Abstract
Image annotation and markup are at the core of medical interpretation in both the clinical and the research setting. Digital medical images are managed with the DICOM standard format. While DICOM contains a large amount of meta-data about whom, where, and how the image was acquired, DICOM says little about the content or meaning of the pixel data. An image annotation is the explanatory or descriptive information about the pixel data of an image that is generated by a human or machine observer. An image markup is the graphical symbols placed over the image to depict an annotation. While DICOM is the standard for medical image acquisition, manipulation, transmission, storage, and display, there are no standards for image annotation and markup. Many systems expect annotation to be reported verbally, while markups are stored in graphical overlays or proprietary formats. This makes it difficult to extract and compute with both of them. The goal of the Annotation and Image Markup (AIM) project is to develop a mechanism, for modeling, capturing, and serializing image annotation and markup data that can be adopted as a standard by the medical imaging community. The AIM project produces both human- and machine-readable artifacts. This paper describes the AIM information model, schemas, software libraries, and tools so as to prepare researchers and developers for their use of AIM.
Key Words: Image annotation, image markup, caBIG, ontology
Background
Images, in particular medical and scientific images, contain vast amounts of information. While this information may include meta-data about the image, such as how, when, or where the image was acquired, the majority of image information is encoded in the pixel data. Observational or computational descriptions of image features (including their spatial coordinates) can be attached to an image, though there is no standard mechanism for doing so in health care. Moreover, the majority of the human observed image feature descriptions are captured only as free text. This free text is often not associated with the spatial location of the feature, making it difficult to relate image observations to their corresponding image locations. It is difficult for both humans and machines to index, query, and search free text in order to retrieve images or their features based on these free text descriptions. This limits the value of image data and its interpretation for clinical, research, and teaching purposes.
The mission of the National Institutes of Health’s (NIH) National Cancer Institute’s (NCI) Cancer Bioinformatics Grid (caBIG™)1 is to provide infrastructure for creating, communicating, and sharing bioinformatics tools, data and research results, using shared data standards and shared data models. Imaging, critical to cancer research, lies at an almost unique juncture in the translational spectrum between research and clinical practice. Image and annotation information obtained, for example, in cancer clinical trial research, is collected in a clinical setting of commercial information systems. The latter adhere to standards, such as DICOM2 and HL73, and technical frameworks such as Integrating the Healthcare Enterprise (IHE)4. Image annotation, in particular, needs to be available in a standard format that is at the same time syntactically and semantically interoperable with the infrastructure of caBIG while supporting the widespread clinical health care standards, such as DICOM and HL7. At the same time, this standard for annotation must support recommendations being developed for the World Wide Web community, for example, the semantic Web, in order to leverage the full spectrum of cancer biomedical image annotation, whether it exists in commercial PACS systems or in cyberspace.
Selecting a single standard format to store image annotations will streamline software development and enable the work to focus on providing rich annotation features and functionality. Designing the tools to be compatible with other standards will enable a high degree of interoperability and allow the incorporation of the annotation standard into commercial, clinical, information systems. Such interoperability has the potential to open many existing resources and databases of cancer-related image data and metadata for exploitation not only by caBIG™, but by the broader research and clinical radiology community.
This paper describes the use cases and requirements for annotation and image markup and the resulting AIM information model. A free and open source software library that can be used to create and work with instances of AIM is available. The software, schemas, and templates necessary to serialize these AIM instances as DICOM S/R, AIM XML and HL7 Clinical Document Architecture (CDA) can be downloaded from http://cabig.nci.nih.gov/tools/AIM.
Methods
We used agile, iterative processes throughout the life cycle of the AIM project. The project began with use case development and requirements elicitation from the In Vivo Imaging Workspace of the caBIG project. The workspace consists of a set of subject matter experts, typically scientists working in imaging informatics related to clinical and clinical trials research. These scientists are supplemented by a large community of other research imaging users, developers, and scientists. Requirements were derived from the use cases and a formal identification and reconciliation with other efforts and standards for image annotation and markup. In particular, we examined the requirements of the Lung Image Database Consortium (LIDC)5, the Response Evaluation Criteria in Solid Tumors (RECIST) standard6, DICOM SR7,8, and HL7 CDA. The requirements for teaching file image annotation were captured from RadPix (Weadock Software, LLC, Ann Arbor, MI, USA) and the Medical Image Resource Center (MIRC)9 from the Radiological Society of North America (RSNA). We also examined the specification for semantic image annotation of the World Wide Web Consortium (W3C)10.
