Abstract
Core binding factor 1 (Cbfa1)/runt-related transcription factor 2 (Runx2) has been identified as a ‘‘master gene’’ in osteoblastic differentiation. In this two-part study, part I of the study was undertaken to test the hypothesis that bone regeneration is compromised in Cbfa1+/− mice. Compared with wild-type mice, wound healing was dramatically delayed in Cbfa1+/− mice characterized by the presence of a small amount of bone near the base of the wounds. The bone defects were largely filled with fibrous connective tissues 3 weeks after surgery. Part II was performed to determine the effects of Cbfa1 in enhancing bone wound healing using a gene-activated matrix (GAM) method. Cbfa1 cDNA was mixed with a biodegradable bovine type I collagen sponge and was inserted into the periodontal window wounds of mice. Control sponges were collagen matrix without Cbfa1 cDNA. Histological analysis and immunohistochemical staining demonstrated that compared with controls, there was increased new bone formation that almost filled the wound defects 14 days after surgery in the Cbfa1-GAM group. The collagen sponge matrix did not seem to elicit significant foreign body reaction in either group. In conclusion, the reduced expression of Cbfa1 interferes with the process of bone wound healing, and local application of Cbfa1 cDNA incorporated into a collagen matrix promotes bone tissue regeneration.
Millions of Americans are afflicted with periodontal diseases that cause destruction of supporting structures of teeth: periodontal ligament (PDL), alveolar bone, and cementum. The process can lead to the loss of attachment, with destruction of the connective tissue matrix and cells. Loosening and eventual loss of teeth may follow. However, despite considerable research efforts in this area, regeneration of the periodontium has still proved to be an elusive goal of periodontal therapy and remains a subject of intense interest to dentists and dental scientists, with the molecular and cellular bases of PDL formation, repair, and regeneration still poorly understood.
In the 1980s, Gould observed that the progenitor cells for cementum, bone, and PDL fibroblasts were all contained in PDL tissue.1 He described generation of a new, functionally oriented PDL when teeth bearing cultured PDL cells were transplanted into bony sites. By contrast, the transplantation of teeth bearing cultured gingival cells did not lead to regeneration of the PDL. These early studies indicated that regeneration of damaged periodontium can be achieved by the activity of the local stromal stem or progenitor cells found in paravascular locations of the PDL. These progenitor cells, which are stimulated to proliferate at wound sites, are derived from adjacent, unwounded PDL. Selective repopulation of the root surface by PDL cells and subsequent differentiation of these cells into fibroblasts, osteoblasts, and cementoblasts is believed to provide the basis for the regeneration of periodontal tissues and for the restoration of tissue domains.2
Similar to adipocyte3 and myoblast4 differentiation, it is apparent that components of the extracellular matrix are also critical for initiating cell differentiation during osteogenesis.5 In the 1990s, independent studies of a transcription factor that regulated the tissue-specific expression of osteocalcin (OCN)6,7 and analysis of transgenic knockouts8,9 resulted in the identification of core-binding factor 1 (Cbfa1), a runt domain transcription factor and its cognate enhancer. This transcription factor, also referred to as polyoma enhancer-binding protein (PEBP2aA), osteo-blast-specific factor (Osf2), and more recently as runt-related transcription factor 2 (Runx2), is identified as a ‘‘master gene’’ required for osteoblastic differentiation.10 Cbfa1 is expressed during the early development of mesenchymal and epithelial tissues destined to form the mineralized tissues of the tooth and periodontal apparatus.11,12 In Cbfa1−/− mice,8,9 there is an almost complete lack of mineralized bone. The absence of the osteodifferentiation markers, alkaline phosphatase and OCN, is consistent with an early block in osteogenic development in these mice. Furthermore, numbers of in vitro studies have shown that Cbfa1 is a positive regulator that can up-regulate the expression of bone matrix genes, including type I collagen, osteopontin (OPN), OCN, fibronectin, and bone sialoprotein (BSP).13–16 And it is reported that various mutations in the human Cbfa1 gene locus, on chromosome 6p21, have been identified to correlate with the disease of cleidocranial dysplasia (CCD). This disease is an autosomal dominant disorder that is characterized by a short stature, varying degrees of loss of the clavicles, open cranial fontanels, and dental manifestations including supernumerary teeth, delayed tooth eruption, tooth hypoplasia, and absence of cellular cementum formation.9,17,18 Although evidences have shown that Cbfa1 has an essential role in osteogenesis, an unambiguous effect of Cbfa1 in promoting differentiation of regenerative cells in periodontium has not yet been established. Therefore, to determine the function of Cbfa1 in bone repair and regeneration, we proposed a loss-of-function study to test the hypothesis that bone regeneration is compromised in Cbfa1+/− mice.
In the second part of this research, a gain-of-function study was performed. It is well known that bone regeneration and tissue engineering are of enormous importance, particularly for reconstruction of oral and maxillofacial tissues that have been lost or damaged by diseases, trauma, or congenital disorders. The ultimate therapeutic goal of corrective surgery is to regenerate tissues to their normal or predisease state. Considering the key role of Cbfa1 in osteoblast differentiation, it can be deduced that Cbfa1 may also play an important role in bone regeneration and may induce an embryonic (regenerative) environment in the injured adult tissues. Here, in the second part of this study, the effects of overexpressed Cbfa1 in enhancing bone wound healing using a gene-activated matrix (GAM) method were determined.
