Abstract
The ICP0 protein of herpes simplex virus type 1 is an E3 ubiquitin ligase and transactivator required for the efficient switch between latent and lytic infection. As DNA damaging treatments are known to reactivate latent virus, we wished to explore whether ICP0 modulates the cellular response to DNA damage. We report that ICP0 prevents accumulation of repair factors at cellular damage sites, acting between recruitment of the mediator proteins Mdc1 and 53BP1. We identify RNF8 and RNF168, cellular histone ubiquitin ligases responsible for anchoring repair factors at sites of damage, as new targets for ICP0-mediated degradation. By targeting these ligases, ICP0 expression results in loss of ubiquitinated forms of H2A, mobilization of DNA repair proteins and enhanced viral fitness. Our study raises the possibility that the ICP0-mediated control of histone ubiquitination may link DNA repair, relief of transcriptional repression, and activation of latent viral genomes.
Keywords: herpes simplex virus type 1, histone H2A, ICP0, irradiation-induced foci, ubiquitin
Introduction
Herpes simplex virus type 1 (HSV-1) is a DNA virus with a biphasic lifecycle. During lytic infection, the immediate early (IE) genes create a favourable cellular environment for the virus. In the latent phase, HSV-1 enters sensory neurons innervating the site of infection, and the genome persists episomally for the lifetime of the host (reviewed in Roizman et al, 2006). Reactivation can occur spontaneously or be induced by ultra-violet (UV) damage or stress. Understanding the mechanisms that control establishment, maintenance, and reactivation from latency remains one of the most controversial and challenging subjects in the field (Efstathiou and Preston, 2005).
The IE protein ICP0 is one of the first viral proteins to be expressed during infection (reviewed in Everett, 2000). ICP0 is non-essential, but its deletion significantly impairs lytic replication, especially at low multiplicity of infection (MOI) (Stow and Stow, 1986; Cai and Schaffer, 1992; Chen and Silverstein, 1992; Everett et al, 2004). Although ICP0 is a promiscuous transactivator of many viral and cellular promoters, it is unable to bind DNA directly and the mechanism of transactivation is unclear. ICP0 is a RING finger E3 ubiquitin ligase that induces proteasome-mediated degradation of several cellular proteins including the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (Parkinson et al, 1999), components of the nuclear domain structures known as ND10 (or PML nuclear bodies) (Everett et al, 1998a; Gu and Roizman, 2003), and centromeric proteins (Everett et al, 1999a; Lomonte et al, 2001; Lomonte and Morency, 2007). ICP0 is also required for efficient production of infectious virus during reactivation from latency (Leib et al, 1989; Cai et al, 1993; Halford and Schaffer, 2001; Thompson and Sawtell, 2006). Despite this plethora of functions, the cellular targets responsible for the role of ICP0 in enhancing lytic growth and controlling reactivation from latency remain a subject of debate (Everett, 2000; Arthur et al, 2001; Everett et al, 2004, 2006, 2008, 2009; Lomonte et al, 2004; Gu et al, 2005; Poon et al, 2006; Gu and Roizman, 2007; Terry-Allison et al, 2007).
The cellular response to DNA damage consists of a complex network of signalling pathways that ultimately co-ordinate DNA repair. The PI3 kinase-like kinases, ataxia-telangiectasia mutated (ATM) and DNA-PK, control the responses to double-strand breaks (DSBs), whereas ATR (ATM and Rad3-related) responds primarily to single-strand DNA damage. The MRN complex, consisting of Mre11, Rad50, and Nbs1 (D'Amours and Jackson, 2002; Petrini and Stracker, 2003; Stewart et al, 2003; Lavin and Kozlov, 2007) senses DSBs and recruits ATM, which then undergoes autophosphorylation (Bakkenist and Kastan, 2003). Activated ATM phosphorylates a number of substrates, including the histone variant H2AX (which becomes γH2AX) (Rogakou et al, 1998). This phosphorylation spreads over an extensive region of chromatin flanking the break (Rogakou et al, 1999) and stabilizes association of ATM and MRN with the DSB. This produces subnuclear structures termed irradiation-induced foci (IRIF) (Fernandez-Capetillo et al, 2003). The organization of IRIF and the mechanisms by which proteins are recruited and retained at these structures are beginning to be elucidated (Huen and Chen, 2008). The mediator protein, Mdc1 (mediator of DNA damage checkpoint protein 1) (Stewart et al, 2003; Stucki and Jackson, 2004) interacts directly with both γH2AX and ATM (Lou et al, 2006). Mdc1 itself is also phosphorylated and serves as a docking station for other repair proteins. Another mediator, 53BP1 (tumour protein p53-binding protein 1) (Schultz et al, 2000; Wang et al, 2002), binds to the constitutively methylated histone H4-K20 dimethyl (Huyen et al, 2004; Mochan et al, 2004; Botuyan et al, 2006). Retention of 53BP1 is dependent on Mdc1, suggesting that masked H4-K20 methylation signals are exposed in response to Mdc1-dependent chromatin restructuring. Other chromatin post-translational modifications, such as non-degradative mono- and poly-ubiquitination, are also emerging as important regulators of IRIF formation. The cellular ubiquitin ligase RNF8 binds phosphorylated Mdc1 and ubiquitinates H2A and H2AX in response to DNA damage (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Sakasai and Tibbetts, 2008). This event is reinforced by another ubiquitin ligase, RNF168, which binds to and further modifies the ubiquitinated histones (Doil et al, 2009; Stewart et al, 2009). Together these events co-ordinate recruitment of downstream DNA repair factors to IRIF, both by interaction with proteins containing ubiquitin interacting motifs and by inducing chromatin restructuring around the break site (Huen and Chen, 2008; Panier and Durocher, 2009).
Viral infection presents cells with large amounts of foreign DNA and many viruses are known to interact with the cellular DNA repair machinery (Lilley et al, 2007). HSV-1 infection activates and exploits aspects of the cellular DNA damage signalling cascade while inhibiting others (Parkinson et al, 1999; Wilkinson and Weller, 2004, 2005, 2006; Lilley et al, 2005). During lytic infection, HSV-1 recruits activated cellular DNA repair proteins into viral replication compartments (Taylor and Knipe, 2004; Wilkinson and Weller, 2004), where they enhance viral replication (Lilley et al, 2005). DNA damaging treatments such as UV can induce reactivation of latent HSV-1, suggesting that there may be a link between DNA damage signalling and viral reactivation. As ICP0 is important during the early stages of lytic infection and is required for efficient escape from latency, we explored whether ICP0 affects activation or localization of DNA repair proteins at sites of damaged DNA. Here, we identify RNF8 and RNF168 as targets for ICP0-mediated degradation, and describe a novel role for ICP0 in controlling histone ubiquitination to prevent DNA repair proteins from accumulating at sites of cellular damage.