The AIM model was created in Unified Modeling Language (UML) using Enterprise Architect (EA; Sparx Systems, Creswick, Victoria, Australia). The UML model was imported into an ontology developed in Protégé (Stanford University, Palo Alto, CA, USA). The ontology included controlled terminology for anatomic entities, imaging observations and imaging observation characteristics from the Radiology Lexicon, RadLex11.
XML schemata for the AIM project were created in a three-step process. When a version of the AIM UML model was considered mature, an XML metadata interchange file (.XMI) file was generated from EA. This file was used as input to the caCORE software development kit (SDK) version 3.2.112 (caBIG Project, National Cancer Institute, Bethesda, MD, USA) which translated it into an XML schema. This schema was modified by hand with Altova XML Spy 2008 (Altova, Beverly, MA, USA) to remove certain incompatibilities generated by the caCORE package. Many of such issues will be addressed in subsequent caCORE software releases so that future translations to XML schema can be simpler.
The AIM DICOM SR template was constructed from the AIM model and using the DICOM Comprehensive SR information object definition1. We also used the Mammography CAD DICOM SR content tree and the DICOM SR Cancer Clinical Trials Results6,7 as guiding examples of how to create the SR object for AIM.
The AIM software library was developed in standard C++ using Microsoft Visual Studio 2005 (Microsoft, Redmond, WA, USA), the Standard Template Library (STL) (Silicon Graphics, Sunnyvale, CA, USA), the Apache Xerces XML library version 2.4.0 (http://xerces.apache.org) and the Boost C++ library version 1.34.1 (http://www.boost.org). DICOM functionality was provided by DCMTK version 3.5.4 (OFFIS E. V., Oldenburg, Germany). Some software components were developed in Java JDK 6 (Sun Microsystems, Santa Clara, CA, USA) using the Eclipse (http://www.eclipse.org) integrated development environment.
The AIM library creates an object model, using a C++ class for each class in the AIM Schema. The class hierarchy closely follows the AIM Schema. Each class in the object model provides mutator methods (Set and Get methods) for every attribute in the corresponding AIM Schema class. All changes to the class’s state are done via those mutator methods. Thus, the whole AIM Schema is represented by the object model through containment and inheritance. The set of object model operations supported by the AIM library includes serializing the model as XML, DICOM SR, or HL7 CDA format. The reverse set of operations of reading XML, DICOM S/R, and HL7 CDA instances into the object model is supported as well.
Use Cases
The design of the AIM information model was motivated by a key use case for the cancer imaging community: the image-based clinical trial.
Prerequisites:
One series of DICOM images from one study of one patient at a first time point.
One series of DICOM images from one study of same patient at a second time point.
Use Case:
Observer 1 annotates observations in images from first time point.
Observer 1 measures each observation.
Software application assigns unique identifier to each image annotation.
Observer 1 assigns human readable name to each image annotation.
For each named image annotation from Observer 1, Observer 2 (blinded to Observer 1’s result or not) annotates the same observation in the images from the second time point.
Observer 2 measures each observation.
Researcher retrieves annotations from Observer1 and Observer 2.
Researcher performs calculations on measurements from both observers.
Researcher creates annotation on the collection of image annotations to document change.
Variations on this use case can meet a lot of different needs. There can be many observers at each of many time points. Adjudicators can be introduced to reconcile truth between different observers at different time points. Image processing and analysis software can replace human observers. Degenerate use cases include a clinician documenting observations in routine clinical work or an instructor documenting observations in a teaching case.
The requirements, derived from this use case and the collective experience of the workspace, were legion. At a highest level, the AIM model encompasses patient, observer, equipment, image, anatomic entities, observations and observation characteristics, two- and three-dimensional coordinates of defined geometric shapes, markup and text annotation, comments, and defined and arbitrary (defined by an application) calculations as well as the ability to store arbitrary calculation results.
The Model
As introduced above, an annotation is explanatory or descriptive information, generated by humans or machines, directly related to the content of a referenced image or images. It describes information about the meaning of pixel information in images. Annotations become a collection of image semantic content that can be used for data mining purposes. Markups can be used to depict textual information and regions-of-interest visually along side of an image. Computation results may be created from image markup or other comparison methods in order to provide more meaningful information about images.
