Abstract
Autophagy is primarily considered a non-selective degradation process induced by starvation. Nutrient-independent basal autophagy, in contrast, imposes intracellular QC by selective disposal of aberrant protein aggregates and damaged organelles, a process critical for suppressing neurodegenerative diseases. The molecular mechanism that distinguishes these two fundamental autophagic responses, however, remains mysterious. Here, we identify the ubiquitin-binding deacetylase, histone deacetylase-6 (HDAC6), as a central component of basal autophagy that targets protein aggregates and damaged mitochondria. Surprisingly, HDAC6 is not required for autophagy activation; rather, it controls the fusion of autophagosomes to lysosomes. HDAC6 promotes autophagy by recruiting a cortactin-dependent, actin-remodelling machinery, which in turn assembles an F-actin network that stimulates autophagosome–lysosome fusion and substrate degradation. Indeed, HDAC6 deficiency leads to autophagosome maturation failure, protein aggregate build-up, and neurodegeneration. Remarkably, HDAC6 and F-actin assembly are completely dispensable for starvation-induced autophagy, uncovering the fundamental difference of these autophagic modes. Our study identifies HDAC6 and the actin cytoskeleton as critical components that define QC autophagy and uncovers a novel regulation of autophagy at the level of autophagosome–lysosome fusion.
Keywords: actin, autophagosome–lysosome fusion, autophagy, HDAC6, neurodegeneration
Introduction
Macro-autophagy—hereafter referred to as autophagy—is the primary degradation pathway responsible for the disposal of long-lived proteins, macromolecular complexes, and organelles (Mizushima et al, 2008). Autophagy consists of two discrete but essential steps: formation of autophagosomes that sequester cytosolic constituents, and delivery of autophagic substrates to lysosomes where the contents are degraded (reviewed by Levine and Klionsky, 2004). The extraordinary ability of autophagosomes to sequester substrates of diverse sizes and origins endows autophagy with a unique degradative capacity that complements the proteasome system. However, autophagosomes lack intrinsic protease activities. Productive autophagy requires efficient fusion of autophagosomes to lysosomes. Although extensive characterization of ATG genes essential for autophagy has yielded critical insight into the mechanism of autophagy activation and autophagosome formation (reviewed by Levine and Klionsky, 2004), whether and how the fusion of autophagosomes to lysosomes is controlled remain poorly understood. Elucidating the molecular machinery that controls this fusion event could be crucial to deciphering the growing complexity of the autophagy system.
The second unresolved issue in autophagy lies with its unique substrate repertoire. Autophagy has been predominantly characterized as a non-selective degradative pathway activated by starvation. In this context, autophagy degrades cytosolic contents and organelles non-discriminatively to supply cells with essential macromolecules and energy for survival (Mizushima et al, 2008). However, it has become apparent that autophagy is not solely dedicated to nutrient management (Mizushima et al, 2004). Although this nutrient-independent ‘basal' autophagy remains poorly defined, evidence suggests that its main function is to enforce intracellular quality control (QC) by selective disposal of protein aggregates and damaged organelles, including mitochondria (reviewed by Mizushima et al, 2008). Such an activity is best illustrated by the finding that neural-specific ablation of atg5 or atg7, two genes essential for autophagy, leads to accumulation of ubiquitin-positive protein aggregates and progressive loss of neurons in mice (Hara et al, 2006; Komatsu et al, 2006). We will refer to this form of autophagy as QC autophagy to better define its specific function. Accordingly, despite their shared dependence on a common ATG machinery and ability to degrade cytosolic constituents via lysosomes, QC autophagy and starvation-induced autophagy are distinct in their function, nature, and substrate specificity. There is, however, little molecular understanding of what distinguishes these two fundamental autophagic modes. Nor is it known how QC autophagy achieves substrate specificity. The identification of specific factors that operate in one but not both forms of autophagy would be crucial to functionally dissect these autophagic pathways.
The protein deacetylase histone deacetylase-6 (HDAC6) has emerged as an important player in the cellular management of protein aggregates. Unique among HDAC family members, HDAC6 has an intrinsic ubiquitin-binding activity and associates with both microtubules and the F-actin cytoskeleton (Seigneurin-Berny et al, 2001; Hubbert et al, 2002; Kawaguchi et al, 2003; Gao et al, 2007; Zhang et al, 2007). While the HDAC6–actin interaction has been primarily linked to regulation of cell motility (Gao et al, 2007; Zhang et al, 2007), its association with the microtubule network and ubiquitinated proteins has led to the surprising finding that HDAC6 is a regulatory component of aggresome, the MTOC (microtubule-organizing centre)-localized inclusion body where excess protein aggregates are deposited (Kawaguchi et al, 2003). The formation of aggresomes, which are related to Lewy bodies found in many forms of neurodegenerative diseases, is proposed to protect cells by concentrating toxic protein aggregates to the MTOC where they are processed by autophagy (Kopito, 2000). Through its ubiquitin-binding BUZ finger, HDAC6 binds to and facilitates the transport of ubiquitinated misfolded proteins through the microtubule network to form an aggresome (Kawaguchi et al, 2003). Interestingly, evidence suggests that HDAC6 also plays a role in the eventual clearance of aggresomes, implying a functional connection between HDAC6 and autophagy (Iwata et al, 2005; Pandey et al, 2007). How HDAC6 intersects with the autophagy machinery to facilitate the clearance of protein aggregates or other autophagic substrates is not known. The answer to this question could provide new insight into the molecular nature of QC autophagy important for protein aggregate clearance.
In this report, we identify HDAC6 as a novel component that controls the fusion of autophagosomes and lysosomes associated with QC autophagy. HDAC6 promotes these fusion events by recruiting a cortactin-dependent, actin-remodelling machinery to ubiquitinated protein aggregates, where assembly of F-actin facilitates autophagosome–lysosome fusion and clearance of autophagic substrates. Remarkably, we found that HDAC6 and actin are completely dispensable for starvation-induced autophagy, thereby uncoupling these two fundamental autophagic modes at the molecular level. Our study identifies the ubiquitin-binding HDAC6 and the actin cytoskeleton as the integral components that define QC autophagy and uncovers a novel mechanism that controls autophagosome–lysosome fusion. These findings provide a molecular framework to understand the complexity of autophagy and implicate autophagosome–lysosome fusion defects in the pathogenesis of neurodegenerative diseases.
