical integrase structural domains are involved in extensive protein-DNA and protein-protein interactions. Binding of strand transfer inhibitors displaces the reactive viral DNA end from the active site, disarming the viral nucleoprotein complex. Our findings define the structural basis of retroviral DNA integration and will allow modeling of the HIV-1 intasome to aid in the development of antiretroviral drugs.
Retroviral integrase (IN) recognizes and acts upon the termini of the linear double-stranded DNA molecule that is produced by reverse transcription1,2. Initially, in a reaction termed 3′-processing, IN removes two or three nucleotides from one or both viral DNA ends to expose the 3′ hydroxyl groups of the invariant CA dinucleotides. Next, following import of the viral DNA into the nucleus, IN inserts both 3′ ends of the viral DNA into opposing strands of cellular genomic DNA. Mechanistically and structurally, IN belongs to a diverse family of polynucleotidyl transferases3, which notably includes RNaseH4 and the transposases from Tn55 and eukaryotic mobile element Mos16 (reviewed in ref7,8). The reactions catalyzed by these enzymes proceed via SN2-type nucleophilic substitution, assisted by divalent metal cofactors4,9. In retroviral IN, a pair of divalent metal cations (Mg2+ or Mn2+) are thought to be coordinated by three carboxylates of the invariant D,D-35-E motif within the catalytic core domain (CCD). To function, IN further requires its N-terminal domain (NTD), a three-helical bundle stabilized through binding a Zn atom, and a C-terminal domain (CTD) that adopts an SH3-like fold10,11. In vivo, IN acts within a large nucleoprotein complex that contains viral DNA and a number of virus-and host cell-derived components. The minimal functional complex involving viral DNA and IN, herein referred to as the intasome, can be assembled in vitro from purified components12.
Despite its acute importance for antiretroviral drug discovery and decades of rigorous research7,13, the complete structure of IN, either as a separate protein or in the context of the functional intasome, is lacking. Accordingly, the structural organization of the enzyme active site, which is believed to adopt its functional state only upon viral DNA binding, is unknown. Because clinically useful HIV-1 IN strand transfer inhibitors14,15 (InSTIs) preferentially bind to and inhibit the intasome complex as compared to free IN16, the mechanism of drug action is poorly understood. We have now obtained diffracting crystals of the full-length IN from the prototype foamy virus (PFV) in complex with its cognate viral DNA. The availability of these crystals enabled us to determine the long-sought structure of the retroviral intasome and explain the mechanism of strand transfer inhibitor action.
Crystallization of the PFV intasome
The majority of characterized INs predominantly promote the insertion of one viral DNA end into one strand of a target DNA duplex in vitro. By contrast, we recently reported that recombinant PFV IN catalyzes efficient concerted insertion of two PFV DNA ends into target DNA17. Herein, we obtained soluble and fully functional PFV intasome preparations using recombinant PFV IN and double-stranded oligonucleotides that mimic the viral DNA ends (Supplementary Fig. 1). To bypass the initial catalytic step, 3′-processing, the IN-DNA complexes were assembled using “pre-processed” oligonucleotides with recessed termini, which model the viral DNA ends prior to their insertion into host chromosomal DNA. The IN-DNA complexes were remarkably stable and did not dissociate or lose activity, even upon prolonged incubations under high ionic strength conditions (Supplementary Fig. 1 and data not shown).
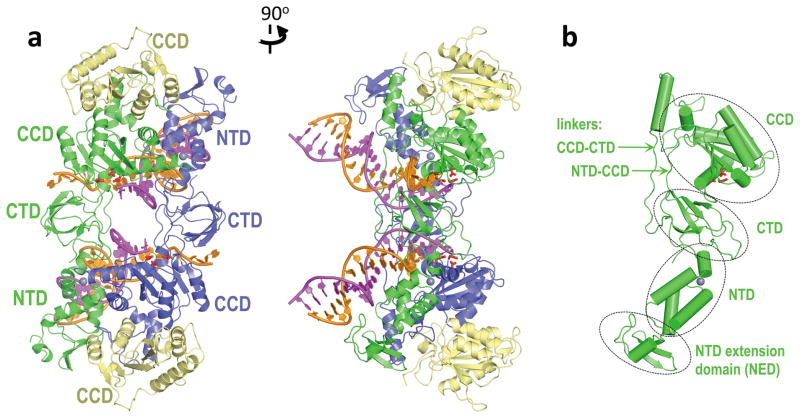
Following extensive crystallization trials we identified a crystal form of full-length wild type PFV IN in complex with a 19-bp donor DNA that diffracted X-rays to 2.9 Å resolution, enabling us to determine the three-dimensional structure of the intasome (Supplementary Table 1). The asymmetric unit contained a single IN dimer with a tightly associated viral DNA molecule, and a pair of symmetry-related IN dimers formed an oblong tetramer (130 Å by 65 Å by 70 Å; Fig. 1a and Supplementary Movie 1). The IN dimer-dimer interface is stabilized by intermolecular NTD-CCD interactions, as previously observed in partial structures of lentiviral INs18,19. The overall shape of the tetramer is reminiscent of the low-resolution envelope obtained by a recent negative stain electron microscopy study of HIV-1 IN complexes20. Nonetheless, its architecture is drastically different from all previously reported models.