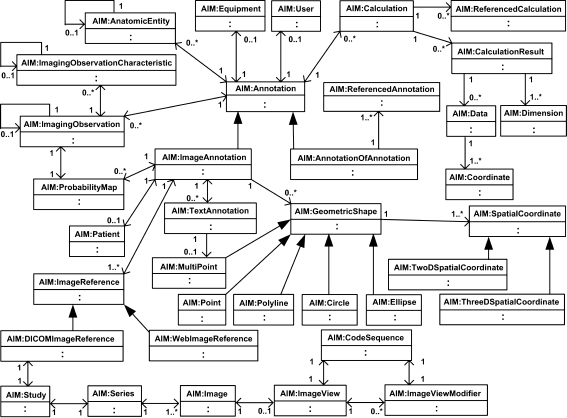
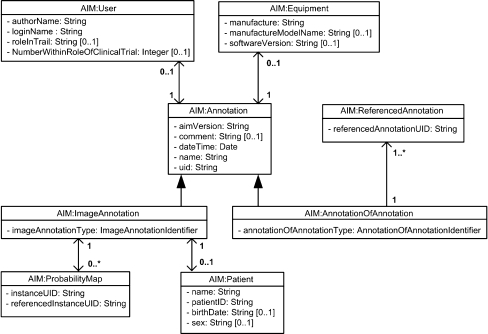
The AIM model is shown in Figure 1. The model is organized into logical groups of classes relating to specific information model components. The Annotation class itself is an abstract class. There are two kinds of annotation that can be instantiated. ImageAnnotation class annotates images. AnnotationOfAnnotation class annotates other AIM annotations for comparison and reference purposes. The latter uses the ReferencedAnnotation class, which has a UID of a referenced ImageAnnotation or AnnotationOfAnnotation. AIM annotations are, therefore, either an instance of ImageAnnotation or AnnotationOfAnnotation. These are the two root objects that inherit all properties of the abstract class Annotation. Each root object has a type attribute that is used to identify what kind of ImageAnnotation or an AnnotationOfAnnotation is being instantiated. The type of annotation is used to retrieve type-specific constraints from the ontology13. The sixteen currently defined types of annotation are listed in Table 1. Annotation class also captures the name and general description of the AIM annotation, the version of AIM schema, creation date and time, and the annotation unique identifier (UID). Both ImageAnnotation and AnnotationOfAnnotation, Figure 2, may have User and Equipment classes. The Patient class may only be associated with an ImageAnnotation.
Fig 1.
AIM UML Class diagram.
Table 1.
Types of Annotations
| ImageAnnotation | RECIST baseline target lesion |
| RECIST baseline non-target lesion | |
| RECIST follow-up target lesion | |
| RECIST follow-up non-target lesion | |
| LIDC chest CT nodule | |
| Brain tumor baseline target lesion | |
| Brain tumor follow-up target lesion | |
| Teaching | |
| Quality control | |
| Clinical finding | |
| Other | |
| AnnotationOfAnnotation | RECIST baseline sum of longest diameter |
| RECIST follow-up sum of longest diameter | |
| Interval change | |
| Summary statistic | |
| Other |
Fig 2.
General information group.
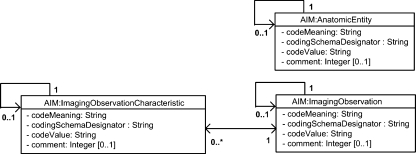
AnatomicEntity, ImagingObservation, and ImagingObservationCharacteristic classes, Figure 3, are the key classes used to gather the respective features of the particular annotation. The possible values that can be populated in these classes come from controlled terminologies and are generated by the AIM library through its API to the AIM Ontology in Protégé. The values returned depend on the type of annotation being made.
Fig 3.
Ontology finding group.
AnatomicEntity is used to store the anatomy of the observation being made. The term must be from a recognized controlled vocabulary (RadLex11, SNOMED-CT14, UMLS15, etc).
ImagingObservation is used for the depiction of “things” in the image. For example, “mass,” “opacity,” ”foreign body,” “artifact” are all examples of ImagingObservation. ImagingObservation can be associated with one or more ImagingObservationCharacteristic, descriptors of ImagingObservation. So, for example, “Spiculated” ontologically is_a kind of “Margin” and it might, therefore, be an ImagingOberservationCharacteristic for an ImagingObservation of “Mass.”
A new class, Inference, is being developed for the next version of the AIM model. Disease entities are more appropriately modeled as inferences in medical imaging. “Pleural Effusion,” for example, is a disease which in imaging is an inference from a collection of ImagingObservations, ImagingObservationCharacteristics, and Anatomy.