Results
Loss of HDAC6 results in defects in protein aggregate clearance but not autophagy activation
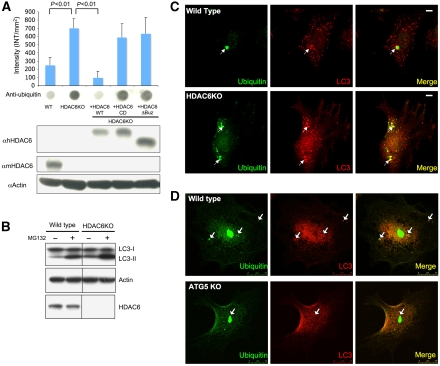
We have previously shown that HDAC6-knockdown cells are deficient in aggresome formation (Kawaguchi et al, 2003). To determine whether HDAC6 is also required for protein aggregate degradation, we treated mouse embryonic fibroblasts (MEFs) derived from HDAC6-knockout (KO) and matching wild-type mice with the proteasome inhibitor MG132. The levels of accumulated ubiquitinated aggregates were then analysed by a filter-trap assay. As shown in Figure 1A, ubiquitinated protein aggregates accumulated by ∼3-fold in HDAC6 KO MEFs as compared with that in wild-type MEFs, indicating that HDAC6 KO MEFs are deficient in protein aggregate degradation. Importantly, abnormal accumulation of protein aggregates is completely reversed in HDAC6 KO MEFs reconstituted with wild-type HDAC6, but not catalytically inactive or ubiquitin-binding-deficient mutants (Figure 1A), indicating that enzymatic activity and ubiquitin-binding activity are both required for HDAC6 to promote the clearance of ubiquitinated protein aggregates.
Figure 1.
HDAC6 KO MEF cells are defective in aggregate clearance but not in autophagosome induction or targeting. (A) Filter-trap analysis of MG132-induced, SDS-insoluble ubiquitinated aggregates generated in wild-type (WT), HDAC6 KO, and KO MEFs reconstituted with different HDAC6 constructs as indicated. The signal intensity from the ubiquitin immunoblot (bottom panel) was quantified and presented as the average of the means from three independent experiments along with the standard deviation (top). Note the significant accumulation of ubiquitin-positive aggregates in HDAC6 KO MEFs and KO MEFs stably expressing HDAC6-CD and ΔBUZ mutants. WT, HDAC6 KO, and HDAC6 KO MEFs stably expressing human HDAC6 (hHDAC6 WT), HDAC6 CD (catalytically inactive mutant), or HDAC6 ΔBUZ (ubiquitin-binding-deficient mutant) were analysed for the level of HDAC6 using an anti-human HDAC6 antibody. Mouse endogenous HDAC6 (mHDAC6) and actin levels were determined by each corresponding antibody. (B) WT and HDAC6 KO MEFs were treated with MG132 and subjected to Western blot analysis for LC3, actin, and HDAC6. (C) Cells were treated with MG132 as described under Materials and methods and immunostained with antibodies to ubiquitin (green) and LC3 (red). Arrows indicate ubiquitin-positive aggregates that colocalize with LC3-positive autophagosomes. Scale bar, 10 μm. (D) WT and ATG5 KO MEFs are treated with 2.5 μM MG132 for 1 day and incubated with normal growth media for 18 h. MEFs are immunostained with anti-LC3 (red) and anti-ubiquitin antibody (green).
Autophagy is considered the dominant machinery for protein aggregate clearance. To unravel the mechanistic link between HDAC6 and the autophagy machinery, we next investigated whether autophagy is properly activated in HDAC6 KO MEFs upon proteasome inhibition. We assessed autophagy activation by first monitoring the conversion of LC3 from the cytosolic form, LC3-I, to the autophagosome-associated form, LC3-II. Unexpectedly, despite the clear defect in protein aggregate clearance, conversion to LC3-II is prominently induced in HDAC6 KO MEFs upon MG132 treatment, suggesting that autophagosomes form normally in the absence of HDAC6 (Figure 1B). To further evaluate autophagosomes involved in protein aggregate clearance, we examined the formation of LC3-positive autophagosomes in wild-type and HDAC6 KO MEFs treated with MG132 and then chased in normal medium for 18 h. As shown in Figure 1C, prominent LC3-positive autophagic vesicles were clearly induced and readily found surrounding ubiquitin-positive protein aggregates in both cell types. Importantly, this staining is not caused by non-specific incorporation of LC3 into protein aggregates (Kuma et al, 2007), as no LC3-positive structures were observed in ATG5-deficient MEFs subject to the same treatment (Figure 1D and Supplementary Figure S1A). These results indicate that autophagosome formation proceeds normally in the absence of HDAC6; however, autophagy-dependent protein aggregate degradation is defective.
HDAC6 is required for fusion of autophagosomes and lysosomes associated with QC autophagy
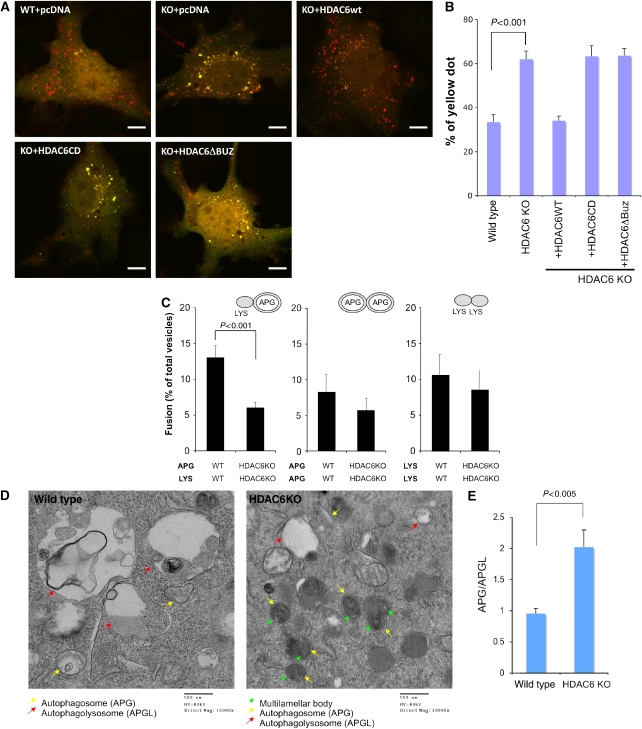
Autophagosomes must fuse to lysosomes to degrade their contents. The inability of HDAC6 KO MEFs to degrade protein aggregates despite an apparent induction of autophagosomes prompted us to ask whether HDAC6 is required for autophagosome–lysosome fusion. To this end, we used a pH-sensitive, double tagged mCherry-GFP-LC3 (dtLC3) reporter to determine the fusion efficiency. As GFP but not Cherry fluorescence is lost in acidic compartments, mCherry-GFP-LC3 labels non-acidic autophagosomes as yellow fluorescence (positive for both green and red), but acidic autophagolysosomes as red fluorescence only (Pankiv et al, 2007). To test the fusion efficiency, we expressed mCherry-GFP-LC3 in wild-type and HDAC6 KO MEFs under normal nutrient conditions. As shown in Figure 2A, we observed a prominent increase in the number of yellow-fluorescence vesicles in HDAC6 KO MEFs as compared with that in wild-type control, indicative of accumulation of autophagosomes caused by a defect in lysosome–autophagosome fusion. Indeed, the double-positive mCherry-GFP-LC3 vesicles are also positive for an autophagosome marker, MDC (mono-dansyl-cadaverine; Supplementary Figure S1B). The observed increase in yellow-fluorescence vesicles in HDAC6 KO MEFs cannot be attributed to loss of lysosomal acidification as the lysosomal pH is in the normal range in both wild-type and HDAC6 KO MEFs (Supplementary Figure S2). Lastly, accumulation of yellow vesicles (i.e. autophagosomes) in HDAC6 KO MEFs is efficiently reversed by reintroduction of wild-type, but not catalytically inactive (CD) or ubiquitin-binding-deficient (ΔBUZ) mutant HDAC6 (Figure 2B). These results strongly indicate that HDAC6 is required for efficient autophagosome–lysosome fusion under normal nutrient conditions.