Figure 1. Architecture of the PFV intasome.
a, Views along (left) and perpendicular (right) to the crystallographic two-fold axis. The inner subunits of the IN tetramer, engaged with viral DNA, are blue and green; outer IN chains are yellow. The reactive and non-transferred DNA strands are magenta and orange, respectively. Side chains of Asp128, Asp185 and Glu221 active site residues are red sticks; gray spheres are Zn atoms. Locations of the canonical IN domains (NTD, CCD and CTD) are indicated; b, inner (green) IN chain with domains and linkers indicated. The orientation is the same as in the right panel of a.
Overall architecture of the PFV intasome
The inner subunits of the tetramer (colored green and blue in Fig. 1) are responsible for all contacts involved in tetramerization and viral DNA binding. The CCDs of the outer subunits (gold in Fig. 1) appear to provide supporting function, their NTDs and CTDs not resolved in the electron density maps. Previous retroviral IN CCD structures revealed a conserved dimeric interface7, and this interface is retained between the inner and outer IN subunits of the intasome. Partial structures of INs from HIV-121, simian immunodeficiency22 and Rous sarcoma23 viruses showed considerable variability of the CCD-CTD linker region. Within the PFV intasome, the CCD-CTD linker adopts an extended conformation for most of its length, tracking parallel to the NTD-CCD linker from the same subunit (Fig. 1b). The interdomain linkers truss both halves of the intasome together, and the structure is further stabilized by a pair of CTDs interacting with both inner CCDs (Fig. 1a), in addition to an extensive network of protein-DNA interactions (see below). Thus, by contrast to previous IN tetramer models based on two domain structures wherein the dimer-dimer interface appeared highly flexible18,19, the overall conformation of the assembled intasome is well constrained. Homology modeling suggests that the notably shorter interdomain linkers in HIV-1 IN (Supplementary Fig. 2) can extend sufficiently to allow a similar overall architecture and topology in the HIV-1 intasome (data not shown). An additional small domain, which we refer to as the NTD extension domain (NED), precedes the PFV IN NTD (Fig. 1b). Based on amino acid sequence comparisons and secondary structure predictions, NEDs are present in other spumaviral and possibly gammaretroviral INs (data not shown).
IN-DNA interactions within the PFV intasome
In total, almost 10,000 Å2 of molecular surface is buried within IN-DNA interfaces of the intasome. The protein-DNA interactions involve amino acid residues from each domain of the inner IN subunits, their inter-domain linkers, and 17 nucleotides from each viral DNA end (Supplementary Fig. 2, 3). Thus, as it was observed in case of Tn5 transposase5, the canonical retroviral IN domains (NTD, CCD and CTD) do not have discrete functions; each contributes to extensive protein-protein and protein-DNA contacts within the functional complex. The most intimate protein-DNA interactions are found within the terminal six nucleotides, where the viral DNA significantly deviates from the ideal B form. Each CTD makes contact with the phosphodiester backbone of both viral DNA molecules, essentially crosslinking the structure. Notably, the NED and NTD of each catalytic subunit interact with the viral DNA molecule engaged at the active site of the opposing CCD (Fig. 1a). The NED interacts with the phosphodiester backbone, while the other elements (NTD, CTD, CCD, NTD-CCD and CCD-CTD linkers) additionally contribute to interactions with DNA bases. These sequence-specific interactions include the main chain carbonyl group of Gly218, which forms a hydrogen bond with guanine 4 of the non-transferred strand (Fig. 2a). The protein-DNA interactions extend into the beginning of CCD α4 helix, which packs into the minor groove at the end of the viral DNA duplex (Fig. 2a and Supplementary Fig. 3). The side chain of Arg222 is involved in hydrogen bonding with T5 and C6 bases of the non-transferred strand (Fig. 2a). Another set of sequence-specific contacts involves residues from the NTD-CCD and CCD-CTD linkers extending from the opposing IN dimer. The side chain of Arg313 intercalates the minor groove of viral DNA, stacking its guanidinium group against adenosine 12 of the reactive strand and forming a hydrogen bond with cytosine 11 (Fig. 2b). Nearby, the side chain of Asn106 interacts with thymine 8 of the non-transferred strand. These interactions cause notable widening of the minor groove of the viral DNA (Fig. 2b and Supplementary Fig. 3b). The overall extent of protein-DNA interaction agrees well with the ~16 bp IN footprint in functional HIV-1 intasomes12.