A probability map is a unique kind of annotation. It allows the user to define an observation and then assign to each pixel in the region of interest, a probability that that pixel represents the observation. For example, consider a rectangular region of interest of tissue and a software application that computes the probability that each pixel is part of the tumor. This result would be stored as a probability map. Another example is a tissue where each pixel is inhomogeneous and an estimate of the contribution of each component is made; a pixel, for example, that is 70% soft tissue and 30% fat. This could be stored as multiple probability maps. The ProbabilityMap class contains references to its own instance UID and referenced instance UID of the image to which the probability map is applied to. An instance of ProbabilityMap must associate with ImagingObservation, which describes what the ProbabilityMap is specifying. An ImageAnnotation may have zero or more ProbabilityMap objects.
The User class represents the observer who creates the annotation. The User class is composed of the user’s full name, a user login name, and an optional role and identifier within a clinical trial (for example, “Reader 37”). The User class could alternatively record information about an algorithm or other automated method that created the annotation. The Equipment class provides information about the system that is used to create the AIM annotations. Equipment class collects manufacture name, model number, and software version.
The Patient class contains basic patient demographic information: patient name, local medical record number, birth date, and sex. These may be de-identified pseudonyms at the discretion of the software application creating the AIM.
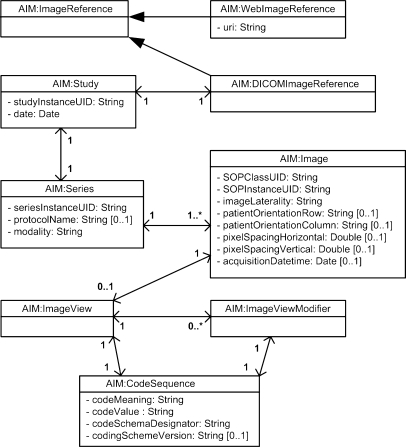
The image reference classes specify the image or collection of images being annotated, which can be seen in Figure 4. AIM annotations may reference DICOM images or web images. By web image, we mean stand-alone images in common lay formats that may or may not be local to the application. Web images, whether local or remote, are referenced by uniform resource identifier (URI).
Fig 4.
Image reference group.
The DICOMImageReference class mimics a subset of the DICOM information model. It has one Study object that has one or more Series objects which in turn have one or more Image objects. Study has study instance UID and date. Series has series instance UID, protocol name, and modality. Image has SOP class UID, SOP instance UID, image laterality, patient orientation, pixel spacing, and acquisition date time. Each Image may have ImageView and ImageViewModifier. ImageView is a sequence that describes the projection of the anatomic region of interest on the image receptor. ImageViewModifier provides modifiers for the view of the patient anatomy.
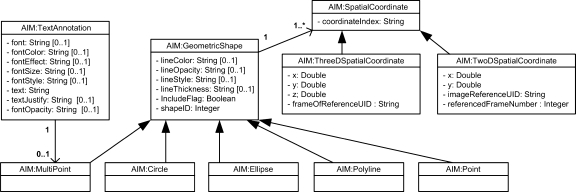
The markup group, Figure 5, captures textual information and the graphical representation of the markup. TextAnnotation is used to label an image. The coordinates of a TextAnnotation are captured as one of the markup graphic types, namely Multipoint. TextAnnotation’s MultiPoint is expected to have no more than two coordinates which represent beginning and ending points of a line connecting TextAnnotation to a point on an image.
Fig 5.
Markup group.
The other available graphic types are Point, Polyline, Circle, and Ellipse, based on existing graphic types in DICOM SR7. These graphic types are used to capture the markup associated with a single annotation. Note that a single annotation may have multiple markups. Two non-adjacent regions of a single image may represent the same thing. Consider also, the case of a donut structure. The markup consists of two concentric circles, the first larger than the second. The observation of interest consists of the intersection of the two shapes. This is supported in the model by having an attribute of each markup specify its inclusion in the annotation.
Each graphic type has appropriate SpatialCoordinate abstract classes, which can be in two- or three-dimensional space. TwoDSpatialCoordinate has x and y coordinates, the SOP Instance UID of the image that contains the pixel and the frame number within the referenced SOP Instance to which the markup applies. The frame number is used as per the DICOM standard. ThreeDSpatialCoordinate has x, y, and z coordinates as well as the DICOM frame of reference for a Series.