Figure 2.
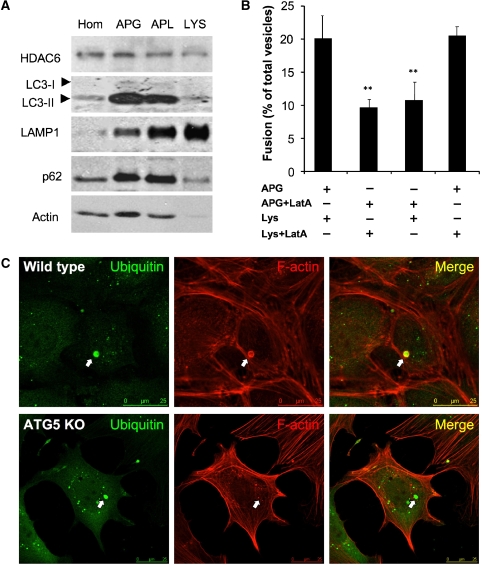
HDAC6 is required for efficient lysosome–autophagosome fusion. (A) Wild-type and HDAC6 KO MEFs were transfected with pcDNA, pcDNA-HDAC6WT, HDAC6CD, or HDAC6ΔBuz, along with mCherry-GFP-LC3 as indicated. Yellow signals indicate non-acidic autophagosomes and red signals indicate acidic autophagolysosomes. Scale bar, 10 μm. (B) The total number of yellow vesicles was quantified from three independent experiments (>12 cells each) and presented as the percentage of total mCherry-GFP-LC3 dots (red plus yellow) along with the standard deviation. (C) In vitro fusion assays. Autophagosomes (APGs) and lysosomes (Lys) purified from wild type and HDAC6−/− MEFs were subjected to heterotypic and homotypic in vitro fusion assays (representative fields are shown in Supplementary Figure S3). Values are means+s.e. of the percentages of fusion from three independent experiments (more than 10 images per each experiment). (D) EM images of wild-type and HDAC6 KO MEFs in normal growth conditions. Yellow arrows, autophagosomes; red arrows, autophagolysosomes; green arrowheads, multilamellar bodies. (E) Quantification of autophagosomes and autophagolysosomes. The error bar represents the standard error.
To gain further evidence that HDAC6 directly regulates autophagosome–lysosome fusion, we purified fractions enriched in autophagosomes or lysosomes from wild-type and HDAC6 KO MEFs and analysed their fusion efficiency in an in vitro assay (H Koga and AM Cuervo, submitted). As shown in Figure 2C and Supplementary Figure S3, lysosomes (Lys) and autophagosomes (APGs) from HDAC6 KO MEFs showed ∼2-fold reduction in fusion as compared with those purified from wild-type MEFs, while homotypic fusion of autophagosomes or lysosomes was not significantly affected. Although the in vitro assay cannot distinguish if fusion defects in the HDAC6 KO MEFs were due to deficiency in docking, tethering, or membrane fusion itself, it provides further evidence that HDAC6 is required for efficient fusion of autophagosomes and lysosomes.
An autophagosome–lysosome fusion deficiency would predict the accumulation of autophagosomes. We therefore analysed the autophagic structures in control and HDAC6 KO MEFs by transmission electron microscopy (EM). In wild-type MEFs, autophagolysosomes were prominent and easily identifiable (Figure 2D, left panel, red arrows, and Supplementary Figure S4). In contrast, very few such structures were observed in HDAC6 KO MEFs (Figure 2D, right panel, and Supplementary Figure S5). Instead, HDAC6-deficient cells accumulated large numbers of double-membrane autophagosome structures (Figure 2D, right panel, yellow arrows), many of which contained multilamellar bodies (MLB, green arrowheads) that were rarely found in wild-type MEFs. The autophagic origin of the vesicular structures that accumulate in HDAC6 KO MEFs was confirmed by immunogold as all them were positive for LC-3 labelling (Supplementary Figure S6). Quantification of autophagic structures confirmed a marked increase (∼2-fold) in the autophagosome/autophagolysosome ratio in HDAC6 KO MEFs (Figure 2E). Thus, fusion assays in vitro and in vivo, and morphometric analysis all support an important role for HDAC6 in the fusion of autophagosomes to lysosomes under basal conditions, a conclusion further confirmed through measurement of autophagic flux analysis and p62 accumulation (Supplementary Figure S7).
HDAC6 is dispensable for non-selective, starvation-induced autophagy
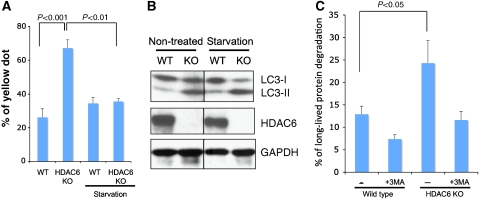
We next determined whether HDAC6 plays a general role in autophagosome–lysosome fusion. Using the mCherry-GFP-LC3 fusion assay we determined whether HDAC6 is required for autophagy induced by starvation. Although HDAC6 KO MEFs were defective in supporting autophagosome–lysosome fusion under normal nutrient conditions (Figure 2), the fusion efficiency was completely normalized and comparable to that of control wild-type MEFs when subjected to starvation (Figure 3A). This fusion defect, however, was not reversed by hypoxia, hydrogen peroxide (H2O2), or mitochondrial complex-I inhibitor (rotenone) treatment, which generated stress that can activate QC autophagy (Supplementary Figure S8). Supporting this observation, starvation-induced LC3-II conversion (Figure 3B) and long-lived protein degradation were normal and even more robust in HDAC6 KO than in wild-type MEFs (Figure 3C). Measurement of LC3 autophagic flux further confirmed normal fusion of autophagosomes to lysosomes in HDAC6 KO MEFs upon starvation (Supplementary Figures S7A and B). Altogether, these results demonstrate that HDAC6 is not required for starvation-induced autophagy.