Figure 2. Sequence-specific protein-DNA interactions.
a, View of the IN active site with bound DNA. Amino acid side chains discussed in text, DNA bases and the main chain carbonyl of Gly218 are shown as sticks; b, interactions of the same DNA molecule with the NTD and NTD-CCD and CCD-CTD linkers from the opposing (blue) IN subunit. Note the widening of the minor groove due to insertion of Arg313 and Asn106 side chains. The Arg69 side chain additionally packs into the major groove, forming hydrogen bonds with guanine 10 of the non-transferred strand.
At the reactive viral DNA terminus, the base pair involving the cytosine of the invariant CA dinucleotide (C16 and A17 of the reactive strand, Supplementary Fig. 3a) is distorted by a buckle of ~30°, while the terminal adenine is completely isolated from its complement on the non-transferred strand (Fig. 2a). The active site loop (PFV IN residues 211–220) is directly involved in separating the viral DNA strands, acting as a plough with highly conserved residues Pro214 and Gln215 (corresponding to HIV-1 Pro145 and Gln146, see Supplementary Fig. 2 for PFV/HIV-1 structure-based alignment) forming its share. In particular, Gln215 displaces thymine 3 of the non-transferred strand, which turns away from the interior of the DNA duplex (Fig. 2a, Supplementary Fig. 3b). The 5′-overhang of the non-transferred strand is threaded between the CCD and interdomain linkers, where it forms extensive contacts with the active site loop and the CTD from the same IN chain (Supplementary Fig. 3c). The involvement of CCD α4 helix and the active site loop in intimate interactions with the viral DNA end agrees well with results of chemical and photo crosslinking of functional HIV-1 IN-DNA complexes24,25,26.
The active site of the functional intasome
The reactive 3′ termini of the donor DNA molecules are positioned within close proximity of the Asp128, Asp185 and Glu221 active site carboxylates (Fig. 2a). Although the crystals could be grown in the presence of MgCl2, which considerably improved their diffraction limit (Supplementary Table 1), data resolution did not allow unambiguous visualization of Mg2+ cations in the active site. Fortuitously, similar to other retroviral INs1, PFV IN can efficiently utilize Mn2+, a more electron-rich element, as a metal ion cofactor (Supplementary Fig. 1d). A difference electron density map calculated using diffraction data collected on crystals soaked in the presence of MnCl2 revealed two strong positive peaks (9.4 and 12.4σ) within the active sites of the inner IN subunits. This result confirmed the expected two-metal binding mode of retroviral INs and revealed the positions of metal ion cofactors within the assembled active site, which could be refined at full occupancy (Fig. 3a, see also Supplementary Fig. 4a for metal atom omit and final electron density maps). Based on the current model for two-metal active site catalysis4, metal atom B, coordinated by the carboxylates of Asp128 and Glu221, is in place to activate the 3′ hydroxyl group of the pre-processed viral DNA for strand transfer while metal A, bound by Asp128 and Asp185, would be expected to destabilize the scissile phosphodiester group in target DNA. Superposition of the Cα atoms of the active site Asp and Glu residues revealed striking conservation between the metal and DNA substrate binding modes of the Tn5 synaptic complex27 and PFV intasome (Supplementary Fig. 5). In addition, the positions of the metal ions are nearly identical to those of Cd2+ and Zn2+ cations observed in structures of the avian sarcoma virus IN CCD28. Of note, soaking crystals in MgCl2 or MnCl2 did not change the organization of the intasome active site (Supplementary Fig. 6). Hence, the positioning of the 3′ end of viral DNA is independent of bound divalent metal ions.
