Abstract
The ability of a viral vector to safely deliver and stably integrate large transgene units (transgenons), which not only include one or several therapeutic genes but also requisite native transcriptional regulatory elements, would be of significant benefit for diseases presently refractory to available technologies. The herpes simplex virus type-1 (HSV-1) amplicon vector has the largest known payload capacity of approximately 130 kb, but its episomal maintenance within the transduced cell nucleus and induction of host cell silencing mechanisms limits the duration of the delivered therapeutic gene(s). Our laboratory developed an integration-competent version of the HSV-1 amplicon by adaptation of the Sleeping Beauty (SB) transposon system, which significantly extends transgene expression in vivo. The maximum size limit of the amplicon-vectored transposable element remains unknown, but previously published plasmid-centric studies have established that DNA segments larger than 6-kb are inefficiently transposed. Herein, we compared the transposition efficiency of SB transposase in the context of both the HSV amplicon vector as well as the HSV amplicon plasmid harboring 7-kb and 12-kb transposable reporter transgene units. Our results indicate that the transposition efficiency of the 12-kb transposable unit via SB transposase was significantly reduced compared to the 7-kb transposable unit when the plasmid version of the HSV amplicon was used. However, the packaged HSV amplicon vector form provided a more amenable platform from which the 12-kb transposable unit was mobilized at a similar efficiency to that of the 7-kb transposable unit via the SB transposase. Overall, our results indicate that SB is competent in stably integrating transgenon units of at least 12 kb in size within the human genome upon delivery of the platform via HSV amplicons.
Keywords: HSV-1 amplicon, Sleeping Beauty, integration capacity
INTRODUCTION
Development of novel viral vector-based therapeutics for presently clinically intractable human diseases may require regulated long-term expression of the therapeutic gene(s) to provide long-lasting benefit. Current strategies have focused on the use of adeno-associated virus (AAV) vectors and lentiviral vectors that intrinsically integrate their genetic payload within the host cell genome, thus facilitating stable genome maintenance and sustained expression of the therapeutic gene. Based upon their inherent biological characteristics, however, AAV and lentiviral viral vectors are limited to genetic segment lengths of 4 and 9 kb, respectively.1,2 These size restrictions unfortunately preclude the development of gene therapeutics for disorders that require the efficient delivery of genes greater than 9 kb. Hence, there is an apparent need in the field to develop and experimentally validate new vector platforms that may significantly raise the size barrier, and at the same time, provide stable therapeutic gene expression in vivo.
The herpes simplex virus (HSV-1) amplicon holds a significant advantage over all other currently available viral vectors due to its ~130 kb payload capacity, thus enabling the delivery of multiple transgene units together with transcriptional regulatory elements for stringent control of gene expression.3 However, long-term expression of transgenes delivered via the HSV amplicon has been challenging due to the episomal existence of the HSV genome within the transduced cell nucleus, its propensity to be segregated mitotically and its susceptibility to host-cell mediated silencing of the amplicon-encoded genetic units.4 Thus, the development of integration competent versions of the HSV amplicon have been undertaken via the use of viral and non-viral elements that facilitate stable integration of the delivered transgene within the transduced cell genome. One such vector is the HSV/AAV hybrid amplicon, which includes the Rep-coding sequence and the inverted terminal repeats (ITRs) of the AAV genome built into the HSV amplicon backbone. This hybrid amplicon system has shown to prolong transgene expression in cell culture and also facilitate site-specific integration of the ITR-flanked transgene unit into the AAVS1 locus in hChr19.5,6 As an alternative strategy, our laboratory previously engineered a bipartite HSV amplicon vector platform using the non-viral elements of the Sleeping Beauty (SB) transposon system,7 which enabled the efficient integration of a transposable transgene unit, termed a transgenon, within the genome of the transduced target cell and extended expression of a reporter transgene in vivo.8,9 The Rous sarcoma virus (RSV) 5’ LTR promoter regulated β-galactosidase/neomycin phosphotransferase fusion reporter transgene unit used in this initial study was 4.6-kb in size and when flanked by the inverted repeat elements (IR/DR) of SB, was efficiently transposed via the “wild-type” SB10 transposase expressed via a co-delivered HSV amplicon. However, the transposable size limit of this platform has yet to be determined.
Previous reports have demonstrated that plasmid-based delivery of SB IR/DR-flanked reporter transgenes are efficiently transposed up to size of 6 kb, with an exponential decrease in SB-mediated transposition efficiency as the size of the transposon is increased to sizes greater than 6 kb.10–12 The generation of the modified ‘sandwich’ transposon system by Zayed and colleagues has been shown to increase the transposable payload limit to ~10-kb from plasmid-based vectors.13 Additionally, the sequences that flank the transposon within the donor plasmid, as well as the efficiency by which the donor construct is delivered into the target cell nucleus, are factors that may significantly affect the transposition efficiency of SB.10 Given the large packaging capacity of conventional HSV-derived vectors, it is possible that the HSV/SB bipartite platform could provide a means to integrate transgene units that exceed the 9-kb limit afforded by lentiviral vectors. Herein, we tested whether a 12-kb transposable unit could be efficiently mobilized by either the “wild-type” SB transposase (SB10) or a hyperactive version of the SB transposase (termed HSB5 14) from the context of an HSV amplicon vector into a human cell genome. Successful mobilization and integration would demonstrate that an integration-competent platform exists that can integrate transgenons up to 12 kb, thereby expanding the number of potential disease targets amenable to HSV amplicon-based therapeutic modalities.