Figure 3.
HDAC6 is not required for starvation-induced autophagy. (A) Autophagosome–lysosome fusion is analysed in wild-type and HDAC6 KO MEFs with or without starvation (6 h) using mCherry-GFP-LC3 as described in Figure 2A. (B) Wild-type and HDAC6 KO MEFs were cultured in Hank's solution for 3 h followed by immunoblotting with an antibody for LC3, HDAC6, and GAPDH. (C) Long-lived protein degradation in wild-type and HDAC6 KO MEF cells. The degradation of [14C]-valine labelled long-lived protein was measured in the presence or absence of 3-methyl adenine (3MA, inhibits the formation of autophagic vacuoles). The average of percentage degradation from three independent experiments is presented. The error bar represents the standard deviation.
HDAC6 recruits an F-actin network to facilitate autophagosome–lysosome fusion
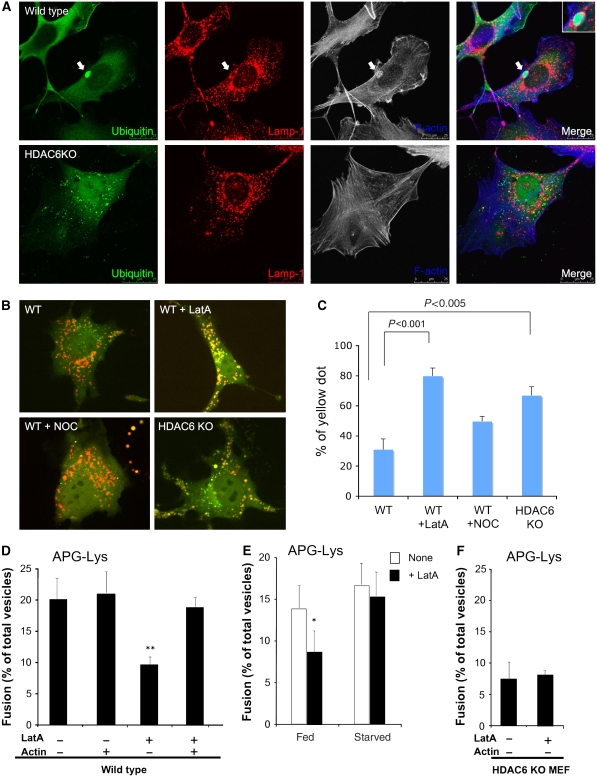
Our analysis showed that HDAC6-deficient autophagosomes and lysosomes remained defective in fusion even when they were mixed in vitro (Figure 2C). This finding suggests that the fusion defect likely involves mechanisms independent of long-range, microtubule-dependent transport, a process known to be regulated by HDAC6. In addition to microtubules, HDAC6 associates with and regulates actin membrane ruffles, a specialized form of the F-actin cytoskeleton (Gao et al, 2007). Interestingly, F-actin remodelling has been reported to promote the fusion of specific vesicular compartments, including those involving lysosomes (Jahraus et al, 2001; Eitzen et al, 2002; Kjeken et al, 2004). These findings prompted us to determine whether the actin cytoskeleton plays a role in QC autophagy associated with protein aggregates. Remarkably, we found that aggresome-like protein aggregates are frequently surrounded by or colocalized with prominent F-actin structures in wild-type MEFs (∼30%; Figure 4A, top panel, and Supplementary Figure S9A). In stark contrast, little F-actin was found around protein aggregates in HDAC6 KO MEFs (Figure 4A, bottom panel). Importantly, formation of the F-actin structure can be efficiently restored by re-introduction of wild-type HDAC6, but not catalytically inactive (CD), or ubiquitin-binding-deficient (ΔBUZ) mutant HDAC6, which is not associated with ubiquitinated protein aggregates (Supplementary Figure S9). These results suggest that HDAC6 promotes the local assembly of an F-actin network at autophagic substrates.
Figure 4.
Actin remodelling is required for autophagosome–lysosome fusion under basal conditions. (A) Wild-type and HDAC6 KO MEFs were treated with MG132 and immunostained with antibodies to Lamp-1 (a lysosome marker, red) and ubiquitin (green) as indicated. F-actin was detected by phalloidin (blue). The arrows indicate ubiquitin-positive aggregates that are surrounded by F-actin and Lamp-1. (B, C) Wild-type and HDAC6 KO MEFs were transfected with mCherry-GFP-LC3, followed by treatment with LatA (100 nM) or nocodazole (250 nM) for 6 h and analysed as described in Figure 3A. (D) Autophagosomes (APGs) and lysosomes (Lys) isolated from fed mouse hepatocytes were treated or not with latrunculin (LatA) as indicated, extensively washed to remove traces of the inhibitor, and then labelled with the antibody and subjected to in vitro fusion assay in the presence or absence of purified actin. The number of total fusion events/total number of vesicles for each condition was as follows: 176/880; 233/1110; 84/930; and 180/950. The differences with untreated samples were significant at **P<0.01. (E) Autophagosomes (APGs) and lysosomes (Lys) isolated from fed or starved mouse hepatocytes were treated or not with latrunculin (LatA) as labelled and subjected to in vitro fusion assay. The differences with untreated samples were significant at *P<0.05. The number of total fusion events/total number of vesicles for each condition was as follows: 169/1250; 87/972; 190/1120; and 175/1165. (F) Autophagosomes (APGs) and lysosomes (Lys) isolated from HDAC6 KO MEFs were treated or not with latrunculin (LatA) as indicated and subjected to in vitro fusion assay.
We next determined whether formation of an F-actin network is required for autophagosome–lysosome fusion. As shown in Figure 4B and C, mCherry-GFP-LC3 reporter assay showed that autophagosome–lysosome fusion was effectively suppressed when cells were treated with latrunculin-A (LatA), which sequesters free G-actin thereby inhibiting F-actin polymerization. Consistent with previous reports (Fass et al, 2006) and our analysis (Figure 2C), treatment with the microtubule-destabilizing agent nocodazole (NOC) has only a mild effect in this assay. To gain further evidence that F-actin polymerization is required for efficient autophagosome–lysosome fusion, we treated the isolated autophagosome (APG) and lysosome (Lys) fractions with latrunculin and assessed the fusion efficiency in vitro. As shown in Figure 4D, latrunculin (LatA) treatment significantly reduced the fusion of purified autophagosomes and lysosomes in vitro. Importantly, after pretreatment of APGs and Lys with latrunculin, the fusion activity can be restored by addition of purified actin (Figure 4D), demonstrating that the effect of latrunculin is actin-dependent.