Results
ICP0 prevents accumulation of activated DNA repair proteins at IRIF
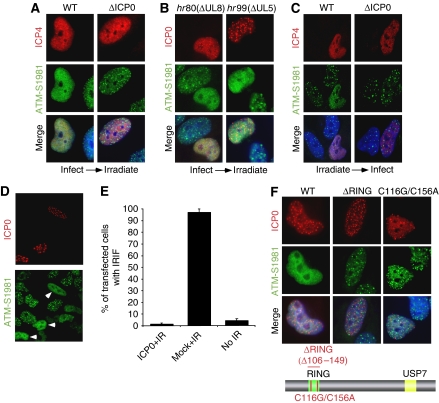
To investigate effects of ICP0 on localization of DNA damage proteins, we infected cells with wild type (WT) or ICP0-null virus for 2 h and then treated cells with 10 Gy ionizing radiation (IR). Cells were allowed to recover for 1 h post irradiation and then processed for immunofluorescence. Infected cells were identified by staining for the viral protein ICP4, and assessed for localization of activated DNA repair proteins at IRIF. Staining with an antibody generated to ATM auto-phosphorylated at residue S1981 showed that accumulation of immuno-reactive proteins at DSBs was prevented in over 90% of cells infected with WT virus (Figure 1A; Supplementary Figure S1A). In contrast, when cells infected with ICP0-null virus were irradiated, accumulation of ATM-dependent phosphorylation events at the DSBs was indistinguishable from uninfected, irradiated cells (Figure 1A). We and others have earlier reported that HSV-1 infection (both WT and ICP0-null) induces ATM activation (Wilkinson and Weller, 2004; Lilley et al, 2005), but the timepoint assessed in Figure 1A (2 h.p.i.) was before that previously used to observe activation of DNA damage signalling by viral infection. As a further control, we infected cells with viruses unable to induce the cellular DNA damage response (Figure 1B). These helicase/primase mutant viruses are unable to replicate, but they express ICP0 and, similar to WT HSV-1, prevented accumulation of ATM-dependent phosphorylated proteins at the DSBs. Early in infection, ICP0 is known to localize to distinct nuclear speckles corresponding to cellular ND10 domains (reviewed in Everett, 2000). The virally infected, irradiated cells react with phospho-specific antibodies, suggesting that ICP0 does not prevent DNA damage signalling, but affects the localization of the activated repair proteins (Figures 1A and B; Supplementary Figure S1B).
Figure 1.
Expression of ICP0 prevents accumulation of DNA repair proteins at IRIF. (A) HeLa cells were infected with WT or ICP0-null HSV-1 at an MOI of 5 for 2 h and treated with 10 Gy IR. Cells were fixed 1 h post IR, stained for ICP4 and assessed for localization of proteins reacting with an antibody generated to phosphorylated ATM. Nuclei were stained with DAPI as shown in the merged image. (B) HeLa cells were infected with ΔUL8 or ΔUL5 mutants of HSV-1 at an MOI of 5 and then treated with 10 Gy IR. Cells were fixed 1 h post IR, and stained with antibodies to ICP0 and phosphorylated ATM. (C) ICP0 can disrupt pre-established IRIF. Uninfected HeLa cells were irradiated at 10 Gy and infected 1 h post IR with WT or ICP0-null HSV-1 at an MOI of 5 for 2 h. Cells were fixed 2 h post infection and stained with antibodies to ICP4 and phosphorylated ATM. (D) HeLa cells were transfected with an ICP0 expression plasmid for 16 h and then treated with 10 Gy IR. Cells were fixed 1 h post IR, and stained with antibodies to ICP0 and phosphorylated ATM. (E) Transfected HeLa cells (200) were scored for the localization at IRIF of proteins reacting with phosphorylated ATM antibody. Data are represented as mean±s.e.m. (F) HeLa cells were transfected with WT or RING mutant ICP0 expression plasmids for 16 h and then treated with 10 Gy IR. Cells were fixed 1 h post IR, and stained with antibodies to ICP0 and phosphorylated ATM.
Damage-induced phosphorylation of ATM and its substrates occurs at the physical site of the DSB (Lukas et al, 2003). ICP0 does not prevent these IR-induced phosphorylation events (Supplementary Figure S1B; data not shown), suggesting that it does not inhibit initial recognition of DNA damage, but instead interferes with events that serve to retain repair factors at breaks. To test this, we assessed the effects of ICP0 on pre-established IRIF. In contrast to Figures 1A and B, in which cells were infected before irradiation, in Figure 1C cells were irradiated before infection. Irradiated cells were allowed to develop IRIF for 1 h and then infected with either WT or ICP0-null virus for 2 h. In cells infected with WT virus, DNA repair proteins became diffusely nuclear, whereas in cells infected with an ICP0-null virus the cellular proteins remained at IRIF (Figure 1C; Supplementary Figures S1C and D). These experiments demonstrate that ICP0 not only prevents stable accumulation of DNA repair proteins at IRIF (Figures 1A and B) but can also disrupt pre-established IRIF (Figure 1C; Supplementary Figure S1D).
We next assessed whether ICP0 expression in the absence of virus is sufficient to prevent accumulation of activated DNA repair proteins at IRIF. Although it has been suggested that ICP0 transfection can activate Chk2 in 293T cells (Li et al, 2008), in our hands expression of ICP0 alone did not induce DNA damage signalling in HeLa cells (Lilley et al, 2005). We transfected HeLa cells with an ICP0 expression plasmid, irradiated at 16 h post transfection and examined IRIF at 1 h post irradiation (Figure 1D). ICP0 can again be seen localized at distinct nuclear speckles corresponding to ND10 domains. We observed that 96% of control cells contained focal accumulations of ATM-dependent phosphorylations, but this was reduced to less than 5% in the ICP0-expressing cells (Figures 1D and E). Together, these data show that ICP0 is necessary and sufficient to prevent accumulation of activated DNA repair proteins at sites of cellular damage.
The RING finger domain of ICP0 is required to prevent accumulation of DNA repair proteins at IRIF
We analysed which domains of ICP0 are important to prevent IRIF formation. ICP0 is a RING finger ubiquitin ligase with defined regions required for localization to ND10 and binding to the deubiquitinase, USP7 (reviewed in Everett, 2000). We tested a panel of ICP0 mutants for their ability to prevent accumulation of local, ATM-dependent phosphorylations at DSBs. Mutants deficient in ND10 localization or USP7 binding were similar to WT ICP0 in their ability to prevent IRIF (Supplementary Figure S2). However, we observed that a ΔRING mutant (Everett, 1988) and a double point mutant of zinc co-ordinating cysteines within the RING finger domain (Panagiotidis et al, 1997) were both unable to prevent accumulation of ATM-dependent phosphorylated proteins at IRIF (Figure 1F). As the ICP0 RING finger is essential for ubiquitin ligase activity (Boutell et al, 2002), these data suggest that ICP0-dependent ubiquitination of cellular substrates is responsible for the observed block to IRIF.
The ICP0-mediated block to IRIF is downstream of γH2AX activation and Mdc1 recruitment, but upstream of 53BP1 accumulation
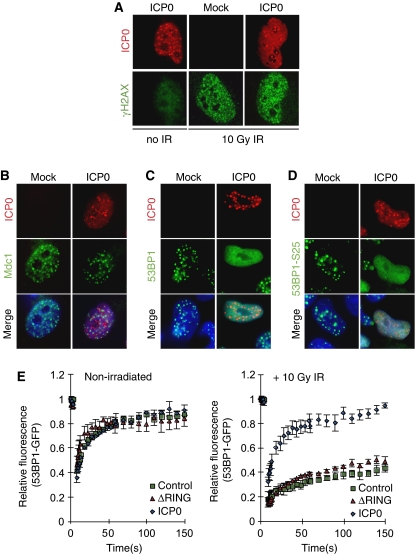
A tightly controlled temporal hierarchy of recognition, recruitment, and amplification signals occurs after the induction of a DSB (Lukas et al, 2004a, 2004b; Bekker-Jensen et al, 2005). We analysed at which point in this hierarchy ICP0 acted to block IRIF. Phosphorylation of the histone variant H2AX is one of the first events to occur after induction of a DSB (Rogakou et al, 1998; Paull et al, 2000). This phosphorylation event is not required for initial recruitment of DNA repair proteins to the break, but is required for sustained accumulation of factors such as 53BP1 at damage sites (Celeste et al, 2003; Ward et al, 2003). We examined the effect of ICP0 on initiation of the γH2AX signal in irradiated cells. HeLa cells transfected with ICP0 were irradiated at 24 h post transfection and processed for immunofluorescence (Figure 2A). We observed that ICP0 alone was not sufficient to activate H2AX (Figure 2A, left panel), and the initial phosphorylation of H2AX in response to IR was not significantly affected by ICP0 (compare Figure 2A right panel to Figure 2A middle panel). Next, cells transfected with ICP0 were irradiated and localization of Mdc1, Nbs1, BRCA1, 53BP1, and 53BP1 phosphorylated at S25 was examined and quantified (Figures 2B–D; Supplementary Figures S3 and S4). We observed that Mdc1 accumulated at sites of damage in the presence of ICP0 (Figure 2B; Supplementary Figure S3A). Similarly, the accumulation of Nbs1 at sites of laser damage (induced by the method of Botvinick and Berns, 2005) was unaffected by the presence of ICP0 (Supplementary Figure 4A). In contrast, ICP0 prevented stable accumulation of 53BP1 and BRCA1 at IRIF (Figure 2C; Supplementary Figures S3B and S4B), but did not prevent the damage-induced phosphorylation of 53BP1 (Figure 2D; Supplementary Figure S3C). We observed that ICP0 also limited the accumulation of 53BP1 at damage-independent foci detected in the absence of IR (Supplementary Figure S5A). Global levels of H4-K20 dimethyl, the binding site for 53BP1, were not affected by ICP0 expression (Supplementary Figure S5B). This indicates that the position of the ICP0-induced block is downstream of initial γH2AX activation and Mdc1 and Nbs1 recruitment, but upstream of 53BP1 and BRCA1 accumulation. Our data suggest that although Mdc1 itself can localize to IRIF in the presence of ICP0 (Figure 2B), it is unable to recruit downstream effectors or induce the chromatin remodelling events that lead to exposure of H4-K20 dimethyl and 53BP1 binding.