MATERIALS AND METHODS
Animals
Six- to 8-week-old mice were maintained and used in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals, prepared by the Institute on Laboratory Animal Resources, National Research Council (DHHS Pub. NIH 86-23, 1985). The animal protocol was approved by the Institutional Animal Use and Care Committee at the Tufts-New England Medical Center in Boston (Massachusetts).
Part I
Wound model
Periodontal window wounds and femoral defects were created in four Cbfa1+/− mice (a generous gift from Dr. Michael Owen, Imperial Cancer Research Fund, London, UK) and four wild mice under general anesthesia (a mixture of ketamine and xylazine, 110 and 10 mg/kg). Periodontal window wounds (1 mm in diameter) were an established model as described before.19,20 In brief, a skin flap was raised along the lower edge of the mandible to expose the underlying bone. The site of the window wound was 1 mm away from the anterior edge of the mandible and between the superior margin of the alveolar bone overlying the mesial root of the lower first molar and the foramen mentale. Under a dissecting microscope at ×10 magnification, a dental burr (#329, Midwest, 0.6 mm in diameter) driven at a low speed by a dental handpiece and cooled with phosphate-buffered saline (PBS) was used to penetrate the overlying alveolar bone. The PDL was removed with a sharpened dental probe. Wounds were untreated and tissues were closed with 4–0 gut suture. Animals were closely observed for 1 hour after surgery. Femoral defects (1 mm in diameter) were created using a 0.6 mm cutting burr through a lateral incision after general anesthesia described above.
Tissue preparation
The animals were sacrificed 3 weeks after wounding. After euthanasia, the part of the mandible between the central incisor and the third molar region was excised and for femoral defects, the whole femur was dissected. The tissues were fixed immediately in periodate-lysine-paraformaldehyde21 for 24 hours. The tissue was demineralized in 0.2 N HCL for 2 days with constant stirring. Demineralized tissues were washed in 0.1 M phosphate buffer (pH 7.2) overnight. The tissues were then dehydrated in an ascending series of ethanol, cleared in xylene, and embedded in paraffin. As described in previous publications,22 tissue sections, 6 μm thick, were mounted on glass slides.
Histomorphometric assessment
For each animal, three hematoxylin & eosin (H&E)-stained sections of each wound were analyzed morphometrically (Carl Zeiss Inc., Hamburg, Germany). With the aid of the computer software package, Image-pro plus (Media Cybernetics), image analysis was performed to assess: (a) the area of the drilled bone by digitizing the reversal line in the bone at the cut margin that provided an estimate of the original wound outline for each animal and the width of closure of the bony defect at the outer surface of the drilled site, (b) the area of the newly formed bone, (c) these two areas were expressed as a percentage (area of newly formed bone/area of defect × 100). Mean ± standard errors of the percentages were computed, and (d) the width of regenerating PDL was compared with the unwounded side (as a control) in the same sample.
Part II
Cbfa1 plasmid DNA preparation
Large-scale DNA preparation of the plasmid pCMV-Osf2/Cbfa1 (containing the complete cDNA of mouse Cbfa1, provided by Dr. Gerard Karsenty, Bayer College of Medicine, Houston, TX) was performed using an EndofreePlasmid Mega kit (Qiagen, Valencia, CA). The plasmid DNA was stored at −20°C in endotoxin-free TE buffer before use.
Assembly of Cbfa1 GAM prototypes
Cell Prime, an acid-soluble form of bovine type I collagen, was purchased from Cohesion Technologies (Palo Alto, CA). To generate GAM, equal volumes of sterile tissue, culture medium (Dulbecco’s modified Eagle’s medium [DMEM]) and bovine collagen were mixed to achieve a pH of approximately 7.0. Sterile, pure CMV-Cbfa1 plasmid DNA in TE buffer was mixed with the neutralized collagen with a concentration of 1.2 mg DNA in 1 mg collagen. The DNA and the collagen were mixed at room temperature until a uniform distribution of plasmid DNA was achieved. The DNA–collagen mixture was then frozen at −86°C and lyophilized.
Transplantation of Cbfa1-GAM into wounds
Periodontal window wounds were created in 22 seven-week-old mice as described in part I. The mice were divided into two groups randomly. In group I, bone defects were filled with Cbfa1 GAM with a dose of 1.2 mg plasmid DNA per wound. Sterile GAM prototypes were placed and held in the defect sites until they were surrounded by clotted blood. Control sponges in group II were comprised of biodegradable, biocompatible collagen without plasmid DNA. Animals were euthanized at 7 and 14 days after the transplantation, respectively. These time points were chosen because previous studies had shown that 1 week was required for initial healing and cell repopulation of wounded connective tissue (PDL in the periodontal wounds), and 2 weeks were required for the initiation of mineralization of the regenerating new bone. After euthanasia, tissue preparation and histomorphometric analysis were performed as described in Part I.
Immunohistochemical staining
The tissue slides were first deparaffinized and rehydrated, and then submerged in hydrogen peroxide for peroxidase quenching. Before using the primary antibodies, the slides were incubated with normal serum to block the nonspecific bindings. Monoclonal antibodies against BSP (LF120 from Dr. Larry Fisher, NIH/NIDCR) were then used at a dilution of 1 : 200. After overnight incubation, the biotinylated secondary antibodies were applied to the slides. Finally, the substrate-chromogen 3-Amino-9-ethylcarbazole (AEC) was applied and the slides were counterstained with hematoxylin and mounted. Control sections were incubated with an irrelevant antibody (anti-human CD4 lymphocyte antigen) to estimate background staining. All slides were randomly coded to prevent examiner’s bias.