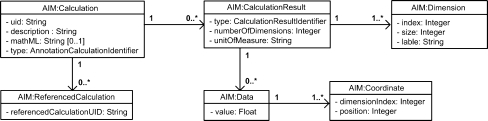
The Calculation group, shown in Figure 6, represents how calculation results are stored in an annotation. Calculation results may or may not be directly associated with graphical symbols or markups. In a simple example, given an image with a single ellipse markup, calculation results might include area in square millimeters, and maximum and minimum pixel values.
Fig 6.
Calculation group.
The AIM model supports both defined and arbitrary calculations. Defined calculations include common linear, area, and volumetric calculations. Arbitrary calculations are defined by the application and free text description of the calculation can be included. A MathML16 representation of the algorithm may also be included in the class. The ReferencedCalculation class is used as a mechanism for referencing other calculations that the current calculation may be based upon.
The CalculationResult class is used to store the results of calculations. In order to store a wide variety of results, it contains the total number of dimensions to the result, a string representation of the universal common units of measurement (UCUM)17 units for each of the dimensions, and the type of result such as scalar, vector, histogram, or array. The Dimension class describes each of the dimensions of the result, the index of the current dimension and the size of the dimension. The Data class is used to store the actual result. The Coordinate class identifies the location within a dimension for the Data class. For example, a CalculationResult of an area would have a scalar result type with one dimension, one “mm2” unit and one Data value.
Discussion
The AIM Model allows for the creation of a broad variety of image annotations. The primary use case is the capture of measurements made by observers in clinical trials to facilitate the use of imaging as a biomarker. The same model can, however, support a broad variety of image annotations for clinical, teaching, and other research purposes. Administrative AIM annotations, such as for documenting imaging artifacts and other quality control issues, are being considered.
The National Cancer Institute makes large collections of research images available to the imaging community through its National Cancer Imaging Archive (NCIA) web site (http://ncia.nci.nih.gov/). We anticipate the development of large collections of AIM annotations to accompany these large image collections.
In leveraging the methodologies of the caBIG project, we raise the bar of interoperability for the AIM model. The AIM model is caBIG Silver-level compliant for interoperability. This means that all appropriate data collection fields and attributes of data objects in AIM use controlled terminologies reviewed and validated by the caBIG Vocabularies and Common Data Elements Workspace (VCDE Workspace). Common Data Elements (CDEs) built from controlled terminologies and according to practices validated by the VCDE workspace are used throughout the AIM model. These CDEs are registered as ISO/IEC 11179 standard metadata components in the caBIG Context of the cancer Data Standards Repository (caDSR)18. The AIM model expressed in UML as class diagrams and as XMI files, have been reviewed and validated by the VCDE Workspace. The model itself is then available for use from the caDSR.
Importantly, since the model was developed using this caBIG methodology, other caBIG tools can be used to develop software that makes use of AIM annotations. For example, a prototype AIM Grid storage service is being developed.
Other caBIG projects, for example, querying of AIM annotations, will leverage this work. In the not too distant future, it will be possible to query a set of distributed caGRID services for, “Find all the CT Images of the lung containing a spiculated mass with a longest diameter between 5 and 7 mm and having decreased in size by at least 10% from prior measurement.” It is the power of this kind of query that motivated the AIM project.
Lastly, the AIM software not only creates AIM instances, but it can serialize them for storage and transmission as XML, DICOM S/R, and HL7 CDA. Most caBIG tools will use the native AIM XML as part of grid service payloads for a variety of purposes. AIM XML can also be easily incorporated into other XML standards such as MIRC documents. Research imaging software such as the caBIG In Vivo Imaging Workspace’s eXtensible Imaging Platform (XIP)19 also makes direct use of the XML schemata. Most clinical and clinical research users do their image annotation and measurement work on commercial clinical and research software. Several vendors have expressed interest in the AIM model and software. In their case, the vendors have the choice of implementing AIM annotations as XML or as DICOM S/R. When balloted by the DICOM standards committee, AIM DICOM S/R will likely become another DICOM S/R object similar to the Mammography or Chest CAD1 objects, DICOM part 17. Vendors of commercial picture archiving and communication systems (PACS) workstations will then have the choice of implementing AIM as either a DICOM S/R or an XML object depending on their integration strategies. Regardless of their choice, our AIM toolkit will enable conversion between these formats and interoperability among imaging systems. It is important for purchasers of clinical imaging equipment to recognize the value in standardizing image annotation formats. As AIM becomes widely adopted for image annotation, the community will benefit if purchasers contractually require support for AIM annotation in new image display and manipulation equipment.