RESULTS & DISCUSSION
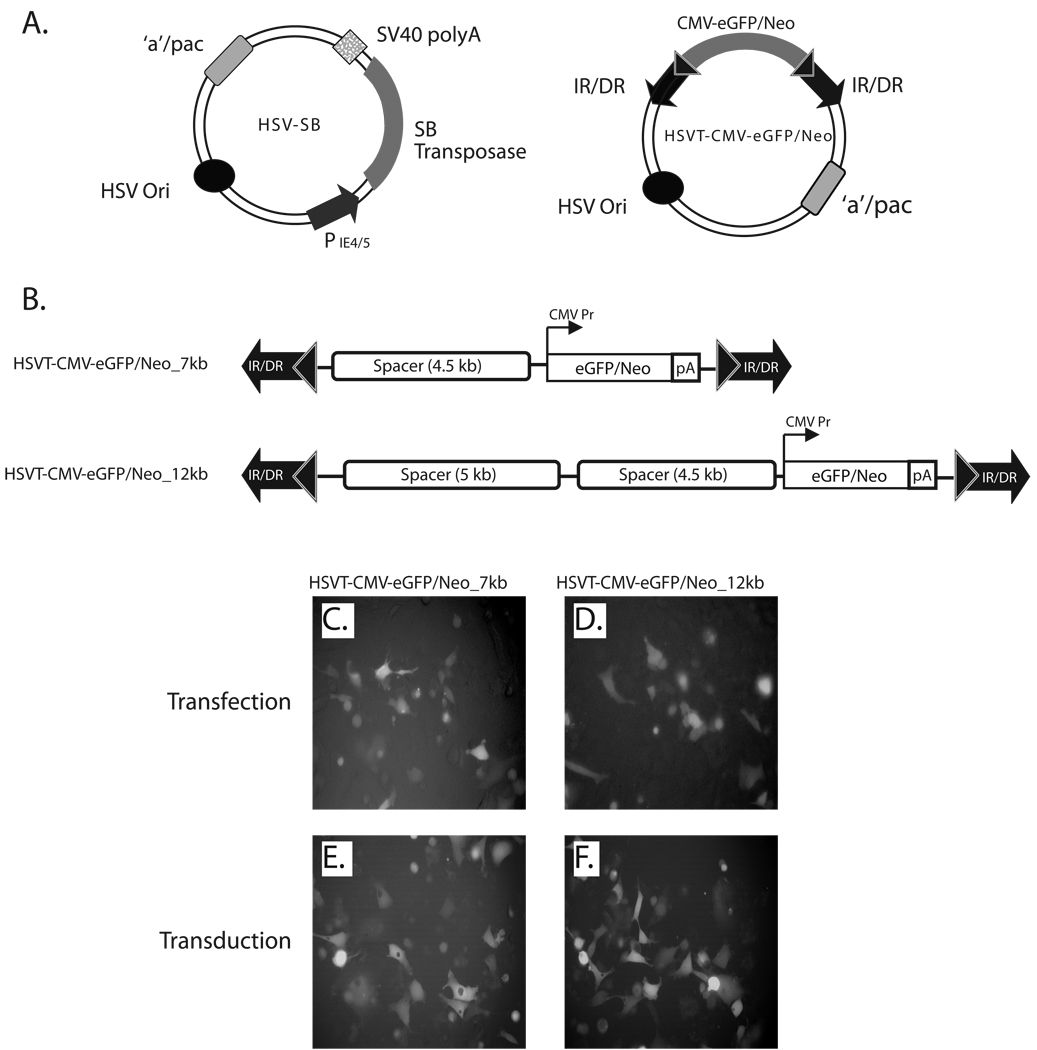
The HSV/Sleeping Beauty bipartite amplicon vector platform consists of an HSV-SB amplicon vector, which expresses in trans the SB transposase gene under the transcriptional regulation of the HSV IE 4/5 promoter and an integration-competent reporter amplicon vector (termed HSVT-CMV-eGFP/Neo) that harbors the SB IR/DR elements, which in turn flank an enhanced green-fluorescent protein fused to a neomycin phosphotransferase gene (eGFP/Neo) under the transcriptional regulation of the cytomegalovirus (CMV) promoter (Figure 1A). To test whether the current HSV/SB amplicon vector platform could deliver and stably integrate large trangene units within the human cell genome, we constructed two differently sized transposable reporter units (7 kb and 12 kb) within the HSVT-CMV-eGFP/Neo vector by incorporating non-coding, spacer DNA fragments 5’ of the CMV-eGFP/Neo transcription unit to generate the pHSVT-CMV-eGFP/Neo_7kb and pHSVT-CMV-eGFP/Neo_12kb amplicon plasmids (Figure 1B). eGFP expression from these HSVT amplicon plasmids was confirmed in HeLa cells 48-h post-transfection by fluorescence microscopy (Figure 1C and D). Subsequently, helper virus-free packaging technology was employed to produce the HSVT-CMV-eGFP/Neo_7kb and HSVT-CMV-eGFP/Neo_12kb amplicon stocks as described previously 15, and eGFP expression was confirmed in HeLa cells by fluorescence microscopy 48-h post-transduction (Figures 1E and F).