Figure 3. PFV IN active site in committed and drug-bound states.
Views without drug (a) and with MK0518 (b) or GS9137 (c) bound. Protein and DNA in upper panels are cartoons, with A17, DNA bases and the side chains of indicated amino acids as sticks. Drug atoms are colored: yellow, C; blue, N; red, O; orange, P; gray, F; green, Cl. The complex is shown as a solvent accessible surface in lower panels, colored by atoms (light gray, C; red, O; blue, N). Gray spheres are Mn2+ (a, labeled A and B) or Mg2+ (b, c) ions.
Model for target DNA binding
The active sites of the inner IN subunits, engaged with the 3′ termini of the viral DNA, are located deep within the dimer-dimer interface. Therefore, the only mode of host chromosomal DNA (target DNA) binding that would not require dramatic rearrangement of the intasome or severe DNA bending is along the cleft between IN dimers (Fig. 4). This target DNA binding mode could not have been predicted based on previous partial IN structures, and starkly differs from what we recently proposed18. Modeling B-form DNA within the cleft results in near perfect alignment of the active sites with opposing target DNA phoshodiester bonds separated by 4 bp, the known spacing of concerted PFV integration17. It is easy to see how mutations within the α2 helix of the CCD, described by Katzman and colleagues29, would prevent target DNA binding (Fig. 4). We tentatively speculate that the NTDs and/or the CTDs of the outer IN subunits, disordered in our structures, could be involved in target DNA capture30. However, this target-binding model requires verification using mutagenesis or crystallographic approaches.
Figure 4. Predicted target DNA binding orientation.
Cartoon representation of the intasome as in Fig. 1a with active site side chains shown as red sticks and 〈2 helices, known to contribute to target DNA binding29, in cyan. The DNA molecule modeled in black and gray shows the most likely orientation for target DNA binding. Note that the CTD, juxtaposed to the target DNA in this model, is known to possess sequence non-specific DNA binding activity35.
Mechanism for strand transfer inhibitor action
We have previously shown that PFV IN is sensitive to HIV-1 IN InSTIs17. These compounds are thought to engage metal ion cofactors in the IN active site through interactions with uniquely positioned oxygen atoms of the pharmacophore31. The role of the remaining common InSTI feature, a fluorobenzyl group, is enigmatic. Intasome structure refinement using diffraction data collected on crystals soaked in the presence of Mg2+ and the clinical InSTI MK0518 (also known as raltegravir)14 or GS9137 (elvitegravir)15 revealed strong additional electron density within the active sites of the inner IN subunits. Structures of MK0518 or GS9137 with pairs of Mg atoms could be easily fitted into the maps and refined to 2.85 and 3.15 Å resolution, respectively (Fig. 3b, c, Supplementary Fig. 4d, f and Supplementary Table 1). In addition, soaking crystals in the presence of the drugs and Mn2+ produced similar results, with manganese atoms and drug molecules refining at almost precisely the same positions (Supplementary Fig. 4b, c, e, g). Concordantly, InSTIs inhibited both the Mg2+ and Mn2+-dependent activities of the PFV intasome (Supplementary Fig. 1d).
Based on the structures, the two InSTIs appear to have very similar modes of binding and action, involving an induced fit mechanism. Their metal chelating oxygen atoms orient towards the metal cofactors of the active site, while their halobenzyl groups fit within a tight pocket created by displacement of the 3′ adenosine (A17). Within it, the drugs make intimate Van der Waals interactions with the bases of the invariant CA dinucleotide, guanine 4 from the non-transferred strand and conserved residues Pro214 and Gln215 (Fig. 3b, c). In addition, the isopropyl and methyl-oxadiazole groups of MK0518 are involved in hydrophobic and stacking interactions with the side chains of Pro214 and Tyr212, respectively (Fig. 3b), further stabilizing this drug in the active site. Through its quinolone base and isopropyl group, GS9137 interacts with Pro214 (Fig. 3c). Crucially, this mode of drug binding results in displacement of the reactive 3′ viral DNA end from the active site (Fig. 3b, c), which can only result in deactivation of the intasome. Thus, upon binding of MK0518, the reactive 3′ hydroxyl group moves away from the active site by more than 6 Å, compared to its positions in the Mg2+ or Mn2+-containing, or apo crystals.