The localization of BSP was studied on transverse sections of the first mandibular molar using previously described semi-quantitative methods.19 In brief, the localization of BSP expression was studied in the newly formed alveolar bone (site 1) and in the unwounded alveolar bone (site 2). The unwounded side serves as an internal control for the assessment of the staining intensity at the wounded side. The relative intensities of BSP staining were classified as intense (3), moderate (2), weak (1), or negative (0). For each slide the intensity of BSP staining was determined by comparing the staining at each wounded site with unwounded alveolar bone in the same section. Unwounded alveolar bone was classified as 2 (or moderate) staining intensity and experimental sites were compared with this internal standard. Stronger staining than internal controls was classified as 3 (intense), lower than internal controls as 1 (low), and no staining as 0 or negative. This method is semi-quantitative and provides meaningful comparative data when differences of staining intensity are large.
Morphometric assessment of immunostained sections
At least three sections of BSP-immunolabeled bone defects in each group were analyzed morphometrically. Slides were coded so that time of killing, type of wound, and type of staining were unknown at the time of measurement. Image analysis (Carl Zeiss Inc., Image-pro plus, Media Cybernetics) was used to assess the area of the drilled bone by digitizing the reversal line in the bone at the cut margin that provided an estimate of the original wound outline for each wound. Second, the area of the BSP-stained tissue in the bone compartment of the wound was digitized. Finally, these two areas were expressed as a percentage (area of BSP staining/area of the defect×100), representing a newly formed bone ratio. Mean ± standard errors of the percentages were computed.
Total RNA extraction and reverse transcriptase polymerase-chain reaction (RT-PCR) analysis
For RT-PCR analysis, the implant samples (3 each time point) were excised under a dissecting microscope to avoid harvesting the host bone. Freshly dissected implant tissues were collected into ‘‘RNAlater’’ solution (Qiagen) at 4°C and homogenized in TRIzol solution (Invitrogen, Carlsbad, CA), followed by the RNA isolation procedure recommended by the manufacturer. Freshly isolated RNA was reverse transcribed with a SuperScript™ first-strand synthesis system (Invitrogen) following the manufacturer’s recommendations. The resulting cDNA was then amplified by PCR with the Platinum PCR supermix (Invitrogen). The sequences of the primers for amplification of mouse Runx2/Cbfa1 were: 5′-GAGGCCGCCGCACG ACAACCG-3′ and 5′-CTCCGGCCCACAAATCTCAGA-3′ (product size: 294 bp); for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCT GTTGCTGTA-3′ (product size: 450 bp). Linearity of the PCR conditions was determined for each primer pair in preliminary experiments (data not shown). Images of the amplified products in 1.5% agarose gels were captured with a UVP bioimaging system and processed by Adobe Photoshop 6.0 (Adobe Systems Incorporated, San Jose, CA) and Scion Image Beta 4.02 (Scion Image, Frederick, MD). The data are presented as the fold changes in gene expression after normalization to the expression of G3PDH.
Statistical analysis
Raw data were kept separate and the means for each animal, type of treatment, postwounding time, and site examined were computed. Mean data from each animal were considered as independent samples. Data were assessed by analysis of variance (ANOVA) to evaluate the differences between the morphometric assessments and immunostaining findings with respect to different sites, type of treatment, i.e., with vs. without Cbfa1 cDNA transplantation, and postwounding time.
RESULTS
In the present two-part study, we used a mouse model to study tissue regeneration and wound repair. Tissue sections were subsequently processed for histomorphometric analyses and immunohistochemical staining of BSP to assess the effect of Cbfa1 on the regeneration of periodontal and femoral defects.
Part I
In wild-type (WT) mice, the relative volume of new bone matrix (Figure 1) was higher than in the Cbfa1+/− mice (p < 0.05). The newly formed bone in WT mice showed a trabecular structure, and the bone was well mineralized and merged with the normal bone at the defect edges. In contrast, wound healing was dramatically delayed in Cbfa1+/− mice characterized by the presence of a small amount of bone near the base of the wounds. The alveolar bone defects were largely filled with fibrous connective tissues 3 weeks after surgery (Figure 2). The bone wounds in femurs in WT mice showed callus formation at the inner side. In some cases, the defects almost healed completely. In Cbfa1+/− mice, the repair occurred at a much slower pace and granulation tissues were seen at the wound sites (Figure 3). In some cases, the femur wound was still widely open without any significant bone formed.
Figure 1.
Graph depicting the newly formed bone area in core binding factor 1 (Cbfa1)+/+ (wild-type [WT]) mice and in Cbfa1+/− mice. Newly formed bone area is expressed as a percentage (area of newly formed bone/area of wound defect × 100). In WT mice, the relative volume of new bone matrix was higher than in the Cbfa1+/− mice (p < 0.05). *p < 0.05 Cbfa1+/+ vs. Cbfa1+/−.
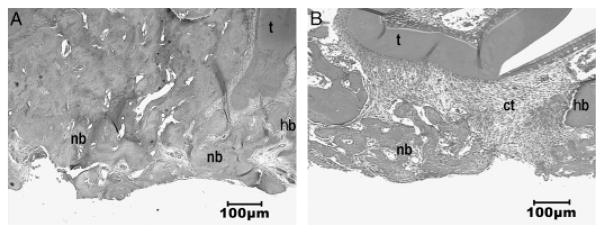
Figure 2.