Figure 1. Schematic representation of the bipartite HSV/Sleeping Beauty amplicon vector platform and expression testing of the 7- and 12-kb transposable reporter transcription units within the plasmid and viral forms of the HSVT amplicon vector.
(A) The two-component HSV/Sleeping Beauty (SB) amplicon vector platform consists of a HSV-SB amplicon vector, which expresses the SB transposase under the transcriptional regulation of the HSV IE 4/5 promoter and an integration-competent reporter amplicon vector, termed HSVT-CMV-eGFP/Neo. The latter harbors the IR/DR DNA elements of SB, which flank an enhanced green fluorescent protein gene fused to a neomycin phosphotransferase gene (eGFP-Neo) under the transcriptional regulation of the cytomegalovirus (CMV) promoter. (B) To generate 7-kb and 12-kb transposable segments, the CMV-eGFP/Neo-SV40 polyA (2.5 kb) transcription unit was excised from the pBSFBR-CMV-eGFP/Neo construct using NotI (New England BioLabs, MA) and cloned into the corresponding NotI site in a previously engineered PmeI site-containing pBSIIKS(+) shuttle vector to generate pBSIIKS/P_CMV-eGFP/Neo. Subsequently, a 4.5-kb non-coding spacer DNA fragment was obtained by digestion of pDelta28E4 (kindly provided by Dr. Brendan Lee) with Hind III, and was cloned into the corresponding Hind III site 5’ to the CMV-eGFP/Neo transcription unit to generate pBSIIKS_CMV-eGFP/Neo_4.5kb. Thereafter, the CMV-eGFP/Neo (2.5-kb) transcription unit plus the 4.5-kb spacer DNA fragment were excised using PmeI. This 7-kb segment was cloned into a blunted NotI site in the pHSVTmcs amplicon to generate the pHSVT-CMV-eGFP/Neo_7kb amplicon plasmid. To extend the size of the transposable segment to 12-kb, a 5-kb non-coding spacer fragment, which was also obtained from the pDelta28E4 plasmid via digestion with FspI, was cloned into a blunted ClaI site located upstream of the 4.5 kb-spacer fragment within pHSVT-CMV-eGFP/Neo_7kb to generate pHSVT-CMV-eGFP/Neo_12kb. Both constructs were verified by DNA sequence analysis and subsequently packaged into helper virus-free HSV amplicon particles and viral titers were determined as described previously.15 (C, D) To verify eGFP expression from both pHSVT-CMV-eGFP/Neo_7kb and pHSVT-CMV-eGFP/Neo_12kb amplicon plasmids, separate cultures of 5 × 104 HeLa cells were transfected with each construct using FuGENE6® transfection reagent (Roche, Germany) and visualized by fluorescent microscopy 48 hrs post-transfection. (E, F) Similarly, eGFP expression derived from packaged HSV amplicon particles were confirmed by transducing HeLa cell monolayers using HSVT-CMV-eGFP/Neo_7kb and HSVT-CMV-eGFP/Neo_12kb virus at an MOI of 0.1 for 1 h at 37° C. After the 1−h incubation, virus-containing media was removed, the cells were rinsed once with media, and subsequently replenished with fresh media. eGFP expression was observed by fluorescent microscopy 48 h post-transduction.