Because the core contact points consisting of invariant nucleotide bases and amino acid residues are conserved in HIV-1, the mode of InSTI binding and action are unlikely to significantly differ. The extensive contacts with the viral DNA end observed in our structures elucidate why the InSTIs preferentially interact with and inhibit the DNA-bound form of HIV-1 IN16. Moreover, the induced fit caused by displacement of the 3′ adenosine by the halobenzyl groups of these compounds explains why the deletion of this base dramatically increased InSTI on- and off- rates for binding to HIV-1 IN-DNA complexes32. Furthermore, mutations of HIV-1 IN residue Tyr143, which, based on our structure, is expected to interact with the methyl-oxadiazole group of MK0518 (Fig. 3b, Supplementary Fig. 2), are known to confer resistance to this drug33. Common InSTI resistance pathways involve mutations of HIV-1 IN Gln148 or Asn15533, which correspond to PFV IN residues Ser217 and Asn224, respectively (Supplementary Fig. 2). Mutations at these positions are likely to interfere with coordination of metal cofactors by the active site carboxylates, as proposed recently34. Conceivably, a slight shift in metal ion cofactor positions might suffice to abrogate drug binding, which relies on its spatially constrained metal chelating groups, albeit at a steep price of impaired viral replication fitness due to detuning of the IN active site structure.
Our findings will allow the generation of reliable HIV-1 IN and InSTI pharamacophore models, which will be invaluable for the development of next generation strand transfer inhibitors. Their design should take advantage of the most conserved elements of the IN active site elucidated here, such as the bases of the invariant CA dinucleotide, positions of the metal co-factors and the main chain atoms of the protein.
METHODS SUMMARY
Protein-DNA complexes were assembled using full-length wild type PFV IN17 and synthetic double-stranded DNA that modeled the viral U5 end (5′-TACAAAATTCCATGACA/5′-ATTGTCATGGAATTTTGTA) by dialysis, reducing NaCl concentration from 500 mM to 200 mM. A typical result of complex assembly is shown in Supplementary Fig. 1a. The intasome was crystallized by vapor diffusion in hanging drops with a reservoir solution containing 1.35 M ammonium sulfate, 25% (v/v) glycerol, 4.8% (v/v) 1,6-hexanediol and 50 mM 2-(N-morpholino) ethanesulfonic acid (MES), pH 6.5. The crystals were soaked in the presence of MK0518, GS9137, Mg2+, and/or Mn2+, as required. Diffraction data were obtained at the beamlines I02 and I04 of the Diamond Light Source (Oxford, UK), and the structure was solved by molecular replacement. Crystallographic and refinement statistics for the seven resulting models are summarized in Supplementary Table 1. The structures, refined to 2.85–3.25 Å resolution, included PFV IN residues 10–374 and 116–278 in chains A and B, respectively, without gaps, and both complete strands of the pre-processed donor DNA molecule (17 and 19 nucleotides in chains C and D, respectively). The models had good geometry with 92.2–96.4 and not more than 0.4 % of amino acid residues in most preferred and disallowed regions of the Ramachandran plot, respectively.
METHODS
Preparation and analyses of the PFV intasome
Full-length PFV IN (corresponding to residues 752 1143 of PFV POL) was produced in Escherichia coli strain PC236 transformed with pSSH6P-PFV-INFL17 and purified as previously described17. The protein was stored in aliquots at −80°C in 0.5 M NaCl, 5 mM dithiothreitol, 10% glycerol, 50 mM Tris-HCl, pH 7.4. Ion exchange HPLC-purified oligonucleotides were purchased from Eurogentec (Seraing, Belgium). Protein-DNA complexes were prepared by dialyses of mixtures containing 120 μM PFV IN, 50 μM synthetic DNA duplex, 500 mM NaCl, and 50 mM BisTris propane-HCl, pH 7.45, against excess 200 mM NaCl, 2 mM DTT, 25μM ZnCl2, 20 mM BisTris propane-HCl, pH 7.45 for 18–24 h at 18 °C. Dialyzed material was supplemented with an additional 120 or 800 mM NaCl (0.32 or 1 M NaCl final), incubated for 1 h on ice and analyzed by size exclusion chromatography (SEC) using a Superdex 200 HR 10/30 column, attached to an ÄKTA Purifier system (GE Healthcare). The column was operated in 0.32 or 1 M NaCl supplemented with 20 mM BisTris propane-HCl, pH 7.45 at 1 ml/min, 20°C. Strand transfer assays with SEC-purified intasome were carried out using established buffer conditions17. A typical reaction contained 300 ng supercoiled pGEM9 target DNA, 12 μOD280 (~30nM) intasome, 125 mM NaCl, 5 mM MgCl2 (or MnCl2), 10 mM dithiothreitol, 4 μM ZnCl2, 25 mM BisTris propane-HCl, pH 7.45, in a final volume of 40 μl. The reaction conditions were modified as required. Following incubation at 37 °C for 30–60 min, the products were deproteinized, separated in 1.5% (w/v) agarose gels and visualized by staining with ethidium bromide, as previously described17.