As HDAC6 is selectively required for fusion associated with QC but not starvation-induced autophagy (Figure 3), we compared the effect of latrunculin on autophagosomes and lysosomes purified under fed or starved conditions. Remarkably, the autophagic compartments purified from cells or mice subject to starvation are much more resistant to LatA than those purified from cells maintained in normal medium or from fed animals (Figure 4E and Supplementary Figures S10A and C). These results indicate that assembly of F-actin is selectively required for QC but not starvation-induced autophagy. Consistent with the proposal that HDAC6 regulates autophagy via promoting F-actin network formation, LatA treatment did not further inhibit the fusion of autophagosomes and lysosomes purified from HDAC6 KO MEFs (Figure 4F and Supplementary Figures S10B and D). Altogether, these findings support the conclusion that HDAC6 promotes autophagosome–lysosome fusion associated with QC autophagy by recruiting an F-actin network to autophagic substrates.
Autophagosomes are associated with actin and required for F-actin network formation at ubiquitinated protein aggregates
Our data indicate that F-actin polymerization assists autophagosome–lysosome fusion (Figure 4). This raises an important question as to the source of actin that feeds into F-actin polymerization. Interestingly, we found that actin is abundantly present in the autophagosomal but almost undetectable in lysosomal fractions (Figure 5A). This spatial distribution suggests a possibility that autophagosome-associated actin might become polymerized by an HDAC6-dependent mechanism to stimulate fusion to lysosomes. Supporting this hypothesis, we found that LatA treatment of purified autophagosomes significantly inhibited autophagosome–lysosome fusion, whereas the same treatment in lysosomes had little effect on fusion (Figure 5B). If autophagosomes were the main sources of actin, one would also predict that autophagosomes would be required to establish the F-actin network at autophagic substrates. Indeed, we found that ATG5 KO MEFs cannot generate an F-actin network at protein aggregates (Figure 5C). Collectively, these data indicate that autophagosomes are associated with actins, which can be assembled into an F-actin network at the autophagic substrates by an HDAC6-dependent mechanism.
Figure 5.
Autophagosomes are required for actin remodelling at protein aggregates. (A) Biochemical characterization of autophagic compartments isolated from HDAC6 KO cells. Different subcellular fractions (75 μg protein) isolated from the wild type (WT) were subjected to SDS–PAGE and immunoblotting for the indicated proteins. Hom, homogenate; APG, autophagosomes; APL, autophagolysosomes; Lys, lysosomes. (B) Autophagosomes (APGs) and lysosomes (Lys) isolated from fed cells were treated or not with latrunculin (LatA) as labelled and subjected to in vitro fusion assay. The differences with untreated samples were significant at **P<0.01. (C) WT and ATG5 KO MEFs were treated with MG132 and immunostained with antibodies to ubiquitin (green) as indicated. F-actin was detected by phalloidin (red). The arrows indicated ubiquitin-positive protein aggregates.
HDAC6 recruits cortactin to assemble an F-actin network essential for autophagosome–lysosome fusion and protein aggregate clearance
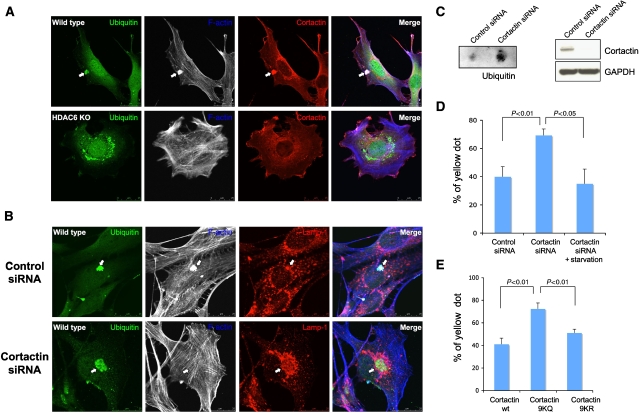
Our results indicate that HDAC6 promotes the formation of an F-actin network at protein aggregates (Figure 4A). Interestingly, cortactin, a key component of the F-actin polymerization machinery, is a substrate of HDAC6 (Zhang et al, 2007). We therefore determined whether cortactin is involved in the HDAC6-dependent F-actin assembly at protein aggregates. We found that cortactin is concentrated at F-actin-positive, aggresome-like protein aggregates in control MEFs (Figure 6A). In contrast, little cortactin was found in HDAC6 KO MEFs, indicating that HDAC6 recruits cortactin to protein aggregates (Figure 6A, bottom panels). Importantly, knockdown of cortactin by siRNA completely prevented the formation of the F-actin network at inclusion bodies, but had little effect on lysosome distribution (Figure 6B, bottom panels). This result demonstrates an essential role of cortactin in the assembly of the F-actin network at protein aggregates.
Figure 6.
Cortactin is recruited to protein aggregates and required for efficient autophagosome–lysosome fusion in homeostatic autophagy. (A) Wild-type and HDAC6 KO MEFs were treated with MG132 and immunostained with antibodies against cortactin (red), ubiquitin (green), and phalloidin for F-actin (blue) as indicated. The arrows indicated ubiquitin-positive aggregates that were colocalized with F-actin and cortactin. (B) Wild-type MEFs were transfected with control or cortactin siRNA, treated with MG132, and stained with antibodies for Lap-1 (red, to label lysosome), or ubiquitin (green) and phalloidin for actin (blue). Note that F-actin staining at protein aggregates was lost, but lysosomes remained concentrated in cortactin knockdown cells (arrow). (C) Wild-type MEFs were transfected with control or cortactin siRNA, treated MG132 2.5 μM for 18 h, and subjected to filter-trap assay using a ubiquitin antibody. The knockdown level of endogenous cortactin was confirmed by immunoblotting using an antibody against cortactin and GAPDH in the right panel. (D) U2OS cells were transfected with control siRNA and cortactin siRNA. Autophagosome–lysosome fusion was analysed with or without starvation (6 h) using the mCherry-GFP-LC3 reporter as described in Figure 3A. (E) The mCherry-GFP-LC3 plasmid was cotransfected with wild type, 9KQ (acetylation-mimic), or 9KR (deacetylation-mimic) cortactin-expressing plasmids into wild-type MEFs. Autophagosome–lysosome fusion was analysed as described in Figure 3A.