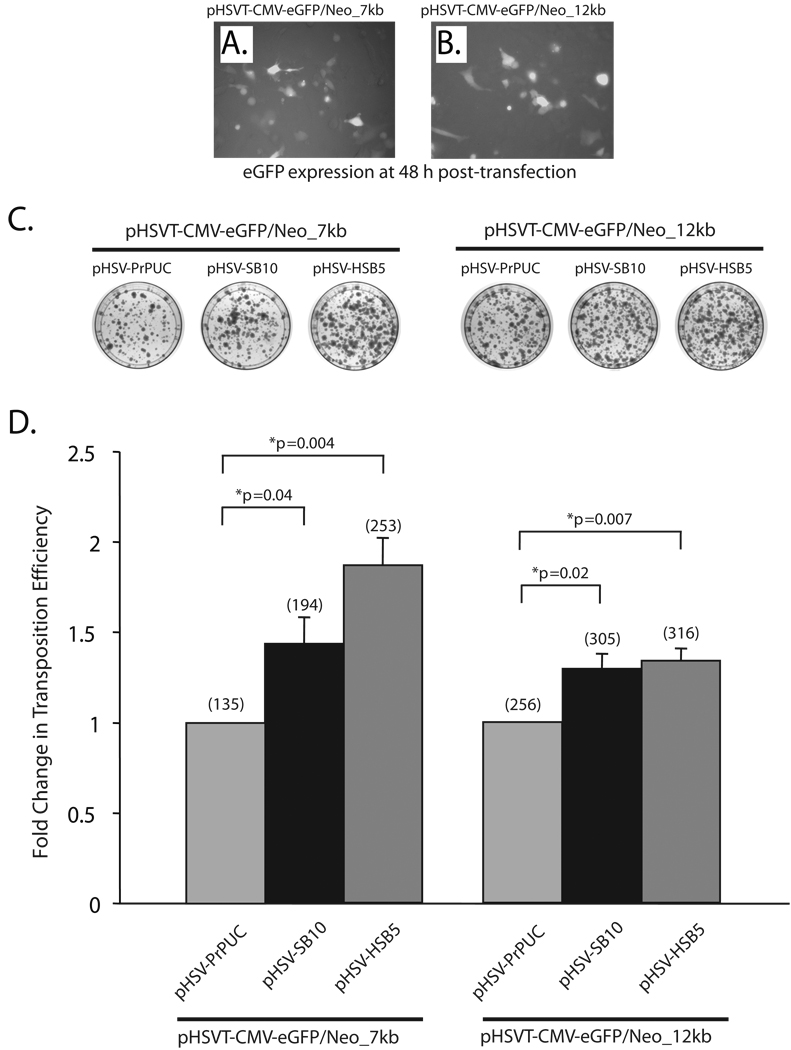
To test the transposition efficiency of the ‘wild-type’ transposase (SB10) as well as a hyperactive version of the SB transposase (HSB514) in the context of the HSV amplicon plasmids harboring the 7-kb and 12-kb transposable units, we performed a colony-forming assay in HeLa cells (Figure 2). An equivalent amount of the SB transposase expressing HSV amplicon plasmid (pHSV-SB10 or pHSV-HSB5) was co-transfected with either the pHSVT-CMV-eGFP/Neo_7kb or pHSVT-CMV-eGFP/Neo_12kb amplicon plasmid in HeLa cells. As depicted in representative images in Figure 2A and B, comparable number of cells were transfected with the differently sized transposable reporter constructs as detected by fluorescence microscopy 48 h post-transfection. Two days post-transfection, the cells were placed under G418 selection for a period of two weeks, at which point the G418-resistant colonies were fixed with 4% paraformaldehyde, stained with methylene blue, and enumerated to determine the transposition level (Figure 2C). The pHSVPrPUC amplicon plasmid served as an empty vector control to assess the number of G418-resistant colonies arising from random integration events of the pHSVT-CMV-eGFP/Neo_7kb and pHSVT-CMV-eGFP/Neo_12kb amplicon plasmids. The efficiency of each SB transposase to mobilize the IR/DR-flanked units was represented as fold-change relative to the pHSVPrPUC empty vector control, wherein SB transposase was absent. As shown in Figure 2D, the SB10 transposase exhibited a 1.43-fold increase in the transposition efficiency of the 7-kb transposable unit compared to the pHSVPrPUC empty vector control. Furthermore, as the size of the transposable unit was increased to 12-kb, the transposition efficiency of SB10 decreased by 10% as shown in Figure 2D. The hyperactive HSB5 transposase, which has been predicted to form a more stable synaptic complex 14, showed a higher fold-change in transposition efficiency with the 7-kb transposable unit compared to the SB10 transposase (Figure 2D). However, the transposition efficiency of HSB5 pertaining to the 12-kb transposable unit was diminished to levels similar to SB10 transposase. These results are consistent with previous studies that assessed the transposable cargo capacity of the “wild-type” SB transposase, where a significant decrease in the transposition efficiency was observed as the size of the transposable segment exceeded 6-kb.10–12 Additionally, our results suggest that the use of hyperactive mutants of the SB transposase may be beneficial in enhancing the transposition efficiency of transgene units ≤7-kb, but does not appear to significantly impact efficiencies of transposing larger transgene segments from the context of plasmid-based vectors. The observed diminution in transposing large DNA segments (>6-kb) via the SB transposase from the context of a plasmid-based vector could be due to several factors: (1) inefficient transport/delivery of large DNA molecules from the plasma membrane into the host cell nucleus; (2) the inability to form a functional synaptic complex due to the large intervening DNA segment between the IR/DR elements 10,12; and/or (3) the length and sequence features of the DNA flanking the IR/DR elements, which comprise the plasmid vector backbone, may unduly influence the transposition process.10,16
Figure 2. Assessing the transposition efficiency of SB transposase in the context of HSV amplicon plasmids harboring differently sized transposable units.