Figure 2.
The level of the ICP0-induced block to IRIF. (A) ICP0 does not affect initial phosphorylation of H2AX. HeLa cells were transfected with an ICP0 expression plasmid for 16 h and then mock treated or treated with 10 Gy IR. Cells were fixed 15 min post IR and stained for ICP0 and γH2AX. (B) ICP0 blocks IRIF formation downstream of Mdc1. HeLa cells were transfected with an ICP0 expression plasmid for 16 h and then treated with 10 Gy IR. Cells were fixed 1 h post IR and stained for ICP0 and Mdc1. (C) ICP0 blocks IRIF upstream of 53BP1. HeLa cells were transfected with an ICP0 expression plasmid for 16 h and then treated with 10 Gy IR. Cells were fixed 1 h post IR and stained for ICP0 and 53BP1. (D) ICP0 expression does not block phosphorylation of 53BP1. HeLa cells were transfected with an ICP0 expression plasmid for 16 h and then treated with 10 Gy IR. Cells were fixed 1 h post IR and stained for ICP0 and 53BP1-S25. (E) ICP0 prevents the IR-induced decrease in 53BP1 mobility. U20S cells stably expressing GFP-tagged 53BP1 were transfected with WT or ΔRING ICP0; 16 h post transfection, cells were subject to 10 Gy IR and 15 min post IR, 53BP1 mobility was assessed through FRAP analysis. Adjusted fluorescence intensity is shown as a fraction of the pre-bleach intensity.
It has been reported earlier that HSV-1 recruits certain DNA repair proteins to replication compartments to enhance viral replication (Taylor and Knipe, 2004; Wilkinson and Weller, 2004; Lilley et al, 2005). For example, proteins phosphorylated by ATM accumulate at viral replication compartments and ATM promotes HSV-1 replication (Lilley et al, 2005). We hypothesized that one reason ICP0 may target IRIF is to keep certain repair proteins mobile, and thus more freely available to be recruited into viral compartments. To investigate this possibility we used 53BP1 as a marker for DNA repair proteins mobilized by ICP0. The dynamics of 53BP1 in the presence or absence of ICP0 were measured by fluorescence recovery after photobleaching (FRAP) assays using GFP-53BP1. We transfected U20S cells expressing GFP-tagged 53BP1 (Bekker-Jensen et al, 2005) with WT ICP0 or the ΔRING mutant and analysed the rate at which GFP-53BP1 re-populated a bleached area (Figure 2E; Supplementary Figure S5C). We observed that in the absence of IR, GFP-53BP1 was highly mobile in all conditions (Figure 2E, left panel). After irradiation, in cells transfected with a control plasmid or the ΔRING mutant of ICP0, GFP-53BP1 became significantly less mobile as it was retained at damage-induced foci (Figure 2E, right panel). In contrast, GFP-53BP1 was not immobilized after IR when WT ICP0 was expressed, displaying kinetics very similar to non-irradiated cells (Figure 2E). These data show that ICP0 prevents the IR-induced decrease in 53BP1 mobility, thereby inhibiting its stable retention at IRIF.
ICP0 expression leads to loss of ubiquitinated forms of H2A, H2AX, and γH2AX
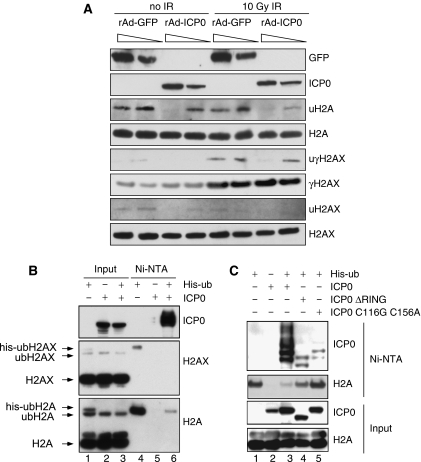
Given the emerging role of chromatin in the DNA damage response (Tsukuda et al, 2005), we examined the effect of ICP0 expression on levels or ubiquitination status of histone proteins. Infections with recombinant adenovirus expressing ICP0 (Halford et al, 2001) showed that histones H2A and H2AX were under-ubiquitinated in a dose-dependent manner in the presence of ICP0 (Figure 3A). At 1 h post irradiation, ICP0 did not significantly affect global levels of H2AX phosphorylation but did reduce damage-induced ubiquitination of γH2AX (Figure 3A). Next, we used a 6xhis-tagged ubiquitin construct (his-ubiquitin) to isolate ubiquitinated proteins under denaturing conditions in the presence or absence of ICP0 (Figure 3B). We observed that H2A and H2AX are normally mono-ubiquitinated, but that their ubiquitination was significantly reduced in the presence of ICP0 (compare lane 6 to lane 4 in Figure 3B). The his-ubiquitinated H2A seemed to be more affected than endogenous H2A (compare lane 1 to lane 3 in Figure 3B), suggesting that de novo ubiquitination events may be preferentially blocked by ICP0. This loss of ubiquitinated H2A was dependent on the RING finger domain of ICP0, as deletion or catalytic mutations in the RING finger significantly reduced the effect (Figure 3C). Auto-ubiquitination of ICP0 served as a positive control in the his-ubiquitin purifications (Figures 3B and C). Immunofluorescence experiments in the presence of excess ubiquitin excluded the possibility that ICP0 was preventing IRIF by sequestering ubiquitin (Supplementary Figure S5D). Damage-induced ubiquitination of H2A and H2AX has recently been shown to have a role in co-ordinating recruitment of DNA repair factors to IRIF (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Sakasai and Tibbetts, 2008). However, we observed that the effects of ICP0 on ubiquitinated H2A and H2AX occurred in the absence of IR treatment (Figure 3A). ICP0 prevented the block to 53BP1 IRIF in cells lacking several known degradation targets, suggesting that their degradation is not necessary for the effects we describe (Supplementary Figures S6 and S7). Together, these data imply that targeting of a novel ICP0 substrate leads to constitutive loss of ubiquitinated H2A and H2AX, and suggest that this is the mechanism by which ICP0 prevents stable accumulation of repair proteins at DSBs.
Figure 3.
ICP0 expression induces loss of ubiquitinated forms of H2A and H2AX. (A) HeLa cells were infected with recombinant adenoviruses expressing either GFP or ICP0, plus recombinant adenovirus expressing the tet activator (1:1) at a total MOI of 50 or 10. Cells were irradiated 23 h.p.i. and then collected 1 h post IR. Lysates and acid soluble chromatin fractions prepared and analysed by immunoblotting for levels and ubiquitination status of H2A, H2AX, and γH2AX. (B) 293T cells were transfected with ICP0 and his-tagged ubiquitin. Cells were lysed in denaturing conditions 24 h post transfection, and his-ubiquitin-conjugated proteins were purified over nickel beads (Ni–NTA). Input (5%) and eluted proteins were analysed by immunoblotting for levels and ubiquitination status of H2A and H2AX. (C) 293T cells were transfected with WT or mutant versions of ICP0 and his-tagged ubiquitin. Samples were purified and blotted as in (B).