Remarkably, the failure to form an F-actin network was accompanied by a prominent defect in protein aggregate clearance (Figure 6C and Supplementary Figure S11) and accumulation of abnormally large aggresomes in cortactin-deficient cells (Figure 6B, bottom panels, and Supplementary Figure S11A). These findings, for the first time, demonstrate that F-actin remodelling is required for autophagy-dependent protein aggregate degradation. Importantly, mCherry-GFP-LC3 reporter assay showed that cortactin knockdown also led to significant inhibition of autophagosome–lysosome fusion under normal but not starvation conditions (Figure 6D). Lastly, to investigate the potential importance of cortactin acetylation, we assessed the effects of wild-type, acetylation-resistant (KR), or acetylation-mimicking (KQ) mutant cortactin on autophagosome–lysosome fusion. As shown in Figure 6E, the expression of the acetylation-mimicking mutant (KQ), but not wild-type or acetylation-resistant mutant (KR), significantly inhibited autophagosome–lysosome fusion, indicating that acetylated cortactin cannot support fusion. Taken together, these results suggest that HDAC6 recruits and deacetylates cortactin, thereby promoting F-actin remodelling important for autophagosome–lysosome fusion and protein aggregate clearance.
HDAC6 deficiency causes ubiquitinated protein aggregate accumulation and neurodegeneration
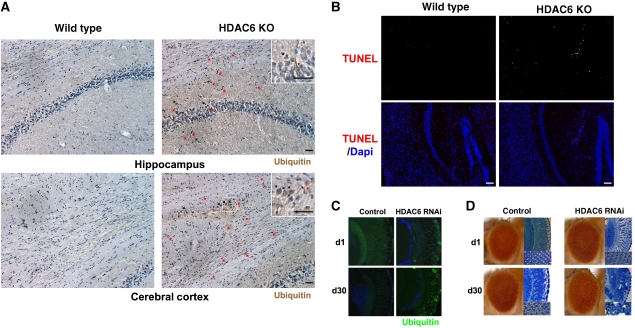
QC autophagy was proposed to remove toxic protein aggregates and protect neurons (Hara et al, 2006; Komatsu et al, 2006). We therefore investigated if there are any neurodegenerative phenotypes in HDAC6-deficient animals. While HDAC6 KO mice are fertile and viable, they developed prominent ubiquitin-positive aggregates in the brain as early as 6 months of age, whereas very few such structures were found in littermate controls (Figure 7A). Importantly, TUNEL assay demonstrated the presence of apoptotic cell death in the brains of HDAC6 KO mice but not in wild-type littermate controls (Figure 7B). Similar neurodegenerative phenotypes were observed in strains of transgenic Drosophila that express an HDAC6 siRNA in photoreceptor neurons (Figure 7C and D). Together, these results demonstrate that HDAC6 deficiency leads to age-dependent neurodegeneration and ubiquitinated protein aggregate accumulation.
Figure 7.
HDAC6 KO mouse and knockdown fly developed spontaneous neurodegeneration and protein aggregate accumulation. (A) HDAC6 KO mice accumulate ubiquitin-positive protein aggregates in the brain. The hippocampus and cerebral cortex regions from 6-month-old wild-type and HDAC6 KO littermates were subjected to immunostaining with a ubiquitin antibody and counterstained with hematoxylin. The red arrows indicate ubiquitin-positive neuritic aggregates and black arrows indicate cytoplasmic aggregates. These ubiquitin-positive structures were rarely observed in control littermates. Scale bar, 50 μm (B) Apoptotic cell death in the cortex and hippocampus region of HDAC6 KO mice as determined by TUNEL staining. Apoptotic cells were only observed in HDAC6 KO mice. Scale bar, 100 μm. (C) HDAC6 depletion in the Drosophila eye leads to ubiquitin-positive pathology. Immunostaining for ubiquitin (green) in frontal eye sections of 1-day-old (d1) and 30-day-old (d30) fly eyes. The eyes of HDAC6-depleted flies (GMR:GAL4/UAS-HDAC6RNAi) developed ubiquitin- positive cytoplasmic inclusions that become more prominent at day 30 (d30). Blue, DAPI. (D) Depletion of HDAC6 in the Drosophila eye leads to age-dependent degeneration. Light micrographs (left) and corresponding Richardson-stained frontal eye sections (right) of 1-day-old and 30-day-old fly eyes. The eyes of control flies (GMR:GAL4/+) and HDAC6-depleted flies (GMR:GAL4/UAS-HDAC6RNAi) show normal highly organized ommatidial array at day 1; 30-day-old control animals also show no defects, but 30-day-old HDAC6-depleted flies show degeneration with disorganization of the ommatidial array and loss of normal eye architecture ( × 40 and × 80).
Discussion
Although autophagy was initially characterized as a non-selective degradation process in response to starvation, nutrient-independent autophagic functions have recently become evident. This so-called ‘basal' autophagy, while not essential for survival, appears to function as an intracellular QC machinery crucial for protecting individuals from devastating disorders, such as neurodegenerative disease. Although the concept of QC autophagy is important for illuminating the complexity of autophagy, the molecular understanding of this process is limited. In this study, we presented evidence that the ubiquitin-binding deacetylase HDAC6 and cortactin-dependent F-actin cytoskeleton are central components that define and distinguish QC from nutrient-regulated autophagy. We further show that this ubiquitin-dependent, actin-remodelling machinery promotes QC autophagy by stimulating the fusion of autophagosomes and lysosomes, thereby uncovering a novel regulation of this poorly understood step in autophagy. These findings provide a molecular framework to understand the distinct nature and function of selective QC autophagy and non-selective nutrient-regulated autophagy, as well as providing new insights into the importance of autophagosome–lysosome fusion events in the development of neurodegenerative diseases.
QC autophagy versus nutrient-regulated autophagy—the role of the actin cytoskeleton
The finding that HDAC6 is required for QC autophagy but dispensable for starvation-induced autophagy clearly shows that these autophagic modes are differentially regulated. HDAC6 has been previously linked to microtubule-dependent transport of protein aggregates and lysosomes to the MTOC (Kawaguchi et al, 2003; Iwata et al, 2005). While this mechanism would concentrate autophagic substrates and lysosomes, it is not sufficient for completion of autophagy (Figure 6B). The current study has identified the actin cytoskeleton as a novel component required for the final step of QC autophagy: fusion of autophagosomes to lysosomes. Our data suggest a model where HDAC6 recruits and activates an actin-remodelling factor, cortactin, to the autophagic substrates, where it stimulates local assembly of an F-actin network (Figure 6B, and see model in Supplementary Figure S13). Supporting this recruitment model, we found that exogenous recombinant HDAC6 did not reverse the fusion defect of autophagosomes and lysosomes purified from HDAC6 KO MEFs (Koga H and Cuervo AM, unpublished data); however, fusion activity was significantly stimulated by addition of cortactin and actin (Supplementary Figure S9B, and Koga H and Cuervo AM, unpublished data). Importantly, cortactin and actin have little effect if autophagosomes and lysosomes were purified from starved cells (Koga H and Cuervo AM, unpublished data), providing further evidence that HDAC6 and actin are not required for starvation-induced autophagy. As F-actin remodelling has been shown to promote fusion of specific vesicular compartments (Jahraus et al, 2001; Kjeken et al, 2004), we propose that the F-actin network recruited by HDAC6 creates a platform where autophagosomes and lysosomes can undergo efficient fusion, leading to autophagic substrate degradation. Supporting this model, preventing F-actin network formation by either cortactin siRNA or latrunculin treatment suppressed autophagosome–lysosome fusion and protein aggregate clearance (Figures 4B and F; 6C and D). These findings indicate that QC autophagy involves both microtubule-dependent transport and actin-dependent fusion. In both steps, HDAC6 appears to play a critical role, although future studies are required to determine its specific involvement in vesicular docking, tethering, or membrane fusion.