HeLa cells (1×105 cells/well) were co-transfected with 165 ng of the plasmid versions of the HSV amplicon vectors expressing either the ‘wild-type’ SB transposase (pHSV-SB10) or the hyperactive HSB5 transposase (pHSV-HSB5), together with either 116 ng of the pHSVT-CMV-eGFP/Neo_7kb amplicon or 165 ng of the pHSVT-CMV-eGFP/Neo_12kb amplicon using the FuGENE6® transfection reagent (Roche, Germany). The pHSVPrPUC empty vector plasmid was transfected with each of the differently sized eGFP/Neo transposon-harbored amplicons to determine the number of G418-resistant colonies that arise due to random integration events in the absence of SB transposase. To ensure equal amounts of DNA were transfected under each condition, the pHSVPrPUC empty vector was used as ”filler” DNA. (A, B) eGFP expression was assessed 48 h post-transfection via fluorescence microscopy to determine the transfection efficiency of the pHSVT-CMV-eGFP/Neo_7kb and the pHSVT-CMV-eGFP/Neo_12kb amplicon plasmids in the presence of the SB-expressing amplicon plasmid. (C) Thereafter, the cells were trypsinized and seeded at a 1:3 dilution on 100-mm dishes and placed under 600 µg/ml G418 selection for a period of 2 weeks. Subsequently, the cells were fixed with 4% paraformaldehyde and stained with 2% methylene blue, washed extensively with dH2O and blue colonies were enumerated. (D) The transposition efficiency of SB10 and HSB5 in the presence of either the 7- or 12-kb transposons is represented as a fold-change in the number of G418-resistant colonies compared to when SB transposase was absent. The average number of G418-resistant colonies (n=3) corresponding to each group is indicated in parentheses above each bar in the histogram. Error bars represent standard error. Statistical analysis was conducted using the student t-test and p values are indicated.
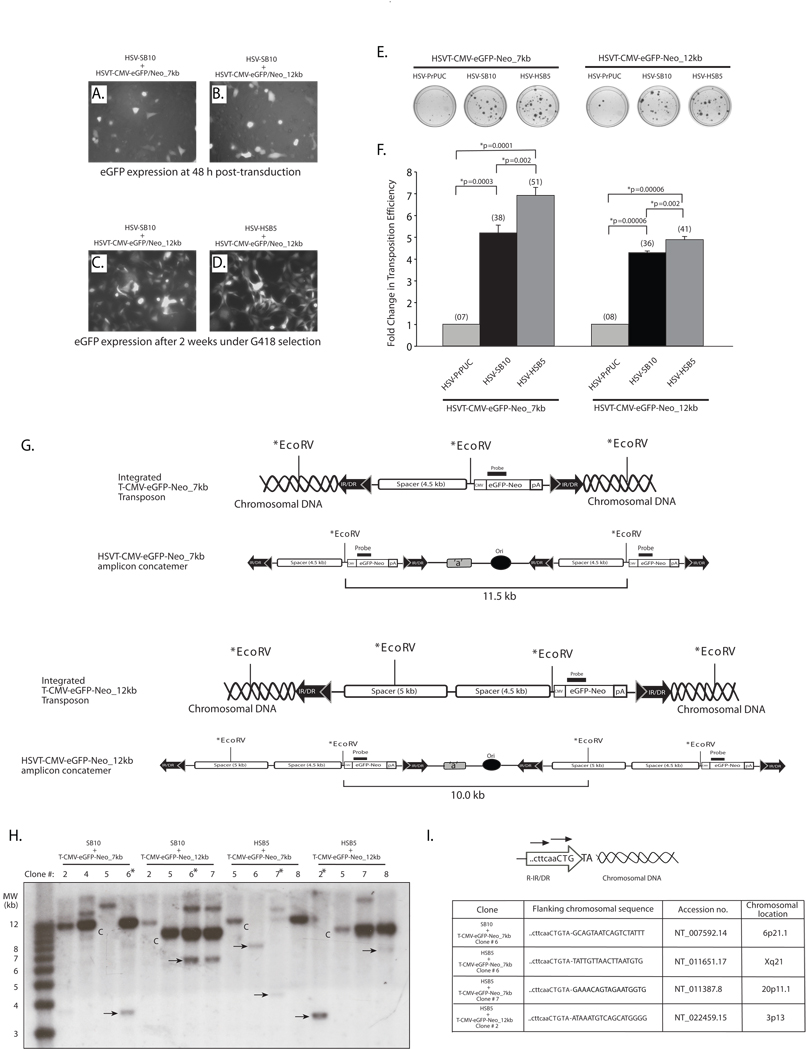
Considering the limitations associated with SB-mediated transposition of large transgene units from plasmids, we assessed whether a transposon-harbored HSV amplicon particle might provide a more amenable platform from which large transgene units can be efficiently mobilized into the host cell genome. To this end, we individually co-transduced either packaged HSV-SB10 or HSV-HSB5 amplicon with the HSVT-CMV-eGFP/Neo_7 kb and HSVT-CMV-eGFP/Neo_12 kb amplicons and performed a colony formation assay as described above. As depicted in representative images in Figures 3A and B comparable number of cells were transduced with the differently sized transposable reporter amplicons in the presence of the SB-expressing amplicons 48 h following transduction. After two weeks of G418 selection, the colonies were analyzed by fluorescence microscopy to confirm eGFP expression (Figures 3C and 3D), and subsequently stained with methylene blue (Figure 3E) and colonies enumerated to assess the transposition efficiency of the 7- and 12-kb transgenon units in the presence of the “wild-type” and hyperactive SB transposases (Figure 3F). Upon enumeration of the G418-resistant colonies, we observed a 5.18-fold increase in the transposition efficiency when the HSVT-CMV-eGFP/Neo_7kb amplicon was co-delivered with the HSV-SB10 amplicon compared to the empty vector control (HSVPrPUC) (Figure 3F). Furthermore, the HSB5 transposase expressed via the HSV amplicon increased the transposition efficiency of the 7-kb transposable unit by 33% over the “wild-type” SB10 transposase. When the HSVT-CMV-eGFP/Neo_12kb amplicon was co-transduced with either the HSV-SB10 or HSV-HSB5 amplicons, we observed a 4.28 and 4.88-fold increase in the transposition efficiency, respectively, compared to when the HSVPrPUC control amplicon was present. Although a decrease in the transposition efficiency was observed with SB10 and HSB5 transposase when the size of the transposon was increased from 7 to 12 kb within the HSV amplicon, it was significantly higher than plasmid-based transposition of identically sized transposable units.