Hematoxylin & eosin staining of periodontal window wounds in wild-type (WT) mice (A) and in core binding factor 1(Cbfa1)+/− mice (B). (A) At 3 weeks after the wound, alveolar bone defects almost healed in WT mice. The newly formed bone showed a trabecular structure and the bone was well mineralized. At defect edges, the newly formed bone was merged with the normal bone in the wound sites. Bone remodeling seemed to take place on the surface of the newly formed bone. (B) In contrast, wound healing was dramatically delayed in Cbfa1+/− mice. There was only a small amount of bone spicules formed near the base of the wounds. Instead, the alveolar bone defects were filled with fibrous connective tissues. The bony cut margin remained sharp without apparent bone growth or deposition 3 weeks after surgery. t, tooth; hb, host bone; ct, connective tissue; nb, newly formed bone. Original amplification, ×40.
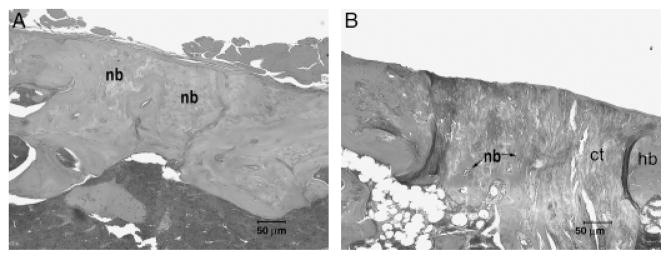
Figure 3.
Hematoxylin & eosin staining of femur defects in wild-type (WT) mice (A) and in core binding factor 1(Cbfa1)+/− mice (B). (A) At 3 weeks after surgery, the bone wounds in the femurs in WT mice (Cbfa1+/+) showed a large amount of callus tissues at the inner side of the wounds. In some cases, the defects almost healed completely. (B) Wound healing was significantly delayed in Cbfa1+/− mice. Although there was newly formed bone present beneath the periostium, repair occurred at a much slower pace and produced a smaller amount of bone compared with that in WT mice at the corresponding time. Granulation tissues were seen at the wound sites. In some cases, the wound was still widely open without any significant bone formed. Original amplification, ×100.
Part II
Morphometric assessment
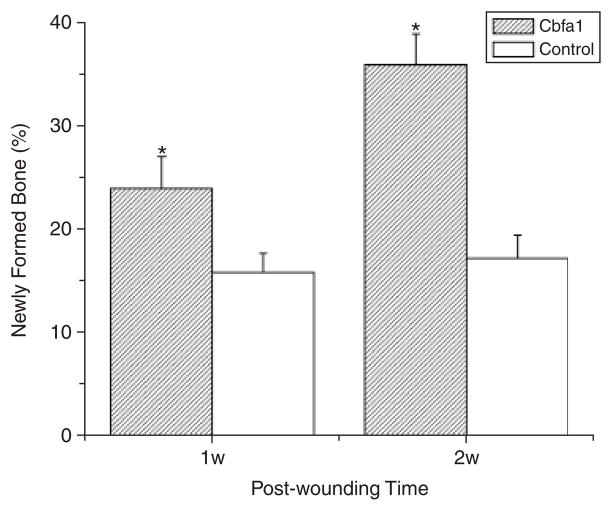
At day 7, small islands of bone matrix could be seen scattered in the bone defects in all the Cbfa1-applied tissues and some of the controls (Figure 4). However, the relative volume of new bone matrix in the defects of the Cbfa1 group was higher than in controls (> 1.5-fold) (p < 0.05). At day 14, bone formation in the Cbfa1 group exhibited an even higher increase ( > twofold) compared with controls (p < 0.05) (Figure 5). And at this time, the newly formed bones almost filled the wound defects in the Cbfa1 group, while in controls, the newly formed bones were poorly organized and weakly stained. In some samples of control tissues at day 14, there were large amounts of granulation tissues in the wound sites.
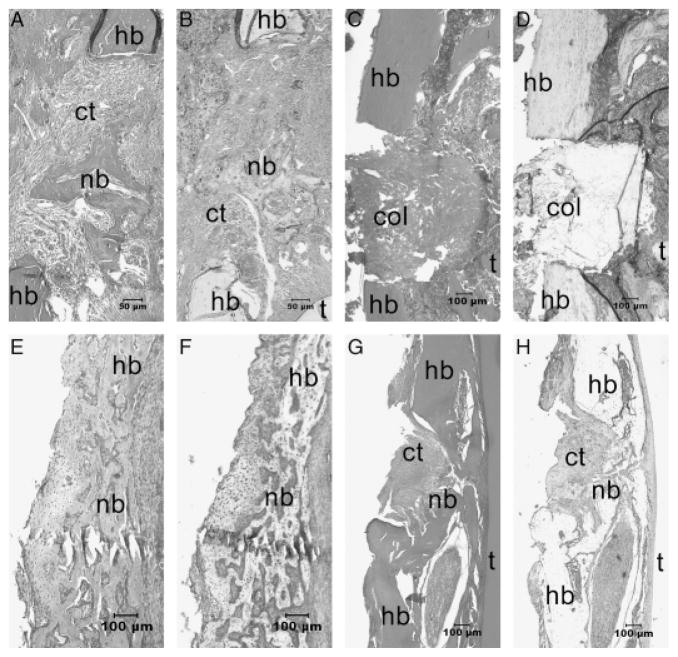
Figure 4.