Crystallization and structure determination
Over 30 DNA constructs were tested in initial crystallization trials with full-length wild type and several mutant PFV IN proteins in ~40,000 initial sparse matrix conditions. Although several crystal forms could be identified and optimized, only one, obtained using a 19-bp mimic of pre-processed U5 end of PFV DNA (5′-TACAAAATTCCATGACA/5′-ATTGTCATGGAATTTTGTA) and wild type full-length IN, diffracted X rays to a resolution better than 6 Å. For crystallization, the protein-DNA complex assembled by dialysis and supplemented with an additional 120 mM NaCl (320 mM NaCl final) was concentrated to 10–14 mg/ml using Amicon Ultra-4 centrifugal devices (Millipore). Intasome preparations were stable for extended periods of time when stored on ice. Crystals were grown by vapor diffusion in hanging drops at 18 °C by mixing 1 μl IN-DNA complex and 1 μl of reservoir solution containing 1.35 M ammonium sulfate, 25 mM MgCl2, 25% (v/v) glycerol, 4.8% (v/v) 1,6-hexanediol, 50 mM MES-NaOH, pH 6.5. MgCl2 was omitted to obtain apo crystals used in Mn2+ and InSTI/Mn2+ soaking experiments. Crystals typically appeared within 24–48 h and grew to a size of ~100 by 100 by 75 μm within 7–10 days. To obtain drug-bound forms of the complex, intasome crystals were incubated in stabilizing solution (reservoir plus 200 mM NaCl and 7 mM DTT), supplemented with 1 mM MK0518 or GS9137 for 60 h at 18°C. To visualize metal atoms in the active sites, crystals grown without MgCl2 were soaked in the presence of 25 mM MnCl2 in the absence or presence of 1 mM MK0518 or GS9137.
The crystals were frozen by rapid immersion in liquid nitrogen, and diffraction data, acquired on I02 and I04 beamlines of the Diamond Light Source (Oxford, UK), were processed using Mosflm37 and SCALA38. Availability of the PFV IN CCD structure17 and the high solvent content of the crystal form (68.5%) made it possible to determine the intasome structure by molecular replacement. The solution was found in Phaser39 in space group P41212 using PFV IN CCD (PDB ID 3dlr)17, HIV-1 IN CTD (residues 223–266 of chain A from PDB ID 1ex4)21 and a generic 7-bp B-form DNA duplex as search models (Supplementary Fig. 7a). The initial phases, modified with prime-and-switch mode of RESOLVE40 (Supplementary Fig. 7b) were used as input for Buccaneer41, which built most of the protein part of the structure, including the NTD, NTD-CCD and CCD-CTD linkers. The models were improved by iterative manual building in Coot42, simulated annealing in Phenix43 and maximum likelihood restrained positional refinement in Refmac44. Non-crystallographic symmetry restraints for CCD regions similar in chains A and B (residues 121–200, 241–257 and 263–273) were applied throughout. Translation, libration and screw (TLS) refinement45 was included in the final stages. Geometry of the final structures was analyzed using Molprobity46. Examples of electron density and omit maps are given in Supplementary Fig. 4 and 7. Supplementary Table 1 lists unit cell parameters and crystallographic statistics. Coordinates and restraints files for the drug molecules were created using the PRODRG2 server47. Figures were prepared using PyMOL (DeLano Scientific, San Carlos, CA, USA) and ESPript48, molecular surface areas were calculated using Areaimol38,49 and protein-DNA contacts were analyzed with help of NUCPLOT50.