Figure 3. The SB transposase is competent in efficiently mobilizing a 12-kb transposable unit from the context of an HSV amplicon vector.
HeLa cells (1×105 cells/well) were co-transduced with HSV amplicon vectors expressing either the “wild-type” SB transposase (HSV-SB10) or the hyperactive HSB5 transposase (HSV-HSB5), together with either the HSVT-CMV-eGFP/Neo_7kb amplicon or the HSVT-CMV-eGFP/Neo_12kb amplicon at an MOI of 1.0. Upon incubation for 1 h at 37°C, the virus-containing medium was removed and cells were washed once and replenished with fresh media. (A–B) Enhance GFP expression was assessed 48 h post-transduction via fluorescence microscopy to determine the transduction efficiency of the HSVT-CMV-eGFP/Neo_7kb and HSVT-CMV-eGFP/Neo_12kb amplicons in the presence of the co-delivered SB-expressing amplicon vector. Subsequently, the cells were trypsinized and seeded at a 1:3 dilution on 100-mm dishes and placed under 600 µg/ml G418 selection for a period of 2 weeks. (C–D) After 2 weeks, G418-resistant colonies were fixed with 4% paraformaldehyde and analyzed via fluorescence microscopy for eGFP expression. Representative images of G418-resistant colonies 2 weeks post-transduction of HSVT-CMV-eGFP/Neo_12kb either with HSV-SB10 or HSV-SB12 are shown. (E) Upon confirmation of eGFP expression, the G418-resistant colonies were stained with 2% methylene blue, washed extensively with dH2O, and blue colonies were enumerated. (F) The transposition efficiency of SB10 and HSB5 in the presence of either the 7- or 12-kb transposon-harboring HSVT amplicons is represented as a fold-change in the number of G418-resistant colonies compared to when SB transposase was absent (HSVPrPUC control amplicon). The average number of G418-resistant colonies (n=3) corresponding to each group is indicated in parentheses above each bar in the histogram. Error bars represent standard error. Statistical analysis was conducted using the student t-test and p values are indicated. (G) Schematic representations of integrated configurations of the T-CMV-eGFP-Neo_7kb and T-CMV-eGFP-Neo_12kb transgenons and their concatemeric amplicon genome counterparts. The relative locations of signature EcoRV restriction enzyme recognition sites and the location of a DNA probe used in subsequent Southern blot analyses are depicted. (H) Similarly, G418-resistant colonies that were co-transduced with either HSV-SB10 or HSV-HSB5 and HSVT-CMV-eGFP/Neo_7kb or HSVT-CMV-eGFP/Neo_12kb were trypsinized and expanded in 60-mm dishes for the purpose of isolating genomic DNA for Southern blot analysis to determine chromosomal integration of the 7- and 12-kb transposons. Once confluent, these transduced HeLa cell monolayers were lysed using lysis buffer (10 mM Tris.Cl, 100 mM NaCl, 25 mM EDTA and 0.5% SDS) and genomic DNA was isolated using phenol:chloroform extraction followed by ethanol precipitation. Ten micrograms of genomic DNA was digested with EcoRV, which cuts once within the 7-kb transposable unit 5’ of the CMV-eGFP-Neo transcription unit and twice within the 12-kb transposable unit. The digested genomic DNA was electrophoresed on a 0.8% TAE agarose gel and transferred onto a nylon membrane, UV cross-linked and probed with a α-32P-dCTP-radiolabeled eGFP/Neo probe (917-bp), which was obtained by excising the eGFP-Neo fragment from the pBSFBR-CMV-eGFP-Neo vector using Pst1 and BamHI. The 1-kb plus DNA ladder (Invitrogen, Carlsbad, CA) was radiolabeled according to manufacturer recommendations (Lane 1). “C” indicates HSV amplicon units generated by EcoRV digestion of the episomal HSV amplicon concatemer or integrated concatemeric transgenon units, while the ‘arrows’ indicate predicted SB-mediated integration events within the HeLa cell genome and “*” demarcates clones later analyzed by integration site mapping. (I) To determine the integration sites of the 7-kb and 12-kb transposable units within the HeLa cell genome, linker-mediated PCR (LM-PCR) analysis was conducted using the GenomeWalker™ Universal Kit (Clontech, Mountain view, CA) according to manufacturer recommendations. Chromosomal sequences flanking the right IR/DR junction of the 7- and 12-kb transposable units were PCR amplified from genomic DNA samples that were analyzed by Southern blotting using a nested primer set described in Largaespada et al.,25 and cloned into the TOPO-TA vector (Invitrogen, Carlsbad, CA) for subsequent sequence analysis.