Hemotoxylin & eosin staining results of the periodontal window wounds in the core binding factor 1(Cbfa1)-gene activated matrix (GAM) group (A, E) and in the control group (C, G). Bone sialoprotein (BSP) staining results in the Cbfa1-GAM group (B, F) and in the control group (D, H) are also shown. (A) At day 7, small islands of bone matrix could be seen scattered in the bone defects in all the Cbfa1-applied tissues. (B) BSP staining in the Cbfa1 group was more intense in the newly formed alveolar bone. (C) In contrast, the newly formed bones were less and (D) the BSP staining was weak in the control group at day 7. (E and F) At day 14, the newly formed bones almost filled the wound defects in the Cbfa1 group. (G and H) In controls, the newly formed bones were poorly organized and weakly stained at day 14. In some samples of control tissues at day 14, there were large amounts of granulation tissues in the wound sites. nb, newly formed bone; hb, host bone; t, tooth; ct, connective tissue; Col, collagen. Original amplification, (A and B), ×100; (C–H), ×40.
Figure 5.
Newly formed bone area in alveolar bone defects. At day 7, the relative volume of new bone matrix in the defects of the core binding factor 1 (Cbfa1) group was higher than in controls (>1.5-fold) (p<0.05). At day 14, bone formation in the Cbfa1 group exhibited an even higher increase (>twofold) compared with controls (p<0.05). *p < 0.05 Cbfa1 vs. control group.
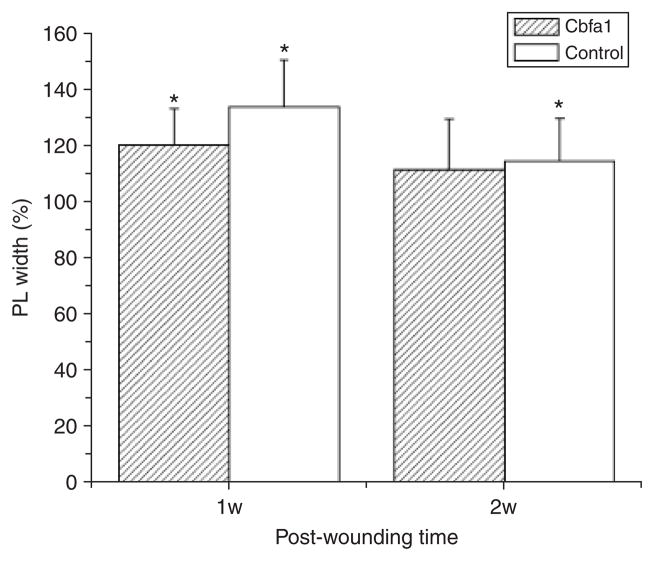
The PDL width was measured to assess the homeostasis of soft connective tissue domains. At day 7, PDL width in the wounded sides significantly increased compared with unwounded sides in both groups (p < 0.05). At day 14, PDL width in the wounded sides decreased and exhibited values similar to unwound sides in the Cbfa1 group (p > 0.4). However, PL width in the wounded sides remained significantly high compared with the unwounded side in the control group at this time (p < 0.05). These measures were expressed as a percentage of the digitized normal values for each time point (mean ± standard errors of the percentages) (Figure 6).
Figure 6.
The periodontal ligament width was measured to assess the homeostasis of soft connective tissue domains. At day 7, periodontal ligament (PDL) width in the wounded sides significantly increased compared with the unwounded sides in both groups (p < 0.05). At day 14, PL width in wounded sides decreased and exhibited values similar to the unwounded sides in the Cbfa1 group (p > 0.4). However, PDL width was still significantly increased compared with the unwounded sides in the control group at this time (p < 0.05). These measures were expressed as a percentage of the digitized normal values for each time point. *p < 0.05, wounded sides vs. unwounded sides.
The collagen sponge matrix did not elicit significant foreign body reaction in either the experimental or control groups. Examination of wound sites showed that implantation of GAM containing Cbfa1 DNA promoted bone formation.
Cbfa1-GAM stimulates expression of BSP through up-regulation of Cbfa1 in periodontal window wounds
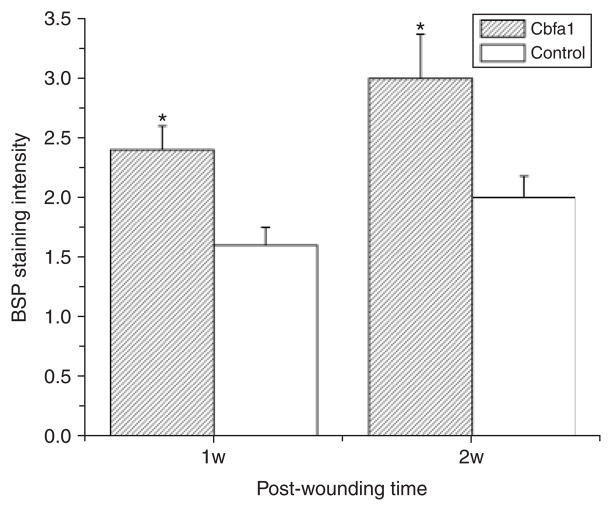
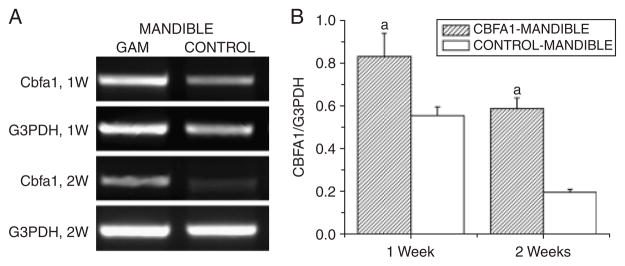
BSP was used as a phenotypic marker of mineralizing connective tissues to study cell differentiation in the periodontium. At day 7, staining for BSP was weak in the newly formed alveolar bone in the control group. In some samples, there was no new bone formed, and the staining for BSP was negative. In contrast, BSP staining in the Cbfa1 group was more intense in the newly formed alveolar bone than in the unwounded alveolar bone area (p < 0.05). At day 14, the staining intensity for BSP became much stronger in both groups in the newly formed alveolar bone, but particularly in the Cbfa1 group (p < 0.05) (Figure 7). Consistently, the data demonstrated that the GAM significantly up-regulated the expression of cbfa1 in submandibular defects as early as day 7, and remained at a high level until 14 days after surgery (Figure 8).
