Abstract
The sequential action of five distinct endosomal-sorting complex required for transport (ESCRT) complexes is required for the lysosomal downregulation of cell surface receptors through the multivesicular body (MVB) pathway. On endosomes, the assembly of ESCRT-III is a highly ordered process. We show that the length of ESCRT-III (Snf7) oligomers controls the size of MVB vesicles and addresses how ESCRT-II regulates ESCRT-III assembly. The first step of ESCRT-III assembly is mediated by Vps20, which nucleates Snf7/Vps32 oligomerization, and serves as the link to ESCRT-II. The ESCRT-II subunit Vps25 induces an essential conformational switch that converts inactive monomeric Vps20 into the active nucleator for Snf7 oligomerization. Each ESCRT-II complex contains two Vps25 molecules (arms) that generate a characteristic Y-shaped structure. Mutant ‘one-armed' ESCRT-II complexes with a single Vps25 arm are sufficient to nucleate Snf7 oligomerization. However, these oligomers cannot execute ESCRT-III function. Both Vps25 arms provide essential geometry for the assembly of a functional ESCRT-III complex. We propose that ESCRT-II serves as a scaffold that nucleates the assembly of two Snf7 oligomers, which together are required for cargo sequestration and vesicle formation during MVB sorting.
Keywords: endosomes, ESCRT, multivesicular bodies (MVB), Snf7, Vps25
Introduction
The evolutionary conserved endosomal-sorting complex required for transport (ESCRT) complexes generate multivesicular bodies (MVBs), assist viral budding and promote the final membrane scission step at the end of cytokinesis. These unrelated processes all require a topologically similar membrane fission event that is distinct from the membrane/vesicle fission associated with the formation of endocytic and secretory vesicles, which bud into the cytoplasm. ESCRT-mediated processes function in a wide variety of cellular and developmental processes and their dysfunction contributes to diseases ranging from cancer to neuro-degeneration (Raiborg and Stenmark, 2009; Saksena and Emr, 2009).
The core of the ESCRT machinery consists of a subset of the vacuolar protein-sorting gene products (VPS genes) that were first identified in yeast. A total of 17 VPS gene products assemble into five distinct sub-complexes: ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III and the Vps4 complex. These complexes are transiently recruited from the cytoplasm to endosomes. Their sequential action directs the sorting of ubiquitinated transmembrane proteins (cargo) and the inward budding of MVB vesicles into the lumen of the endosome, thereby generating MVB vesicles that are filled with cargo. Fusion of MVBs with the lysosome delivers the cargo-laden MVB vesicles into lysosomes, where they are finally degraded. ESCRT-0, I and II function early in the pathway and are recruited to endosomes as stable heteromeric complexes. Their distinct ubiquitin interacting modules bind to, collect and concentrate ubiquitinated transmembrane proteins (Hurley and Emr, 2006; Williams and Urbe, 2007). In contrast, ESCRT-III subunits are inactive monomers in the cytoplasm. ESCRT-III consists of four-core subunits: Vps20, Snf7, Vps24 and Vps2 (Babst et al, 2002a). On endosomes, they assemble in a highly ordered manner to generate the transient ESCRT-III complex. Vps20 nucleates the oligomerization of Snf7. Approximately 10–20 Snf7 molecules could form a ‘ring-like' filament that is capped by Vps24 (Teis et al, 2008). The capping of Snf7 filaments by Vps24 recruits Vps2, which in turn recruits the Vps4 AAA–ATPase complex to disassemble ESCRT-III before MVB vesicle formation. Genetic and biochemical experiments suggest that ESCRT-II functions directly upstream of ESCRT-III during MVB sorting and is required for the recruitment and assembly of the ESCRT-III complex on endosomes (Babst et al, 2002b). Moreover, the interaction of Vps25 with Vps20 was found to induce conformational re-arrangements in Vps20 on membranes in vitro (Teo et al, 2004; Saksena et al, 2009). As Vps20 is required to nucleate Snf7 oligomerization, the interaction of Vps25 with Vps20 is likely to be a key step that regulates the ordered assembly of ESCRT-III.
From yeast to human beings, ESCRT-II forms a trilobal complex and consists of Vps36, Vps22 and two Vps25 molecules (Babst et al, 2002b; Hierro et al, 2004; Teo et al, 2004; Im and Hurley, 2008). Vps36 contains a GLUE domain, a ‘hub' that mediates interaction with ESCRT-I (Vps28) (Teo et al, 2006) and binds to PI(3)P (phosphatidyl-inositol-3-phosphate) on endosomes and to ubiquitinated cargo (Alam et al, 2004, 2006; Slagsvold et al, 2005; Hirano et al, 2006). The GLUE domain is connected through a long flexible linker to the core of ESCRT-II. A tight interaction of Vps36 and Vp22 provides the scaffold for the assembly of two Vps25 molecules. One Vps25 binds to Vps36 and the other binds to Vps22 and thereby generates the characteristic Y-shape of the ESCRT-II complex. Despite our knowledge of the molecular architecture of ESCRT-II and the emerging assembly reaction of the transient ESCRT-III complex, the molecular mechanism for the ESCRT-II-dependent assembly of ESCRT-III is not understood.