To determine the fold-increase in transposition efficiency of a 12-kb transgenon from the HSV amplicon as compared to the plasmid version of the amplicon, we calculated the transposition efficiency per 12-kb unit delivered via each modality. We initially calculated the numbers of 12-kb transgenon copies applied during monolayer transduction by multiplying the numbers of transduced viral particles by the factor: 150 kb (wild-type HSV genome size) divided by the total size of the transgenon-harbored amplicon in kilobases. This number was subsequently divided into the number of G418-resistant colonies resulting from SB-mediated transposition events (total number of G418-resistant colonies minus number of background G418-resistant colonies resulting from SB-independent integration). The frequency of transposition per 12-kb transposable unit was determined to be 4.94×10−4. Similarly, the frequency of transposition from the plasmid version of the HSV amplicon was calculated using the numbers of 12-kb transgenon units contained within 165 ng of transfected pHSVT-CMV-eGFP/Neo_12kb plasmid DNA and the numbers of G418-resistant colonies resulting from SB transposition. The frequency of transposition from the amplicon plasmid was 3.01×10−11 per 12-kb transposable unit. Hence, our results indicate that the frequency of SB-mediated transposition of the 12-kb transgenon from the HSV amplicon was approximately 1.7×107-fold higher than plasmid-based transposition.
To confirm SB-mediated integration of the eGFP/Neo-marked 7- and 12-kb transposable units in the HeLa cell genome, Southern blot analysis was conducted using an eGFP/Neo-specific radiolabeled DNA probe (917-bp) on genomic DNA isolated from G418-resistant colonies arising from co-transduction of the SB transposase expressing HSV amplicons with either HSVT-CMV-eGFP-Neo_7kb and HSVT-CMV-eGFP-Neo_12kb amplicons. The EcoRV-generated ~11.5-kb and ~10-kb eGFP/Neo-positive radiolabeled bands that arise respectively in the HSVT-CMV-eGFP-Neo_7kb and HSVT-CMV-eGFP-Neo_12kb amplicon samples result from detection of either the retained episomal amplicon genomes within the transduced cell or integrated concatemeric copies of the transgenon resulting from the usage of non-consecutive IR/DR sites by SB transposase during transposition (see schematic in Figure 3G). Our results indicate that the “wild-type” SB10 transposase and the hyperactive HSB5 transposase mediated an average of 1–2 integration events per G418-resistant colony for each of the 7- and 12-kb transposable units (Figure 3H). The additional eGFP-Neo-positive bands in each lane result from EcoRV restriction digest patterns consistent with integration of either the 7-kb or 12-kb transposable units within the HeLa cell genome. Furthermore, in certain clones, such as HSV-HSB5 + HSVT-CMV-eGFP/Neo_7kb clone #6 and HSV-HSB5 + HSVT-CMV-eGFP/Neo_12kb clone #2, it is evident that the hyperactive HSB5 transposase integrated single transgenon units from the HSV amplicon. Linker-mediated PCR analysis was also conducted on a subset of genomic DNA samples from G418-resistant colonies to determine flanking genomic sequences of the integrated 7-kb and 12-kb transposable units within the HeLa cell genome (Figure 3I). The presence of the cttcaaCTGTA signature sequence of the right IR/DR junction flanked by chromosomal DNA sequences indicated that SB transposase followed its characteristic ‘cut-and-paste’ mechanism of transposition with the integration of large transposable units into chromosomal TA-dinucleotide sites, which are duplicated following SB-mediated integration. Overall, these results indicate that delivery of large transgenon units via HSV amplicon particles provides a more amenable substrate for the SB transposase for integration into the human genome than plasmid vectors harboring the same transposable segments delivered via currently available lipid-based DNA delivery methods.