We found it remarkable that autophagosome–lysosome fusion proceeded normally under starvation conditions in the absence of HDAC6 or cortactin (Figures 3, 4E and 6D). These findings suggest that autophagosomes induced to degrade protein aggregates or damaged organelles, and those induced to recycle nutrients, are intrinsically distinct in their fusion property. We speculate that starvation leads to the formation of autophagosomes with different membrane properties, which are capable of efficient fusion without the assistance of an HDAC6-dependent actin network. The starvation-induced autophagosomes might also be able to fuse to pre-existing QC autophagosomes, permitting HDAC6-independent autophagosome–lysosome fusion. While the biochemical basis for this differential fusion capacity remains to be fully characterized, it is interesting to note that actin is co-purified with autophagosomes, raising the possibility that actin might be an integral component of autophagosomes involved in QC autophagy (Figure 5A). Consistent with this idea, autophagosomes induced by starvation appear to contain less cortactin and actin than those associated with QC autophagy (Koga H and Cuervo AM, unpublished data). In fact, our data show that autophagosomes are required for the formation of F-actin network at protein aggregates (Figure 5C). Although we do not know if incorporation of actin into autophagosomes is a feature unique to QC autophagy, these findings uncover a novel and important link of the actin cytoskeleton to autophagy.
QC autophagy versus nutrient-regulated autophagy—the ubiquitin connection
The distinct requirement for HDAC6 and actin cytoskeleton may reflect the function unique to different autophagic modes. For example, as nutrient-regulated autophagy serves to replenish macromolecules to sustain cell survival under starvation, it follows that autophagosomes formed under starvation are not endowed with selectivity so that they can efficiently recycle macromolecules by non-discriminatively sequestering cytosolic contents and fusing to lysosomes with great efficiency. In contrast, QC autophagy targets aberrant protein aggregates and damaged organelles, but spares their normal counterparts. Thus, this form of autophagy must be equipped with built-in ‘selectivity'. The specific requirement of a ubiquitin-binding deacetylase, HDAC6, for QC but not starvation-induced autophagy suggests that the substrate selectivity involves ubiquitin modification. Supporting this hypothesis, the ubiquitin-binding-deficient HDAC6-ΔBUZ mutant does not associate with protein aggregates (Supplementary Figure S8B) and cannot support the autophagosome–lysosome fusion (Figure 2A and B). Interestingly, under basal conditions, autophagosomes were reported to contain ubiquitinated proteins (Pankiv et al, 2007). We also found that purified autophagosome fractions are enriched for ubiquitin-positive protein aggregates (Supplementary Figure S12). These results support the idea that ubiquitinated proteins are a major class of substrate for QC autophagy.
Although HDAC6 is required for efficient QC autophagy, it is important to point out that recruitment of autophagosomes to protein aggregates is independent of HDAC6 (Figure 1C). This activity is likely mediated by another ubiquitin-binding protein, p62, which binds to LC3 with high affinity and promotes protein aggregate clearance (Seibenhener et al, 2004; Bjorkoy et al, 2005). Thus, HDAC6 and p62 are both required for protein aggregate clearance, but they regulate different components in autophagy. We propose that HDAC6 and p62 independently recognize and bind to specific ubiquitin moieties that mark protein aggregates or other QC autophagic substrates, where they recruit and assemble the components essential for autophagy: autophagosomes, lysosomes, and the actin network. By concentrating autophagic components to the substrates and stimulating the fusion of autophagosomes with lysosomes, such an arrangement would enable QC autophagy to achieve specific and efficient removal of protein aggregates or damaged organelles that arise sporadically in the cytoplasm (Supplementary Figure S13). This mechanism, however, would provide less advantage to the non-selective bulk degradation induced by starvation, whose substrates are abundant, less geographically constrained, and readily accessible to lysosomes. Interestingly, degradation of defective mitochondria, another key substrate of QC autophagy (mitophagy), was recently shown to be mediated by parkin, a ubiquitin E3-ligase (Narendra et al, 2008). Importantly, HDAC6 and cortactin are also required for parkin-dependent clearance of damaged mitochondria (JY Lee and TP Yao, in preparation). Our results support a central role for ubiquitin modification and HDAC6-regulated F-actin remodelling in establishing QC autophagy.
The regulation of autophagosome–lysosome fusion—its implication in neurodegenerative disease
The importance of autophagy in preventing toxic protein aggregate accumulation and neurodegeneration has been unambiguously established in mice lacking ATG5 or ATG7, both of which are required for autophagy activation (Hara et al, 2006; Komatsu et al, 2006). We found that HDAC6 KO mice and HDAC6-knockdown Drosophila transgenic flies accumulated ubiquitin-positive aggregates and developed spontaneous neurodegeneration (Figure 7). However, HDAC6 deficiency did not affect autophagy activation (Figure 1B); rather, it impaired autophagosome–lysosome fusion (Figure 2). This surprising finding highlights the potential importance of fusion defects in the development of neurodegenerative disease (Figure 7).
In HDAC6 KO MEFs, defects in fusion to lysosomes led to a build-up of autophagosomes. EM analysis revealed that many of these autophagosomes showed abnormal morphology and contents (Figure 2D). Interestingly, similar abnormal autophagic structures were also found prominently accumulated in dystrophic axons in Alzheimer's disease (AD) patients (Nixon et al, 2005) and in a mouse model for frontotemporal dementia (FTD), the second most common form of presenile dementia (Leroy et al, 2007). These similarities suggest an interesting possibility that neurons affected by AD or FTD, and cells deficient in HDAC6, might share a common defect: autophagosome–lysosome fusion. In fact, it has been speculated that abundant autophagic structures observed in AD patients could involve a failure to form functional autophagolysosomes (Nixon et al, 2005). Thus, a defect in lysosome fusion, rather than autophagy activation per se, might be a key factor contributing to the pathogenesis of certain neurodegenerative diseases. Interestingly, it was recently reported that inactivation of components of the ESCRT complex, which regulates late endocytic multiple vesicular bodies (MVB), can lead to autophagosome–lysosome fusion defect, protein aggregate accumulation, and FTD (Filimonenko et al, 2007; Lee et al, 2007). Together, these observations suggest a causative relationship between autophagosome–lysosome fusion defects and development of neurodegenerative disease. If this hypothesis is correct, simply activating the formation of autophagosomes, the step targeted by most of the commonly used autophagy-activating drugs, might not be the most effective therapeutic approach to neurodegenerative disease. Supporting this view, in the Drosophila SBMA model, the neuroprotective effect of rapamycin, which potently activates autophagy, was abrogated in the HDAC6-mutant background (Pandey et al, 2007). Instead, we speculate that agents that stimulate autophagosome–lysosome fusion might provide an alternative and effective way to enhance the degradative capacity of autophagy, thereby protecting the neurons. Thus, the autophagosome–lysosome fusion machinery could be an attractive therapeutic target for developing novel therapies for neurodegenerative diseases.