Figure 7.
Immunohistochemical staining for bone sialoprotein (BSP). At day 7, BSP staining in the newly formed alveolar bone in the core binding factor 1 (Cbfa1) group was more intense than in the unwounded alveolar bone area (p < 0.05). But in the control group, staining for BSP was weak in the newly formed alveolar bone. At day 14, the staining intensity for BSP was much stronger in both groups compared with the internal control areas, but particularly in the Cbfa1 group (p < 0.05).
Figure 8.
The Core binding factor 1 (Cbfa1)-gene activated matrix (GAM) system induced high levels of expression of cbfa1 in subman-dibular implant areas. (A) Representative image of semiquantitative reverse transcriptase polymerase-chain reaction (RT-PCR) analyses of cbfa1 expression. (B) Levels of Cbfa1 were normalized with those of the loading control G3PDH in three independent experiments. ap < 0.05 vs. non-GAM implant control.
DISCUSSION
Runx2/Cbfa1 is a transcription factor that belongs to the runt domain gene family23 and functions by forming a heterodimer with Cbfb (core-binding factor beta).24 Cbfa1−/− mice completely lack bone formation because of the maturational arrest of osteoblasts and die soon after birth, indicating that Cbfa1 is an essential factor for osteoblast differentiation.8,9 Cbfa1 heterozygous knockout mice (Cbfa1+/−) stay alive but show morphological defects in the skeletal system as observed in CCD in humans. In addition, chondrocyte maturation is also disturbed in Cbfa1−/− mice,25 and Cbfa1 has been shown to be an important factor for chondrocyte maturation.26 Although chondrocytes had matured, and the matrix was mineralized in restricted parts of the skeleton of Cbfa1−/− mice, osteoclasts were completely absent, and no vascular invasion into the calcified cartilage occurs.8 Therefore, Cbfa1 plays important roles in multiple processes of endochondral ossification, including chondrocyte maturation, vascular invasion into the cartilage, osteoclast differentiation, and osteoblast differentiation.27
In the first part of this study, data demonstrated that in contrast to the WT mice, wound healing was dramatically delayed in Cbfa1+/− mice characterized by the presence of only a small amount of bone near the base of the wounds. The alveolar bone defects were largely filled with fibrous connective tissues 3 weeks after surgery. Bone repair occurred at a much slower pace in these mice and granulation tissues were seen at the wound sites. In some cases, the femur wound was still widely open without any significant bone formed. This loss-of-function study proved that bone regeneration is compromised in Cbfa1+/− mice, thus indicating that Cbfa1 also plays an important role in the differentiation of osteogenic progenitor cells during bone regeneration.
Considering the essential role of Cbfa1 in osteogenesis and bone formation, several studies have applied this transcription factor for tissue engineering. In a recent study, Byers et al.28 reported that primary rat bone marrow stromal cells transduced with Runx2 retroviral vector were seeded onto three-dimensional fused deposition-modeled polycaprolactone scaffolds. These Runx2-modified cells produced biologically equivalent mineralized matrices at nearly twofold higher rates than control cells. Zhao et al.29 and Kojima et al.30 reported that primary osteoprogenitor cells transduced with adenoviral vectors encoding Runx2 formed substantially more bone than cells transduced with control virus. However, there are safety concerns about the clinical use of the viral method. Proviral DNA derived from a retrovirus can be randomly integrated into the target cell genome.31 Thus, the target cell’s genotype is permanently altered. Moreover, during integration of the proviral DNA, insertional mutagenesis can occur if the inserted DNA disrupts housekeeping genes or activates other genes, e.g., an oncogene.31 Furthermore, local toxicity can result from chronic overexpression of the expression cassette product. In addition, most retrovirus can only infect, integrate, and express in dividing target cells. Other limitations include low titers and sensitivity to inactivation.31 As for adenovirus vectors, although problems associated with insertional mutagenesis are avoided, they are still limited in use by the nonspecific immunologic reactions they elicit, as well as the potential for autoimmune reactions to the transgene-encoded proteins. Patient safety is a paramount issue,32 which considerably restricts the clinical application of the virus-based method.