ICP0 expression leads to a decrease in the levels of RNF8 and RNF168
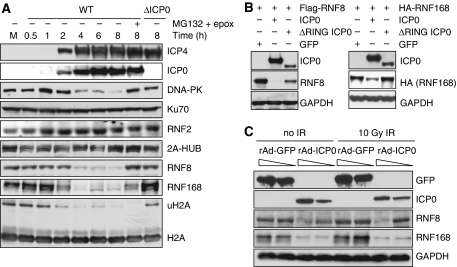
To determine how ICP0 leads to under-ubiquitination of H2A and H2AX, we investigated whether ICP0 negatively affects the level of specific H2A ubiquitin ligases. The main ubiquitin ligases implicated in the modification of H2A are RNF2 (also known as Ring2 or Ring1b), 2A-HUB, RNF8, and RNF168 (Wang et al, 2004; Huen et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Zhou et al, 2008; Doil et al, 2009; Stewart et al, 2009). We studied the levels of these H2A ubiquitin ligases during a timecourse of viral infection in HeLa cells (Figure 4A) and IMR90 cells (data not shown). There was no significant effect of infection on RNF2 or 2A-HUB levels (Figure 4A), but we observed an ICP0-dependent and proteasome-mediated decrease in RNF8 and RNF168 levels (Figure 4A). RNF8 and RNF168 were not degraded during a timecourse of infection with ICP0-null virus (Supplementary Figure S8A). The kinetics of loss of RNF8 and RNF168 mirrored that of DNA-PKcs, a known substrate of ICP0 (Parkinson et al, 1999), although degradation of DNA-PKcs was not required for the degradation of RNF8 and RNF168 (Supplementary Figure S9C). Although the addition of proteasome inhibitors rescued RNF8 and RNF168 degradation, ubiquitinated H2A levels were not rescued (Figure 4A), presumably because of depletion of free nuclear ubiquitin induced by proteasome inhibition (Dantuma et al, 2006; Mailand et al, 2007). We also found that transfection of excess WT but not ΔRING ICP0 was sufficient to degrade RNF8 and RNF168 as both tagged (Figure 4B) and endogenous proteins (Supplementary Figures S9A and B). As with the ICP0-induced loss of ubiquitinated H2A, degradation of RNF8 and RNF168 occurred independently of IR treatment (Figure 4C). RNF8 has also been recently reported to have a role in the ubiquitination of H2B (Wu et al, 2009). Although we observed a decrease in ubiquitinated forms of H2B during HSV-1 infection, this decrease was not dependent on ICP0 or degradation of RNF8 (Supplementary Figure S8B). RNF8 and RNF168 have been reported to use the ubiquitin-conjugating enzyme, Ubc13, to ubiquitinate H2A. We did not detect any decrease in the levels of Ubc13 during the timecourse of viral infection (data not shown), suggesting that ICP0 targets the pathway at the level of E3 ligases rather than E2 enzymes. Together, these data show that ICP0 expression, either from plasmid transfection or viral infection, is necessary and sufficient to induce proteasome-mediated degradation of RNF8 and RNF168 that is independent of DNA damage. We propose that degradation of these H2A ubiquitin ligases explains the loss of ubiquitinated H2A and H2AX observed in the presence of ICP0.
Figure 4.
ICP0 induces degradation of RNF8 and RNF168. (A) HeLa cells were infected with WT or ΔICP0 HSV-1 at an MOI of 5 and lysates prepared for immunoblotting over a timecourse of infection. Degradation of RNF168 and loss of ubiquitinated H2A were detectable at 2 h post infection, whereas DNA-PKcs and RNF8 degradation were detectable at 4 h post infection. Levels of all degradation substrates were rescued by proteasome inhibitors added at 2 h (compare levels of RNF8 and RNF168 at 2 h to levels at 8 h in the presence of MG132 and epox). Ku70 served as a loading control. (B) HeLa cells were transfected with Flag-RNF8 or HA-RNF168 and either ICP0 WT or the ΔRING mutant in a ratio of 3:1, where ICP0 was in excess. Cells were collected at 20 h post transfection, lysates were prepared for immunoblotting and assessed for levels of RNF8 and RNF168. GAPDH was used as a loading control. (C) HeLa cells were infected with recombinant adenoviruses expressing either GFP or ICP0, plus recombinant adenovirus expressing the tet activator (1:1) at a total MOI of 50 or 10. Cells were irradiated 23 h.p.i. and then collected 1 h post IR. Lysates were analysed by immunoblotting and assessed for levels of RNF8 and RNF168. GAPDH was used as a loading control.
ICP0 co-localizes with, binds, and ubiquitinates RNF8
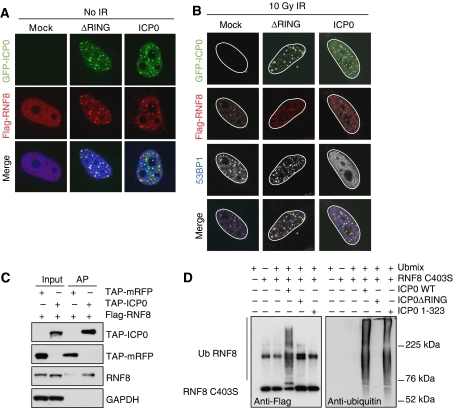
To visualize RNF8 and ICP0 in the same cell, we transfected an excess of tagged RNF8 relative to ICP0. In these co-transfection studies, we observed that both WT and ΔRING ICP0 co-localized with over-expressed RNF8 in the absence of IR (Figure 5A). After irradiation, Flag-RNF8 is recruited to IRIF in the presence of ΔRING ICP0 (Figure 5B). However, in the presence of WT ICP0, the over-expressed RNF8 remains associated with the viral protein, presumably because IRIF are not properly formed in this case (Figure 5B). ICP0 itself does not seem to be recruited to IRIF, suggesting that ICP0 and RNF8 interact at nuclear sites of ICP0 expression rather than sites of damage. This co-localization suggested that ICP0 may bind RNF8 directly. To test this, Flag-RNF8 was co-transfected with tandem affinity purification (TAP)-ICP0 or TAP-mRFP in the presence of proteasome inhibitors. Cells were collected 24 h post transfection, lysates incubated with streptavidin beads, and purified proteins analysed by immunoblotting. We observed that TAP-ICP0 but not TAP-RFP co-purified with Flag-tagged RNF8 (Figure 5C). We repeated the same experiment with tagged RNF168 but were unable to detect a direct interaction (data not shown). As we had detected an interaction between RNF8 and ICP0, we wished to determine whether ICP0 was sufficient to mediate RNF8 ubiquitination directly in an in vitro assay. We purified his-tagged RNF8 and his-tagged full-length or mutant versions of ICP0 and used them in reactions containing ubiquitin, E1, and the ubiquitin-conjugating enzyme UbcH5a. We observed that WT but not the C403S mutant of RNF8 could induce auto-ubiquitination in vitro in the presence of UbcH5a (data not shown), consistent with previous results (Ito et al, 2001). We therefore used the catalytically inactive C403S RNF8 in our in vitro assays to eliminate the possibility of auto-ubiquitination. We observed striking poly-ubiquitination of RNF8 C403S in vitro in the presence of WT but not a ΔRING version of ICP0 (Figure 5D). This shows that ICP0 is sufficient to ubiquitinate RNF8 in vitro and that the RING finger of ICP0 is necessary for this effect. To determine whether the RING finger domain of ICP0 was sufficient to induce the ubiquitination of RNF8, we performed reactions with an N-terminal fragment of ICP0 (amino acids 1–323) containing the RING finger domain. This mutant has efficient E3 ligase activity in vitro (Figure 5D, right panel) but was unable to ubiquitinate RNF8 (Figure 5D, left panel). This indicates that ubiquitination of RNF8 by ICP0 occurs in a substrate-specific manner. Our data show that ICP0 directly binds RNF8 and leads to the proteasome-mediated degradation of RNF8 and RNF168. We predicted that this degradation of RNF8 and RNF168 is responsible for the loss of H2A ubiquitination and disruption of IRIF we observe in the presence of ICP0. To test this directly, we over-expressed WT or RING mutant versions of RNF8 and RNF168 in the presence of WT ICP0, and then irradiated cells, and assessed and quantified the localization of 53BP1 (Figures 6A and B). We observed that the combined over-expression of RNF8 and RNF168 was necessary and sufficient to rescue the ICP0-induced block to 53BP1 IRIF in the majority of cells. Importantly, neither ligase alone nor one WT ligase combined with one catalytically inactive ligase was sufficient. This experiment demonstrates that degradation of RNF8 and RNF168 is directly responsible for the ICP0-induced block to IRIF.