Materials and methods
Cell lines and plasmids
Wild-type and HDAC6 KO MEFs reconstituted with various HDAC6 constructs were prepared as described previously (Gao et al, 2007). The mCherry-GFP-LC3 construct is a generous gift from Dr Terje Johansen. Lipofectamine LTX (Invitrogen) was used for transfection.
Antibodies and reagents
Anti-mouse HDAC6 antibody was generated against amino acids 991 to 1149. The following antibodies/reagents were also used: anti-HDAC6 (H-300; Santa Cruz Biotechnology), anti-acetyl-α-tubulin (Sigma), anti-ubiquitin (Biomol and Calbiochem), anti-p62 (Santa Cruz Biotechnology), anti-LAMP-1 (Hybridoma Bank, IA, USA), anti-LC3 (a generous gift from Dr Ron R Kopito), LatA (Sigma), NOC (Sigma), and phalloidin–Alexa Fluor 647 and phalloidin–rhodamine (Molecular Probes).
Immunofluorescence microscopy
Immunostaining was performed as described previously (Hubbert et al, 2002; Lee et al, 2004). Cells were cultured on glass coverslips, followed by treatment with MG132 at 2.5 μM for 24 h. Cells were washed with phosphate-buffered saline (PBS), incubated with full growth media for 18 h, and then processed for immunostaining. Images were acquired with a spinning-disk confocal microscope (Olympus ZX-70 or Leica DMI6000C) equipped with an ORCA ER charge-coupled-device camera.
Filter-trap assay
Filter-trap assay for aggregates was performed as described by Scherzinger et al (1997). The total protein load was normalized to the volume of the soluble fraction.
Long-live protein degradation assay
Long-lived protein assay was performed according to the published protocol (Pattingre et al, 2004, and detailed protocol in the Supplementary data).
In vitro fusion assay
Autophagosomes, autophagolysosomes, and lysosomes were isolated from cultured cells using a protocol that was modified from a previous reference (Marzella et al, 1982, and detailed protocol in the Supplementary data).
EM image analysis
The following criteria have been used to assign autophagosome versus autophagolysosomes:
APG (autophagosome): The presence of double membrane is the defining component. The density inside the autophagosomes is similar to that of the surrounding cytosol.
APGL (autophagolysosome): Vesicle with single membrane (sometimes there is residual double membrane, but very little) and containing amorphous materials; the lumen of the vesicle is usually of lesser density than the surrounding cytosol (a detailed protocol for immunogold staining can be found in the Supplementary data).
Fly stocks
All Drosophila stocks were maintained at 25°C on standard yeast agar media except where indicated. The UAS-dHDAC6 and UAS-dHADC6RNAi strains were described previously (Pandey et al, 2007). The GMR-GAL4 strain was obtained from the Bloomington Stock Center (Bloomington, IN, USA). For each genotype and condition, at least 100–300 flies were evaluated.
Immunohistochemical analysis of mouse sections
Mice were transcardially perfused with 4% paraformaldehyde in phosphate buffer (pH 7.4). Brains were post-fixed in the same fixative for 3 h in 4°C, incubated with 30% sucrose in 4°C for overnight, and embedded in O.C.T. compound. For immunohistochemical analysis, frozen brain tissue sections (20 μm) were subjected to immunostaining with anti-ubiquitin antibody followed by the Vectastain ABC kit (Vector Laboratory). After immunostaining, the sections were counterstained using Meyer's hematoxylin. Apoptotic neurons were visualized by TUNEL assay using an in Situ Cell Death Detection kit (TMR red; Roche).
Statistical analysis
Two-tailed Student's t-test was conducted for statistic analysis of quantitative data.
Supplementary Material
Acknowledgments
We thank Drs T Johansen, N Mizushima, Y He, RR Kopito, E Seto, and M Zhang for sharing reagents and Dr H Hasagawa for technical advice. We are grateful to Drs B Harvat, D Thiele, C Nicchita, ZM Pei, XN Dong, and AR Means for critically reading the paper. This work is supported by NS053825 (NIH) and Muscular Disease Association to JPT, AG031782-01(NIH) to AMC, and NS054022 (NIH) to TPY.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen G, Wang L, Thorngren N, Wickner W (2002) Remodeling of organelle-bound actin is required for yeast vacuole fusion. J Cell Biol 158: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z (2006) Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 281: 36303–36316 [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179: 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP (2007) Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol 27: 8637–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280: 40282–40292 [DOI] [PubMed] [Google Scholar]
- Jahraus A, Egeberg M, Hinner B, Habermann A, Sackman E, Pralle A, Faulstich H, Rybin V, Defacque H, Griffiths G (2001) ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol Biol Cell 12: 155–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kjeken R, Egeberg M, Habermann A, Kuehnel M, Peyron P, Floetenmeyer M, Walther P, Jahraus A, Defacque H, Kuznetsov SA, Griffiths G (2004) Fusion between phagosomes, early and late endosomes: a role for actin in fusion between late, but not early endocytic organelles. Mol Biol Cell 15: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530 [DOI] [PubMed] [Google Scholar]
- Kuma A, Matsui M, Mizushima N (2007) LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3: 323–328 [DOI] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB (2007) ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 17: 1561–1567 [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim H, Ryu CH, Kim JY, Choi BH, Lim Y, Huh PW, Kim YH, Lee KH, Jun TY, Rha HK, Kang JK, Choi CR (2004) Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem 279: 30265–30273 [DOI] [PubMed] [Google Scholar]
- Leroy K, Bretteville A, Schindowski K, Gilissen E, Authelet M, De Decker R, Yilmaz Z, Buee L, Brion JP (2007) Early axonopathy preceding neurofibrillary tangles in mutant tau transgenic mice. Am J Pathol 171: 976–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H (1982) Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol 93: 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 64: 113–122 [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447: 859–863 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Petiot A, Codogno P (2004) Analyses of Galpha-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy. Methods Enzymol 390: 17–31 [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549–558 [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24: 8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol 21: 8035–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, Yao TP, Lane WS, Seto E (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell 27: 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.