The GAM method offers an alternative and safer approach to virus-based tissue-engineering applications.33,34 This method was designed specifically to provide an ideal environment for tissue regeneration and engineering. In some animal studies, plasmid genes can be delivered, in a polymer GAM, directly to actually injured bone, muscle, and ligament.35,36 The GAM could serve as a platform technology for local gene delivery in various tissues and organs. A GAM consists of two ingredients: plasmid DNA and a biodegradable structural matrix carrier, e.g., a collagen sponge or gel. The carrier serves as a scaffold that holds DNA in situ until endogenous wound-healing cells arrive. Up to 50% of available healing repair cells would be transfected.37 Wound repair cells possess relatively high levels of pinocytosis and potocytosis.38 This inherent ability may facilitate plasmid gene transfer and help explain the high level of gene transfer efficiency associated with the GAM. The cells in the matrix carrier act as local in vivo bioreactors, producing plasmid-coding proteins that augment tissue repair and regeneration. GAM implantation at sites of bone injury was associated with retention and expression of plasmid DNA for at least 6 weeks.34 In this study by Bonadio et al., GAM implants with a pMat-1 plasmid DNA that encodes a secreted peptide fragment of human parathyroid hormone (hPTH 1–34) induced predictable formation of centimeters of normal new bone in a time-, plasmid dose-, and bone gap size-dependent manner.34
In the second part of the present study, Cbfa1 cDNA was locally delivered to the periodontal window wound using the GAM method to assess the effects of this transcription factor on the PDL, a tissue that is thought to contain the progenitor cells for cementum, bone, and PDL fibroblasts. At day 7, we observed that the GAM porous architecture provided scaffolding to promote cell ingrowth. The local granulation tissue fibroblasts, along with capillaries, migrated into the GAM. The osteogenic progenitor cells within the tissue uptook the local plasmid DNA and transiently expressed the gene. The transfected reactive cells then secreted the plasmid-encoded proteins to stimulate and augment the bone regenerative cascade, which has been observed as the expression of both Cbfa1 and BSP increased in our study. As the GAM system delivers genes instead of recombinant proteins, researchers have reported longer-time expression of genes of interest (weeks vs. hours).34,39 In the present study, the relative volume of newly formed bone was higher in the Cbfa1-GAM group than in the control group. At day 14, the newly formed bones almost filled the wound defects in the Cbfa1 group, while in controls, the newly formed bones were poorly organized and weakly stained. Furthermore, Cbfa1 and BSP expression was stronger in the Cbfa1 group than in the control group, which indicated that Cbfa1 cDNA taken up by the PDL cells can be expressed in situ in an autocrine/intracrine manner40 that promoted phenotypic differentiation. As a result, expression of bone and cementum-specific proteins, e.g., BSP could be stimulated. This would lead to an increase in periodontal tissue regeneration.
The PDL width was measured to assess the homeostasis of soft connective tissue domains. At day 7, PDL width in the wounded sides significantly but slightly increased compared with the unwounded sides in both groups. However, at day 14, PDL width in the wounded sides decreased and exhibited values similar to the unwounded sides in the Cbfa1 group. These measures demonstrate that locally delivered Cbfa1 did not cause ankylosis in spite of inducing new bone formation.
In conclusion, bone regeneration was compromised in Cbfa1+/− mice, and locally delivered Cbfa1 using the GAM approach promoted bone regeneration in the periodontal window wound. This study indicates that Cbfa1 also plays an important role in the differentiation of osteogenic progenitor cells during bone regeneration.
Acknowledgments
This work was supported by NIH grants DE11088, DE14537 to JC.
References
- 1.Gould TR, Melcher AH, Brunette DM. Migration and division of progenitor cell populations in periodontal ligament after wounding. J Periodontal Res. 1980;15:20–42. doi: 10.1111/j.1600-0765.1980.tb00258.x. [DOI] [PubMed] [Google Scholar]
- 2.Gould TR, Melcher AH, Brunette DM. Location of progenitor cells in periodontal ligament of mouse molar stimulated by wounding. Anat Rec. 1977;188:133–41. doi: 10.1002/ar.1091880202. [DOI] [PubMed] [Google Scholar]
- 3.Loftus TM, Lane MD. Modulating the transcriptional control of adipogenesis. Curr Opin Genet Dev. 1997;7:603–8. doi: 10.1016/s0959-437x(97)80006-8. [DOI] [PubMed] [Google Scholar]
- 4.Arnold HH, Winter B. Muscle differentiation: more complexity to the network of myogenic regulators. Curr Opin Genet Dev. 1998;8:539–44. doi: 10.1016/s0959-437x(98)80008-7. [DOI] [PubMed] [Google Scholar]
- 5.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–30. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 6.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, Stein J, Stein GS. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/Runt domain transcription factor family. Biochemistry. 1995;34:13123–32. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 8.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 9.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 10.Rodan GA, Harada S. The missing bone. Cell. 1997;89:677–80. doi: 10.1016/s0092-8674(00)80249-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Jiang H, Sodek J, Karsenty G, Thomas H, Ranly D. Comparison of expression patterns of core binding factor (Osf2/Cbfa1) and bone sialoprotein (BSP) In: Goldberg M, Boskey A, Robinson C, editors. Chemistry and biology of mineralized tissue. Chapter 24. Rosemont: American Academy of Orthopaedic Surgery; 2000. pp. 149–54. [Google Scholar]