Figure 5.
ICP0 co-localizes with, binds, and ubiquitinates RNF8. (A) HeLa cells were co-transfected with Flag-RNF8 and GFP-ICP0 or GFP-ΔRING ICP0 for 16 h (3:1 ratio of Flag-RNF8:GFP-ICP0). The co-localization of ICP0 and Flag-RNF8 was assessed by immunofluorescence. (B) HeLa cells were co-transfected with Flag-RNF8 and GFP-ICP0 or GFP-ΔRING ICP0 at the same ratio as in (A) for 16 h and then treated with 10 Gy IR. Localization of ICP0, Flag-RNF8, and 53BP1 were assessed by immunofluorescence 1 h post IR. For four-colour immunofluorescence, nuclei (as visualized by DAPI staining) are outlined in white. (C) Flag-RNF8 was co-transfected with TAP-ICP0 or TAP-mRFP into 293T cells and proteasome inhibitors were added 16 h post transfection to prevent degradation of RNF8. At 24 h post transfection, lysates were immunoprecipitated with streptavidin–sepharose beads overnight at 4°C and purified proteins were analysed by immunoblotting. Inputs (5%) and affinity purifications (AP) are shown. GAPDH was used as a loading control. (D) ICP0 ubiquitinates purified RNF8 in vitro. Ubiquitination reactions were carried out in the presence of purified Flag-RNF8 C403S and either WT, ΔRING, or RING-only (1–323) versions of ICP0. The ubiquitination status of RNF8 was analysed by immunoblotting with anti-Flag antibody (left panel). The band present in all RNF8-containing lanes is either a non-specific band or an SDS-resistant dimer of RNF8 C403S. The ability of ICP0 mutants to form poly-ubiquitin chains was assessed by immunoblotting with anti-ubiquitin antibody (right panel).
Figure 6.
Rescue of the ICP0-induced block to IRIF and negative effect of RNF8 on plaque-forming efficiency of an ICP0-null virus. (A) Combined over-expression of RNF8 WT and RNF168 WT rescues the ICP0-induced block to IRIF. HeLa cells were transfected with GFP-ICP0 in the presence of Flag-RNF8 WT or C403S and Flag-RNF168 WT or C19S. The cells were exposed to 10 Gy IR at 24 h post transfection, fixed at 1 h post IR, and imaged for ICP0, Flag, and 53BP1 (three left columns), or ICP0, γH2AX, and 53BP1 (right column). For four-colour immunofluorescence, nuclei (as visualized by DAPI staining) are outlined in white. In the merged images of the cells in which 53BP1 IRIF are rescued, the Flag signal (RNF8 and RNF168) co-localizes with both ICP0 and 53BP1. (B) Transfected cells (200) from (A) were scored for presence or absence of 53BP1 at IRIF. (C) RNF8 inhibits plaque-forming efficiency of ICP0 mutant viruses. RNF8−/− MEFs or RNF8−/− MEFs reconstituted with RNF8 were infected with WT or ICP0 mutant virus. Relative probabilities of plaque formation were calculated by comparing the numbers of plaques on the different cell lines at each separate dilution of virus. The decrease in ICP0-null virus plaque-forming ability in cells expressing RNF8 ranged from 2.5- to 6-fold, with an average of 3.7-fold. The decrease in ΔRING ICP0 plaque-forming ability in cells expressing RNF8 ranged from three- to five-fold, with an average of 4.2-fold. All infections were carried out at least in duplicate and the experiment was repeated four times. Data are represented as mean±s.e.m. (D) Model for the role of ICP0 in degrading RNF8 and RNF168 to limit H2A ubiquitination and prevent the formation of IRIF. ICP0 targets both RNF8 and RNF168 for proteasome-mediated degradation. The resultant loss of ubiquitinated H2A on cellular DNA prevents the accumulation of downstream repair factors at IRIF. The potential links between ICP0, H2A ubiquitination, and transctiptional silencing are depicted.
RNF8 inhibits the plaque-forming efficiency of an ICP0-null virus
We predicted that if ICP0-mediated degradation of RNF8 were beneficial for the virus, the plaque-forming efficiency of ICP0-null but not WT HSV-1 would be inhibited by RNF8. To test this hypothesis, we took advantage of murine embryonic fibroblasts (MEFs) from C57Bl/6 mice in which both copies of the RNF8 gene had been disrupted (Minter-Dykhouse et al, 2008). We transduced the RNF8−/− MEFs with empty retrovirus or retrovirus expressing human WT RNF8 (Supplementary Figure S10A). We confirmed that RNF8−/− MEFs are unable to form IRIF, and that reintroduction of RNF8 rescued this defect (Supplementary Figure S10B). The dose of ICP0-null virus used for infections was selected to give equivalent numbers of plaques as WT virus on cells which complement ICP0 function (Supplementary Figure S10C). We compared the plaque-forming deficiency of WT and ICP0-null viruses in the presence and absence of RNF8 (Figure 6C; Supplementary Figure S10D). The probability of WT virus to form plaques in the presence or absence of RNF8 was not significantly different (Figure 6C, left bars). However, ICP0-null or RING mutant viruses were approximately four-fold less likely to form plaques in the presence of RNF8 (Figure 6C, middle and right bars). This indicates that RNF8 contributes to the plaque-forming deficiency of an ICP0 mutant virus. These data show that RNF8 is inhibitory to HSV-1 plaque formation, and suggest that this inhibition can be overcome by the expression of ICP0 during WT virus infection.
Discussion
We suggested earlier a link between DNA damage signalling and the HSV-1 lifecycle (Lilley et al, 2005). As ICP0 is important during lytic replication and is required for efficient exit from latency, we examined whether ICP0 affects activation or localization of DNA repair proteins at sites of damaged DNA. We discovered that ICP0 expression results in degradation of the cellular ubiquitin ligases RNF8 and RNF168, leads to downregulation of H2A ubiquitination, and prevents accumulation of cellular DNA repair proteins at IRIF. We also found that the loss of RNF8 enhances viral fitness of an ICP0-null virus. This shows that a viral ubiquitin ligase can target cellular ubiquitin ligases to promote viral growth, and suggests that degradation of RNF8 and RNF168 has physiological relevance to the functions of ICP0 in the HSV-1 lifecycle.