Conclusion
The AIM project defines an information model for image annotation and markup in health care. Although useful when annotating images for clinical and teaching purposes, standardized image annotation and markup is most critical in clinical trials. This is especially true in multicenter clinical trials where data collection and analysis spans multiple investigators and institutions. As a Silver compliant product of the caBIG methodology, the AIM model provides syntactic and semantic interoperability with other caBIG activities. The AIM software provides developers a toolkit that will enable them to adopt AIM in their applications. AIM provides a foundation for standardized image annotation practice in the clinical, research, and translational communities.
Acknowledgements
This work was supported as a task order under contract 79596CBS10 as part of the National Cancer Institute’s caBIG™ program to The Robert H. Lurie Cancer Center of Northwestern University.
References
- 1.Eschenbach AC, Buetow K. Cancer Informatics Vision: caBIG™. Cancer Informatics. 2006;2:22–24. [PMC free article] [PubMed] [Google Scholar]
- 2.Digital Imaging and Communications in Medicine (DICOM) Standard. PS 3 2008. Washington DC: National Electrical Manufacturer’s Association (NEMA). http://medical.nema.org/dicom/2008/. Accessed November 17, 2008
- 3.Health Level 7. Ann Arbor Michigan. Health Level 7, Inc. http://www.hl7.org/ Accessed November 17, 2008
- 4.Integrating the Healthcare Enterprise: IHE Technical Frameworks. Chicago, IL: American College of Cardiology, Health Information Management Systems Society, and the Radiological Society of North America. Available at: http://www.ihe.net/Technical_Framework/. Accessed June 24, 2008
- 5.Lung Image Database Consortium. National Institutes of Health National Cancer Institute, Cancer Imaging Program. http://imaging.cancer.gov/programsandresources/InformationSystems/LIDC. Accessed November 17, 2008
- 6.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Glabbeke M, Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 7.Clunie DA: DICOM Structured Reporting and Cancer Clinical Trials Results. Cancer Informatics, 2007, pp. 33–56 [DOI] [PMC free article] [PubMed]
- 8.Clunie DA: DICOM Structured Reporting. PixelMed Publishing, 2000
- 9.Medical Image Resource Center. Radiological Society of North America. http://rsna.org/mirc. Accessed November 17, 2008
- 10.World Wide Web Consortium Incubator Group Report: Image Annotation on the Semantic Web. World Wide Web Consortium. http://www.w3.org/2005/Incubator/mmsem/XGR-image-annotation-20070814/. August 17, 2007. Accessed November 17, 2008
- 11.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 12.Komatsoulis GA, Warzel DB, Hartel FW, Shanbhag K, Chilukuri R, Fragoso G, Coronado SD, Reeves DM, Hadfield JB, Ludet C, Covitz PA. caCORE version 3: Implementation of a model driven, service-oriented architecture for semantic interoperability. J Biomed Inform. 2008;41(1):106–23. doi: 10.1016/j.jbi.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS: Medical Imaging on the Semantic Web: Annotation and Image Markup, Association for the Advancement of Artificial Intelligence, 2008 Spring Symposium Series, Stanford, CA, 2008
- 14.SNOMED-CT. International Health Standards Development Organization. http://www.ihtsdo.org/snomed-ct/. Accessed November 17, 2008
- 15.Unified Medical Language System. National Institutes of Health National Library of Medicine. http://www.nlm.nih.gov/research/umls/. Accessed November 17, 2008
- 16.World Wide Web Consortium MathML Recommendation. http://www.w3.org/Math/. Accessed November 17, 2008
- 17.Schadow G., McDonald CJ. The Unified Code for Units of Measure. Regenstrief Institute, Indianapolis, IN. http://unitsofmeasure.org/. Accessed November 17, 2008
- 18.Cancer Data Standards Repository (caDSR). National Institutes of Health National Cancer Institute Center for Bioinformatics. http://ncicb.nci.nih.gov/NCICB/infrastructure/cacore_overview/cadsr. Accessed November 17, 2008
- 19.Prior FW, Erickson BJ, Tarbox L. Open source software projects of the caBIG In Vivo Imaging Workspace Software special interest group. J Digit Imaging. 2007;20(Suppl 1):94–100. doi: 10.1007/s10278-007-9061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]