- 12.Jiang H, Sodek J, Karsenty G, Thomas H, Ranly D, Chen J. Expression of core binding factor Osf2/Cbfa1 and bone si-aloprotein in tooth development. Mech Dev. 1999;81:169–73. doi: 10.1016/s0925-4773(98)00232-9. [DOI] [PubMed] [Google Scholar]
- 13.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, Ochi T, Kitamura Y, Ito Y, Nomura S. Transcriptional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene. 1998;17:1517–25. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 14.Xiao ZS, Hinson TK, Quarles LD. Cbfa1 isoform overex-pression upregulates osteocalcin gene expression in non-osteoblastic and pre-osteoblastic cells. J Cell Biochem. 1999;74:596–605. [PubMed] [Google Scholar]
- 15.Kern B, Shen J, Starbuck M, Karsenty G. Cbfa1 contributes to the osteoblast-specific expression of type I collagen genes. J Biol Chem. 2001;276:7101–7. doi: 10.1074/jbc.M006215200. Epub December 5, 2000. [DOI] [PubMed] [Google Scholar]
- 16.Prince M, Banerjee C, Javed A, Green J, Lian JB, Stein GS, Bodine PV, Komm BS. Expression and regulation of Runx2/Cbfa1 and osteoblast phenotypic markers during the growth and differentiation of human osteoblasts. J Cell Biochem. 2001;80:424–40. doi: 10.1002/1097-4644(20010301)80:3<424::aid-jcb160>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16:307–10. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham ML, Seto ML, Hing AV, Bull MJ, Hopkin RJ, Leppig KA. Cleidocranial dysplasia with severe parietal bone dysplasia: C-terminal RUNX2 mutations. Birth Defects Res A Clin Mol Teratol. 2006;76:78–85. doi: 10.1002/bdra.20231. [DOI] [PubMed] [Google Scholar]
- 19.Lekic P, Sodek J, McCulloch CA. Osteopontin and bone si-aloprotein expression in regenerating rat periodontal ligament and alveolar bone. Anat Rec. 1996;244:50–8. doi: 10.1002/(SICI)1097-0185(199601)244:1<50::AID-AR5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 20.Lekic P, Rubbino I, Krasnoshtein F, Cheifetz S, McCulloch CA, Tenenbaum H. Bisphosphonate modulates proliferation and differentiation of rat periodontal ligament cells during wound healing. Anat Rec. 1997;247:329–40. doi: 10.1002/(SICI)1097-0185(199703)247:3<329::AID-AR4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 21.McLean IW, Nakane PK. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077–83. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 22.Sasaguri K, Jiang H, Chen J. The effect of altered functional forces on the expression of bone-matrix proteins in developing mouse mandibular condyle. Arch Oral Biol. 1998;43:83–92. doi: 10.1016/s0003-9969(97)00075-7. [DOI] [PubMed] [Google Scholar]
- 23.Komori T, Kishimoto T. Cbfa1 in bone development. Curr Opin Genet Dev. 1998;8:494–9. doi: 10.1016/s0959-437x(98)80123-8. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8. doi: 10.1038/ng1015. Epub November 18, 2002. [DOI] [PubMed] [Google Scholar]
- 25.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–90. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275:8695–702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- 27.Westendorf JJ. Transcriptional co-repressors of Runx2. J Cell Biochem. 2006;98:54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- 28.Byers BA, Guldberg RE, Hutmacher DW, Garcia AJ. Effects of Runx2 genetic engineering and in vitro maturation of tissue-engineered constructs on the repair of critical size bone defects. J Biomed Mater Res A. 2006;76:646–55. doi: 10.1002/jbm.a.30549. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z, Zhao M, Xiao G, Franceschi RT. Gene transfer of the Runx2 transcription factor enhances osteogenic activity of bone marrow stromal cells in vitro and in vivo. Mol Ther. 2005;12:247–53. doi: 10.1016/j.ymthe.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Kojima H, Uemura T. Strong and rapid induction of osteo-blast differentiation by Cbfa1/Til-1 overexpression for bone regeneration. J Biol Chem. 2005;280:2944–53. doi: 10.1074/jbc.M311598200. Epub November 10, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Crystal RG. 1995 Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–10. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 32.Winn SR, Hu Y, Sfeir C, Hollinger JO. Gene therapy approaches for modulating bone regeneration. Adv Drug Deliv Rev. 2000;42:121–38. doi: 10.1016/s0169-409x(00)00057-0. [DOI] [PubMed] [Google Scholar]
- 33.Fang J, Zhu YY, Smiley E, Bonadio J, Rouleau JP, Goldstein SA, McCauley LK, Davidson BL, Roessler BJ. Stimulation of new bone formation by direct transfer of osteogenic plasmid genes. Proc Natl Acad Sci USA. 1996;93:5753–8. doi: 10.1073/pnas.93.12.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:753–9. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 35.Labhasetwar V, Bonadio J, Goldstein S, Chen W, Levy RJ. A DNA controlled-release coating for gene transfer: transfection in skeletal and cardiac muscle. J Pharm Sci. 1998;87:1347–50. doi: 10.1021/js980077+. [DOI] [PubMed] [Google Scholar]
- 36.Ochiya T, Takahama Y, Nagahara S, Sumita Y, Hisada A, Itoh H, Nagai Y, Terada M. New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet. Nat Med. 1999;5:707–10. doi: 10.1038/9560. [DOI] [PubMed] [Google Scholar]
- 37.Bonadio J. Tissue engineering via local gene delivery. J Mol Med. 2000;78:303–11. doi: 10.1007/s001090000118. [DOI] [PubMed] [Google Scholar]
- 38.Martin P. Wound healing–aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 39.Giannobile WV, Hernandez RA, Finkelman RD, Ryan S, Kiritsy CP, D’Andrea M, Lynch SE. Comparative effects of platelet-derived growth factor-BB and insulin-like growth factor-I, individually and in combination, on periodontal regeneration in Macaca fascicularis. J Periodontal Res. 1996;31:301–12. doi: 10.1111/j.1600-0765.1996.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 40.McCauley LK, Somerman MJ. Biologic modifiers in periodontal regeneration. Dent Clin North Am. 1998;42:361–87. [PubMed] [Google Scholar]