Ubiquitination and IRIF
Our studies highlight the importance of H2A and H2AX ubiquitination in stabilizing DNA repair proteins at IRIF, and coordinating the DNA damage signal in mammalian cells. The recent identification of RNF8 as a damage-induced ubiquitin ligase for H2A and H2AX provided the missing link between histone phosphorylation and ubiquitination (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Sakasai and Tibbetts, 2008). RNF8 binds phosphorylated Mdc1, which in turn is anchored to γH2AX. RNF8 then ubiquitinates H2A and H2AX and this event triggers binding of RNF168, reinforcing ubiquitination at the DSB and signalling chromatin remodelling events that expose the H4-K20 dimethyl mark to which 53BP1 binds (Doil et al, 2009; Stewart et al, 2009). In this study, we show that in the presence of ICP0, ATM is still activated, H2AX is phosphorylated, and Mdc1 and Nbs1 are recruited to the DSB. However, ICP0 blocks the pathway at this level by inducing proteasome-mediated degradation of RNF8 and RNF168, preventing ubiquitination of H2A and H2AX at the break. Although downstream repair factors may transiently associate (to be phosphorylated) they are not stably maintained and remain highly mobile. As ICP0-mediated disruption of H2A and H2AX ubiquitination after DNA damage results in loss of IRIF, our data suggest that foci are reversible and dependent on the continued ubiquitination of H2A by RNF8 and RNF168. This is consistent with earlier studies in which depleting the nuclear pool of ubiquitin after irradiation disrupted 53BP1 IRIF (Mailand et al, 2007).
Targeting both RNF8 and RNF168
Until recently, it was assumed that the primary effect of RNF8 and RNF168 on H2A was the initiation and amplification of damage-induced ubiquitination (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Doil et al, 2009; Stewart et al, 2009). However, studies on RNF8 knockout mice have now shown that basal H2A ubiquitination is reduced by 80% in the absence of RNF8 (Wu et al, 2009). This is consistent with our data showing that the ICP0-induced degradation of RNF8 and RNF168 correlates with damage-independent loss of the majority of the cellular pool of ubiquitinated H2A. However, depletion of RNF8 and/or RNF168 by shRNA does not recapitulate the extensive loss of H2A seen in the presence of ICP0 (CEL, MSC, and MDW, unpublished observations), raising the possibility that ICP0 may also be targeting another as-yet-unidentified ubiquitin ligase for H2A. It has been shown earlier that loss of either RNF8 or RNF168 is sufficient to prevent 53BP1 IRIF and limit H2A ubiquitination (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Sakasai and Tibbetts, 2008; Doil et al, 2009; Stewart et al, 2009). Our observation that ICP0 expression leads to the degradation of both RNF8 and RNF168 raises the possibility that there may be separate RNF8- and RNF168-dependent functions. For example, ICP0 targeting of RNF168 could guard against amplification of a mono-ubiquitination signal set up by an RNF8-independent mechanism. As we could not detect an interaction between RNF168 and ICP0, it is possible that RNF168 is not directly targeted by ICP0. However, we observed an increase in ubiquitination of both WT and RING mutant versions of RNF168 in vitro in the presence of ICP0 and absence of RNF8 (unpublished observations). Furthermore, the fact that RNF8 levels are unaffected by the absence of RNF168 (Doil et al, 2009; Stewart et al, 2009) suggests that the two ligases are independently regulated.
The mechanistic details of how histone ubiquitination is involved in the maintenance of DNA repair factors at IRIF and whether H2A and H2AX are the only substrates for RNF8 and RNF168 remain to be determined. Viral proteins often show key regulators of cellular pathways by the proteins they target. It is possible that ICP0 is mimicking the activity of a cellular E3 ligase that normally serves to control the physiological levels of RNF8 and RNF168. ICP0 provides a useful tool to deplete RNF8, RNF168, and ubiquitinated forms of H2A and H2AX in cells of any genetic background to explore the functions of these cellular proteins in the DNA damage response.
HSV-1 and the cellular response to DNA damage
The interface between HSV-1 infection and the cellular DNA damage response is multi-faceted. We have reported earlier that ATM-mediated DNA damage signalling was beneficial to HSV-1 replication (Lilley et al, 2005). However, other cellular repair proteins such as DNA-PKcs (Parkinson et al, 1999), Ku70 (Taylor and Knipe, 2004), and ATR (Wilkinson and Weller, 2006) may be unfavourable for HSV-1. Our studies identify RNF8 and RNF168 as novel targets for ICP0-mediated degradation. Cellular consequences of loss of RNF8, RNF168, and disruption of IRIF could include decreased capacity to repair DNA, enhanced radiosensitivity, and increased apoptosis; these phenotypes have all been observed in the presence of ICP0 (Hadjipanayis and DeLuca, 2005; Sanfilippo and Blaho, 2006). Plaque-forming efficiency of an ICP0-null virus was partially rescued in RNF8-deficient cells, suggesting that degradation of this cellular ligase is beneficial for HSV-1 replication. Whether RNF8 and RNF168 themselves are detrimental for HSV-1 replication, or whether ICP0 targets these cellular proteins to limit ubiquitinated H2A levels remains to be seen.
Implications for transcriptional silencing
Ubiquitinated H2A is linked to transcriptional repression with roles in silencing the inactive X chromosome (Xi) in mice (Fang et al, 2004; de Napoles et al, 2004) and the promoters of polycomb-target genes such as Hox (Wang et al, 2004). Mutant viruses unable to express ICP0 are transcriptionally silenced until reactivation by superinfection with WT HSV or by providing ICP0 in trans (Preston and Nicholl, 1997; Everett et al, 1998b; Samaniego et al, 1998; Hobbs et al, 2001; Preston, 2007; Terry-Allison et al, 2007). The mechanisms underlying repression and ICP0-induced reactivation of quiescent viral genomes are not fully understood, but it is known that RING finger integrity is required. In this study, we show that ICP0 leads to a striking loss of ubiquitinated forms of H2A and H2AX, and we speculate that one reason ICP0 may control the levels of ubiquitinated H2A is to limit transcriptional repression (Figure 6D).
HSV-1 and chromatin
During lytic infection, incoming HSV-1 genomes induce a cellular response that leads to recruitment of a number of proteins, including histones, presumably in an attempt to silence expression of viral genes (Everett and Murray, 2005; Knipe and Cliffe, 2008). Among the proteins recruited are repressive components of ND10, and this event is prevented by ICP0-mediated disruption of ND10 and degradation of the main component, PML (Everett et al, 1998a, 2006, 2007; Everett and Murray, 2005). It is interesting to note the potential parallels between the ICP0-mediated disruptions of these two sub-cellular structures (IRIF and ND10) to promote virus infection. During latency, the viral genome is nucleosome bound in a form resembling facultative heterochromatin, and lytic promoters are marked with histone modifications characteristic of silenced genes (reviewed in Knipe and Cliffe, 2008). ICP0 has been recently shown to lead to an increase in the proportion of viral genome-associated histone H3 that is acetylated (Cliffe and Knipe, 2008; Coleman et al, 2008). Future studies will explore further whether the latent viral genome is marked with ubiquitinated H2A and whether targeting of cellular H2A ubiquitin ligases provides a functional link between the role of ICP0 in the viral lifecycle and the cellular mechanisms of transcriptional silencing. Transcriptional control of viral genomes by ubiquitination is a novel concept that may prove to be a paradigm for persistent and latent viruses.
Materials and methods
Cell lines
HeLa, Vero, U20S, IMR90, and HEK-293T and HEK-293 cells were purchased from the American Tissue Culture Collection. U20S cells expressing GFP-53BP1 were obtained from J Bartek and J Lukas (Bekker-Jensen et al, 2005). RNF8−/− MEFs were obtained from J Chen (Minter-Dykhouse et al, 2008). The HCT116-based cell line lacking DNA-PKcs was provided by E Hendrickson (Ruis et al, 2008). HepA cells expressing shRNA to PML or Sp100 have been described earlier (Everett et al, 2008). All cells were maintained in Dulbecco modified Eagle's medium (DMEM) or McCoys 5A medium containing 100 U/ml of penicillin and 100 μg/ml of streptomycin, and supplemented with 10% foetal bovine serum (FBS) and selection antibiotics as appropriate. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Viruses and infections
Parental virus HSV-1 strain was 17 syn+ and the matched ICP0 deletion mutant was dl1403 (Stow and Stow, 1986). The ICP0 RING deletion virus was FXE (Everett, 1989). All viruses were grown in Vero cells and titered by plaque assay in U2OS cells, in which ICP0 is not required for efficient plaque formation. Deletion mutants hr80 (ΔUL8) and hr99 (ΔUL5) (Carmichael and Weller, 1989; Zhu and Weller, 1992) were provided by S Weller. Recombinant adenoviruses expressing ICP0 or GFP were obtained from P Schaffer (Halford et al, 2001) and J Wilson, respectively, and grown in 293 cells. Infections were performed on monolayers of cells in DMEM supplemented with 2% FBS (rAdenoviruses) or 0% FBS (HSV-1). After 1 h at 37°C serum was added to a total of 10%. For complementation of RNF8−/− MEFs, retroviruses were prepared and infections performed as described earlier (Carson et al, 2003). For HSV-1 plaque assays on RNF8-deficient cells, 24-well dishes were infected with three-fold dilutions of WT or ICP0-null HSV-1. After virus adsorption, the cells were overlaid with medium containing 0.8% carboxymethylcellulose. Plaques were stained with crystal violet 36 h post infection.