We hypothesize that the increased ability of the HSV/SB amplicon vector platform to facilitate integration of large transgenon units within the human genome could occur for several reasons. First, the enclosure of the transposon-encoded HSV amplicon genome within the nucleocapsid leads to efficient delivery of its genetic cargo into the transduced cell nucleus. Second, certain viral tegument proteins as well as packaging cell line-derived proteins co-packaged within the HSV virion 15 may enhance the transposition process of large transposable units from the HSV amplicon. It has been previously reported that SB-mediated transposition involves host cell factors such as the high mobility group DNA binding protein (HMGB1) 13,17 and DNA-PKcs/Ku, which are members of the non-homologous DNA repair pathway 18,19, and transcription factors that regulate the cell cycle, such as Miz-1.20 Zayed et al, demonstrated that HMGB1 interacted with SB transposase and increased its binding affinity to the inner DR element, possibly facilitating the formation of the synaptic complex for efficient transposition.17 Furthermore, transient over-expression of HMGB1 resulted in an enhancement in the transposition efficiency of SB in mouse embryonic fibroblasts 17 and HeLa cells.13 Thus, the presence of packaging cell line-derived HMGB1 protein in HSV viral stocks could inadvertently set up an optimized platform for mobilizing large transgenons from the HSV amplicon vector. Third, the circularization of the HSV amplicon vector genome upon entry into the transduced cell nucleus could facilitate efficient synaptic complex formation between the IR/DR elements that flank these large transgene units and the SB transposase. Finally, SB-mediated transposition has been shown to be more efficient from a chromosomal context harboring a multi-copy transposon donor substrate in comparison to a single-copy donor locus.21,22 Thus, the head-to-tail configuration of the concatemeric transposon-encoded HSV genome may provide a better substrate for the SB transposase to catalyze its excision and integration events. However, further investigations are required to confirm the characteristics of the HSV amplicon-vector genome that facilitate efficient transposition of large transposable units via the SB transposon system.
The integration-competent HSV/SB amplicon vector platform is a versatile gene delivery modality for genetic diseases that require long-term expression of the therapeutic gene(s) within the affected target cell population. For the first time we have demonstrated that the this gene delivery platform is capable of effectively mobilizing transgene segments up to at least 12 kb into the human genome, thereby exceeding the current integration capacity of adeno-associated viral vectors and lentiviral vectors. This allows the delivery of multiple therapeutic genes, as well as native transcriptional regulatory elements, to increase the efficacy and cell specificity of treatment strategies for multi-factorial genetic diseases. We also hypothesize that further optimization of the HSV/SB amplicon vector platform with the use of more hyperactive versions of the SB transposase (such as the SB100X 22) coupled with the incorporation of more optimized IR/DR elements (such as the T2 23 and ‘sandwich-based’ IR/DR elements 13) into the HSV amplicon vector may further extend the upper bounds of SB-mediated transposable capacity. While we have demonstrated that the current bipartite HSV/SB amplicon vector platform mobilizes large transgenon segments into the human genome, it still manifests a random integration profile characteristic of SB with the attendant potential risk of insertional mutagenesis.8,24 Thus, it would be advantageous to target the integration of transgenons to a “safe” chromosomal location within the human genome, which would increase the safety profile of the HSV/SB amplicon vector platform and expedite its translation to clinical testing.
Acknowledgements
We would like to thank Dr. Brendan Lee (Baylor College of Medicine) for kindly providing the DNA spacer elements. NIH U54-NS045309 (HJF/WJB) supported this work.
Reference
- 1.Hermonat PL, Quirk JG, Bishop BM, Han L. The packaging capacity of adeno-associated virus (AAV) and the potential for wild-type-plus AAV gene therapy vectors. FEBS Lett. 1997;407:78–84. doi: 10.1016/s0014-5793(97)00311-6. [DOI] [PubMed] [Google Scholar]
- 2.Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 3.Senior SL, Wade-Martins R. Herpes simplex virus type 1 amplicon vectors for the infectious delivery and expression of genomic DNA loci. Curr Opin Mol Ther. 2005;7:337–345. [PubMed] [Google Scholar]
- 4.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, et al. Herpes simplex virus type 1/adeno-associated virus rep(+) hybrid amplicon vector improves the stability of transgene expression in human cells by site-specific integration. J Virol. 2002;76:7150–7162. doi: 10.1128/JVI.76.14.7150-7162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heister T, Heid I, Ackermann M, Fraefel C. Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19. J Virol. 2002;76:7163–7173. doi: 10.1128/JVI.76.14.7163-7173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 8.Bowers WJ, et al. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Peterson EB, Mastrangelo MA, Federoff HJ, Bowers WJ. Neuronal specificity of HSV/sleeping beauty amplicon transduction in utero is driven primarily by tropism and cell type composition. Mol Ther. 2007;15:1848–1855. doi: 10.1038/sj.mt.6300267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 11.Karsi A, Moav B, Hackett P, Liu Z. Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol (NY) 2001;3:241–245. doi: 10.1007/s101260000072. [DOI] [PubMed] [Google Scholar]
- 12.Geurts AM, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 13.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 14.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowers WJ, et al. Expression of vhs and VP16 during HSV-1 helper virus-free amplicon packaging enhances titers. Gene Ther. 2001;8:111–120. doi: 10.1038/sj.gt.3301340. [DOI] [PubMed] [Google Scholar]
- 16.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zayed H, et al. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yant SR, Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izsvak Z, et al. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 20.Walisko O, et al. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc Natl Acad Sci U S A. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusa K, Takeda J, Horie K. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol Cell Biol. 2004;24:4004–4018. doi: 10.1128/MCB.24.9.4004-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mates L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 23.Cui Z, et al. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 24.Yant SR, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]