Supplementary Material
Acknowledgments
We thank Dr. Fred Dyda (National Institutes of Health) for critical reading of the manuscript, Dr. Reginald Clayton and Dr. Maxwell Cummings (Tibotec Pharmaceuticals) for generous gift of InSTIs and helpful discussions, Dr. Thomas Sorensen and the staff of the I02 and I04 beamlines of the Diamond Light Source for assistance with X-ray data collection. P.C. and co-workers are funded by the UK Medical Research Council and A.E. by the US National Institutes of Health.
Footnotes
Full Methods and any associate references are available in the online version of the paper at www.nature.com/nature.
Author Contributions
E.V. and P.C. carried out initial trials with truncated PFV IN constructs; S.S.G. and P.C. obtained full-length PFV IN-DNA complexes, carried out crystallization screening and optimization; S.H. soaked and prepared crystals for data collection; S.H. and P.C. collected diffraction data and solved the structures; S.H. refined the final models; S.H. and S.S.G. carried out gel filtration and activity assays, P.C., S.H. and A.E. wrote the paper.
Author Information Atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 3L2Q, 3L2R, 3L2S, 3L2T, 3L2U, 3L2V and 3L2W for Apo, Mg, Mn, Mg/MK0518, Mg/GS9137, Mn/MK0518 and Mn/GS9137 structures, respectively. Raw diffraction images are available upon request. Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to P.C. (p.cherepanov@imperial.ac.uk).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Craigie R. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; 2002. pp. 613–630. [Google Scholar]
- 2.Lewinski MK, Bushman FD. Retroviral DNA integration -mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 3.Dyda F, et al. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 4.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 6.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276:2926–2946. doi: 10.1111/j.1742-4658.2009.07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 10.Cai M, et al. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 11.Eijkelenboom AP, et al. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchand C, Maddali K, Métifiot M, Pommier Y. HIV-1 IN inhibitors: 2010 update and perspectives. Curr Top Med Chem. 2009;9:1016–1037. doi: 10.2174/156802609789630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summa V, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 15.Sato M, et al. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 16.Espeseth AS, et al. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci U S A. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valkov E, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare S, et al. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel F, et al. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009;28:980–991. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JC, et al. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, et al. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50–293)--an initial glance of the viral DNA binding platform. J Mol Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 23.Yang ZN, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivativeof Rous sarcoma virus integrase. J Mol Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 24.Alian A, et al. Catalytically-active complex of HIV-1 integrase with a viral DNA substrate binds anti-integrase drugs. Proc Natl Acad Sci U S A. 2009;106:8192–8197. doi: 10.1073/pnas.0811919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition. Curr Opin Struct Biol. 2004;14:50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Bujacz G, et al. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J Biol Chem. 1997;272:18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 29.Nowak MG, Sudol M, Lee NE, Konsavage WM, Jr, Katzman M. Identifying amino acid residues that contribute to the cellular-DNA binding site on retroviral integrase. Virology. 2009;389:141–148. doi: 10.1016/j.virol.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Miller MD, Bor YC, Bushman F. Target DNA capture by HIV-1 integration complexes. Curr Biol. 1995;5:1047–1056. doi: 10.1016/s0960-9822(95)00209-0. [DOI] [PubMed] [Google Scholar]
- 31.Grobler JA, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langley DR, et al. The terminal (catalytic) adenosine of the HIV LTR controls the kinetics of binding and dissociation of HIV integrase strand transfer inhibitors. Biochemistry. 2008;47:13481–13488. doi: 10.1021/bi801372d. [DOI] [PubMed] [Google Scholar]
- 33.Buzon MJ, et al. The HIV-1 integrase genotype strongly predicts raltegravir susceptibility but not viral fitness of primary virus isolates. AIDS. 2010;24:17–25. doi: 10.1097/QAD.0b013e328331c81e. [DOI] [PubMed] [Google Scholar]
- 34.Grobler JA, Stillmock K, Miller MD, Hazuda DJ. Mechanism by which the HIV integrase active-site mutation N155H confers resistance to raltegravir. Antiviral Ther. 2008;13(Suppl 3):A41. [Google Scholar]
- 35.Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 [Google Scholar]
- 38.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 41.Cowtan K. The Buccaneer software for automated model building. 1 Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 48.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 50.Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.