Plasmids and transfections
Expression vectors for WT and mutant ICP0 were from S Silverstein or described earlier (Everett et al, 1999b). The ΔRING version of ICP0 was FXE, which lacks amino acids 106–149. Flag-RNF8 was obtained from J Chen and J Lukas. Vectors expressing his(6)-ubiquitin or HA(4)-ubiquitin were obtained from T Hunter and R Greenberg, respectively. For complementation of RNF8−/− MEFs, RNF8 WT was cloned into pLPC (Serrano et al, 1997) between EcoRI and XhoI. For affinity purifications, the CMV promoter and a TAP tag consisting of a calmodulin-binding peptide and streptavidin-binding peptide were excised from pNTAP (Stratagene) using AseI and BglI and inserted into pEGFP-ICP0 (Lomonte and Everett, 1999) between the same sites. mRFP was excised from pcDNA3.1(+) and inserted into pNTAP using EcoRI. For bacterial expression, RNF8 WT or C403S was excised from pcDNA4/TO (Mailand et al, 2007) and cloned into pET28a between EcoRI and XhoI. Sequences encoding ICP0 amino acids 1–323 were amplified from p110E53-1 (Everett, 1987) and cloned into pET28a as an NcoI/EcoRI fragment. The resulting plasmids expressed proteins with either N- or C-terminal in frame 6xhis tags. Mammalian cells were transfected with Lipofectamine 2000 (Invitrogen) or polyethylenimine (PEI, Polysciences Inc.) according to manufacturer's protocol.
Antibodies
Primary antibodies were purchased from Novus Biologicals Inc. (NBS1 S343), Rockland (ATM S1981-P), Santa Cruz (53BP1, Ku70, Ku86, ubiquitin), Millipore/Upstate (H2AX S139, H2A, uH2A, H2B), Calbiochem (BRCA1), Chemicon (GFP), Research Diagnostics Inc. (GAPDH), Covance (HA), Transduction Laboratories (DNA-PKcs), Abcam (RNF20 and RNF40), Medimabs (H2B), Sigma (Flag), and Cell Signaling (H2AX). The antibody to phosphorylated 53BP1 was from P Carpenter, the antibody to ICP0 was Mab11060 (Everett et al, 1993), rabbit antisera to Mdc1 was from J Chen, and rabbit antisera to RNF8 was from J Chen or J Lukas. The 58S monoclonal antibody to ICP4 was generated from an ATCC hybridoma cell line. All secondary antibodies were from Jackson Laboratories or Invitrogen.
Immunoblotting and immunofluorescence
For immunoblotting, infected or transfected cells were collected and lysates prepared by standard methods. Chromatin fractions were isolated by extracting the insoluble pellet with 0.1 M HCl for 30 min at 4°C. For immunofluorescence, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min and extracted with 0.5% Triton X-100 in PBS for 10 min. Nuclei were visualized by staining with DAPI. Images were acquired using a Leica TCS SP2 confocal microscope. Quantification was performed by counting 200 cells per condition.
Affinity purifications
Flag-RNF8 was co-transfected with TAP-ICP0 or TAP-mRFP into 10 cm plates of 293T cells. Proteasome inhibitors (10 μM MG132, 1 μM epoxomycin) were added 16 h post transfection. Cells were lysed 24 h post transfection in 500 μl IP buffer (50 mM Tris 7.5, 150 mM NaCl, 1 mM EDTA, 1%NP-40, 2 mM PMSF, 0.5 mM Idoacetimide, 0.5 mM NEM, and protease inhibitors). An amount of 100 μg protein was incubated with 40 μl streptavidin–sepharose beads in 500 μl IP buffer overnight at 4°C. Beads were washed 3 × in IP Buffer and boiled in SDS loading dye. Purified proteins were analysed by immunoblotting.
In vivo ubiquitination assay
A total of 5 × 106 293T cells were transfected with 10 μg of his(6)-ubiquitin plus 10 μg effector plasmid. Cells were collected after 24–48 h and processed for immunoblotting (input) and ubiquitination assays using nickel–nitrilotriacetic acid (Ni–NTA) beads (Qiagen). For the ubiquitination assays, the cell pellets were lysed in buffer A (6 M guanidinium–HCl, 0.1 M Na2HPO4–NaH2PO4, 10 mM Tris–HCl [pH 8.0], 10 mM β-mercaptoethanol) and incubated with Ni–NTA beads overnight at 4°C. Beads were washed once with buffer A and buffer B (8 M urea, 0.1 M Na2HPO4–NaH2PO4, 10 mM Tris–HCl [pH 8.0], 10 mM β-mercaptoethanol), and washed twice with buffer C (8 M urea, 0.1 M Na2HPO4–NaH2PO4, 10 mM Tris–HCl [pH 6.3], 10 mM β-mercaptoethanol) containing 0.2 and then 0.1% Triton X-100. Proteins were eluted from beads by boiling in NuPAGE LDS sample buffer (Invitrogen). The eluted proteins were analysed by immunoblotting.
In vitro ubiquitination assay
Human ubiquitin-activating enzyme (E1), full-length ICP0, the RING finger deletion mutant, and the RING-only 1–323 fragments were purified as described earlier (Boutell et al, 2002). Ubiquitin was purchased from Sigma. Clones expressing recombinant UbcH5a or RNF8 were expressed in BL21 and purified by nickel affinity chromatography. In vitro ubiquitination assays were performed in the presence of 50 ng of purified RNF8 C403S. Reactions were carried out in buffer D (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM ATP) with 20 ng of E1, 50 ng of E2, 2.5 μg of ubiquitin, and 50 ng of purified full-length ICP0, RING finger mutant, or ICP0 1-323 truncation mutant. Reactions were carried out in a final volume of 10 μl for 1 h at 37°C and terminated by the addition of SDS–PAGE boiling mix buffer containing 8 M urea and 100 mM DTT. The reaction products were analysed by immunoblotting.
FRAP analysis
U2OS cells stably expressing GFP-53BP1 (Bekker-Jensen et al, 2005) were grown on glass bottom 35 mm dishes with 14 mm glass microwells (MatTeK Corp.). Before imaging, cells were supplemented with fresh phenol-red free medium with 10% FBS and 10 μM HEPES. Images were acquired using a Leica TCS SP2 confocal microscope. After acquisition of five pre-bleach images, a defined region of interest (ROI) of 1 μM square was bleached five times by the 488-nm argon laser set to 100% transmission. Post-bleach images were acquired at 1-, 5-, and 10-s intervals for a total of 125 s at 10% transmission. The adjusted fluorescence intensity at each timepoint is represented as the fraction of the pre-bleach intensity at the ROI.
Supplementary Material
Acknowledgments
We thank S Silverstein, S Weller, J Chen, M Rosenfeld, H Koseki, J Wilson, T Hunter, R Greenberg, C Preston, E Hendrickson, J Lukas, and J Bartek for generous gifts of reagents. We thank colleagues at the Salk Institute and members of the Weitzman lab for helpful discussions. We thank Andrew Basilio, Linda Shi, and Michael Berns for help with the laser experiment in Supplementary Figure S4A. We acknowledge the James B Pendleton Charitable Trust for providing the Pendleton Microscopy Facility. Work in the Weitzman laboratory is partially supported by a Pioneer Developmental Chair. This work was supported by NIH Grant CA97093 and the Joe W and Dorothy Dorsett Brown Foundation (MDW), a Wellcome Trust International Research Fellowship (CEL), the HA and Mary K Chapman Charitable Trust (MDW/MSC), and a pre-doctoral Ruth L Kirschstein National Research Service Award (MSC).
Footnotes
The authors declare that they have no conflict of interest.
References
- Arthur JL, Scarpini CG, Connor V, Lachmann RH, Tolkovsky AM, Efstathiou S (2001) Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro. J Virol 75: 3885–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J (2005) Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol 170: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinick EL, Berns MW (2005) Internet-based robotic laser scissors and tweezers microscopy. Microsc Res Tech 68: 65–74 [DOI] [PubMed] [Google Scholar]
- Boutell C, Sadis S, Everett RD (2002) Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol 76: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA (1993) The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol 67: 7501–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Schaffer PA (1992) Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66: 2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael EP, Weller SK (1989) Herpes simplex virus type 1 DNA synthesis requires the product of the UL8 gene: isolation and characterization of an ICP6∷lacZ insertion mutation. J Virol 63: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD (2003) The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 22: 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chen J, Silverstein S (1992) Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol 66: 2916–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe AR, Knipe DM (2008) Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol 82: 12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HM, Connor V, Cheng ZS, Grey F, Preston CM, Efstathiou S (2008) Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J Gen Virol 89: 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, Neefjes J (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol 173: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Efstathiou S, Preston CM (2005) Towards an understanding of the molecular basis of herpes simplex virus latency. Virus Res 111: 108–119 [DOI] [PubMed] [Google Scholar]
- Everett RD (1987) A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J 6: 2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD (1988) Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol 202: 87–96 [DOI] [PubMed] [Google Scholar]
- Everett RD (1989) Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol 70: 1185–1202 [DOI] [PubMed] [Google Scholar]
- Everett RD (2000) ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22: 761–770 [DOI] [PubMed] [Google Scholar]
- Everett RD, Boutell C, Orr A (2004) Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol 78: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Cross A, Orr A (1993) A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197: 751–756 [DOI] [PubMed] [Google Scholar]
- Everett RD, Earnshaw WC, Findlay J, Lomonte P (1999a) Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J 18: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J (1998a) The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol 72: 6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A (1999b) The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol 73: 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Murray J (2005) ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol 79: 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Murray J, Orr A, Preston CM (2007) Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol 81: 10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Orr A, Preston CM (1998b) A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J 17: 7161–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Parada C, Gripon P, Sirma H, Orr A (2008) Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82: 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Parsy ML, Orr A (2009) Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol 83: 4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A (2006) PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80: 7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y (2004) Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 279: 52812–52815 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Celeste A, Nussenzweig A (2003) Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle 2: 426–427 [PubMed] [Google Scholar]
- Gu H, Liang Y, Mandel G, Roizman B (2005) Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA 102: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B (2003) The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA 100: 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B (2007) Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA 104: 17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjipanayis CG, DeLuca NA (2005) Inhibition of DNA repair by a herpes simplex virus vector enhances the radiosensitivity of human glioblastoma cells. Cancer Res 65: 5310–5316 [DOI] [PubMed] [Google Scholar]
- Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA (2001) ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol 75: 6143–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford WP, Schaffer PA (2001) ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J Virol 75: 3240–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs WE, Brough DE, Kovesdi I, DeLuca NA (2001) Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J Virol 75: 3391–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Chen J (2008) The DNA damage response pathways: at the crossroad of protein modifications. Cell Res 18: 8–16 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Ito K, Adachi S, Iwakami R, Yasuda H, Muto Y, Seki N, Okano Y (2001) N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur J Biochem 268: 2725–2732 [DOI] [PubMed] [Google Scholar]
- Knipe DM, Cliffe A (2008) Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6: 211–221 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Kozlov S (2007) ATM activation and DNA damage response. Cell Cycle 6: 931–942 [DOI] [PubMed] [Google Scholar]
- Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, Knipe DM, Tyler KL, Schaffer PA (1989) Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol 63: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Baskaran R, Krisky DM, Bein K, Grandi P, Cohen JB, Glorioso JC (2008) Chk2 is required for HSV-1 ICP0-mediated G2/M arrest and enhancement of virus growth. Virology 375: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD (2005) DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci USA 102: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Schwartz RA, Weitzman MD (2007) Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol 15: 119–126 [DOI] [PubMed] [Google Scholar]
- Lomonte P, Everett RD (1999) Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G(1) into S phase of the cell cycle. J Virol 73: 9456–9467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte P, Morency E (2007) Centromeric protein CENP-B proteasomal degradation induced by the viral protein ICP0. FEBS Lett 581: 658–662 [DOI] [PubMed] [Google Scholar]
- Lomonte P, Sullivan KF, Everett RD (2001) Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem 276: 5829–5835 [DOI] [PubMed] [Google Scholar]
- Lomonte P, Thomas J, Texier P, Caron C, Khochbin S, Epstein AL (2004) Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J Virol 78: 6744–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J (2006) MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21: 187–200 [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J (2004a) Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J (2004b) Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 3: 997–1007 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Minter-Dykhouse K, Ward I, Huen MS, Chen J, Lou Z (2008) Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J Cell Biol 181: 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochan TA, Venere M, DiTullio RA Jr, Halazonetis TD (2004) 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst) 3: 945–952 [DOI] [PubMed] [Google Scholar]
- Panagiotidis CA, Lium EK, Silverstein SJ (1997) Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J Virol 71: 1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Durocher D (2009) Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 8: 436–443 [DOI] [PubMed] [Google Scholar]
- Parkinson J, Lees-Miller SP, Everett RD (1999) Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol 73: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH (2003) The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 13: 458–462 [DOI] [PubMed] [Google Scholar]
- Poon AP, Gu H, Roizman B (2006) ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc Natl Acad Sci USA 103: 9993–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM (2007) Reactivation of expression from quiescent herpes simplex virus type 1 genomes in the absence of immediate-early protein ICP0. J Virol 81: 11781–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM, Nicholl MJ (1997) Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol 71: 7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ (2006) Herpes simplex viruses. In: Fields Virology, Knipe DM, Howley P (eds), Vol. 2, pp 2501–2602. Philadelphia: Lippincott Williams and Wilkins [Google Scholar]
- Ruis BL, Fattah KR, Hendrickson EA (2008) The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol 28: 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakasai R, Tibbetts R (2008) RNF8-dependent and RNF8-independent Regulation of 53BP1 in Response to DNA Damage. J Biol Chem 283: 13549–13555 [DOI] [PubMed] [Google Scholar]
- Samaniego LA, Neiderhiser L, DeLuca NA (1998) Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol 72: 3307–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo CM, Blaho JA (2006) ICP0 gene expression is a herpes simplex virus type 1 apoptotic trigger. J Virol 80: 6810–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stow ND, Stow EC (1986) Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol 67: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP (2004) MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst) 3: 953–957 [DOI] [PubMed] [Google Scholar]
- Taylor TJ, Knipe DM (2004) Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol 78: 5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-Allison T, Smith CA, DeLuca NA (2007) Relaxed repression of herpes simplex virus type 1 genomes in Murine trigeminal neurons. J Virol 81: 12394–12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RL, Sawtell NM (2006) Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J Virol 80: 10919–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA (2005) Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, Jorda KG, Chen J (2003) Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem 278: 19579–19582 [DOI] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK (2004) Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol 78: 4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK (2005) Inhibition of the herpes simplex virus type 1 DNA polymerase induces hyperphosphorylation of replication protein A and its accumulation at S-phase-specific sites of DNA damage during infection. J Virol 79: 7162–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK (2006) Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci 119: 2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X (2009) Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol 29: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG (2008) Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LA, Weller SK (1992) The UL5 gene of herpes simplex virus type 1: isolation of a lacZ insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J Virol 66: 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.