Here, we show that the ESCRT-II subunit Vps25 induces a conformational change in Vps20. This conformational switch activates (‘opens') Vps20, which is essential to nucleate Snf7 filaments. A mutant ESCRT-II complex with a single Vps25 molecule is sufficient for the formation of Snf7 oligomers, but not for the formation of a functional ESCRT-III complex that can sequester cargo and form MVB vesicles. Only the Y-shaped ESCRT-II complex with two Vps25 molecules (arms) provides the required geometry to induce the assembly of a functional ESCRT-III complex. It seems that during MVB sorting, ESCRT-II could simultaneously nucleate at least two Snf7 oligomers, which together execute the last steps of MVB sorting, cargo sequestration and MVB vesicle formation.
Results
ESCRT-III assembly affects MVB vesicle size
In vivo and in vitro experiments have shown that the ordered assembly of the ESCRT-III complex constitutes the machinery that executes the last steps of MVB sorting, cargo sequestration and MVB vesicle formation (Teis et al, 2008; Saksena et al, 2009; Wollert et al, 2009). It seems probable that the molecular architecture of ESCRT-III determines how MVB vesicles are formed. The first indication came from over-expression of hSnf7/hVps32 (Chmp4), which assembled into large polymeric circular arrays that deformed the plasma membrane away from the cytoplasm (Hanson et al, 2008). Larger ESCRT-III complexes could possibly alter the size and morphology of MVB vesicles. To directly test this idea, we modified the architecture of ESCRT-III by over-expression of Snf7/Vps32, because Snf7 seems to be key for the formation of MVB vesicles in vivo and in vitro. (Babst et al, 2002a; Hanson et al, 2008; Wollert et al, 2009). Over-expression of Snf7 (∼20-fold, Supplementary Figure 1A) resulted in the accumulation of larger and heterogeneous Snf7 oligomers on endosomes (Supplementary Figure 1B), which caused a delay, but not a block, in MVB sorting (Supplementary Figure 1C). In wild-type (WT) cells, addition of Methionine resulted in the efficient transport of Mup1-GFP from the plasma membrane into vacuole through the MVB pathway. Interestingly, in cells that over-express Snf7, the transport of Mup1-GFP to the vacuole was delayed. Mup1-GFP first accumulated on endosomes and was then transported into the vacuole (Supplementary Figure 1C; Teis et al, 2008). To address how ESCRT-III with larger Snf7 oligomers would alter MVB vesicle morphology, Snf7 was over-expressed in vma4Δ mutant cells. These cells cannot assemble the V-ATPase complex and thus fail to acidify their vacuoles (Morano and Klionsky, 1994). As vacuolar hydrolases are less active in vma4Δ mutants, the contents of their vacuoles are less efficiently degraded. Unlike WT yeast cells, which have electron dense vacuoles when examined by transmission electron microscopy, the vacuoles of vma4Δ mutants are more transparent and MVB vesicles (15–76 MVB vesicles/vacuole/section, mean=37±15, n=30 vacuoles) can be detected in the lumen (Wurmser and Emr, 1998) (Figure 1A and B). The average vesicle diameter was 34±11 nm (n=504 vesicles, from 10 vacuoles) (Supplementary Figure 1D and E), which is consistent with earlier reports, suggesting an average vesicle diameter of 20–40 nm (Wurmser and Emr, 1998; Nickerson et al, 2006; Richter et al, 2007). In contrast, in vma4Δ mutants that over-express Snf7, fewer (2–15 vesicles/vacuole/section, mean=7±3, n=30 vacuoles) and more heterogeneous vesicles with a single limiting membrane were detected (Figure 1A and B). The average vesicle diameter was 75±38 nm (n=315 vesicles from 30 vacuoles) (Supplementary Figure 1D and E). To directly compare the size of MVB vesicle from vma4Δ mutants with vma4Δ mutants that over-express Snf7, the size distribution of 300 vesicles was plotted in 10 nm steps (Figure 1C). In vma4Δ mutants, there was a clear preference for the formation of relatively uniform vesicles with diameters ranging from 20 to 50 nm. In contrast, in the vma4Δ mutants that over-express Snf7, this preference was lost. The smallest vesicle had a diameter of 20 nm and the largest 360 nm. In between, the size of the vesicle diameters was evenly distributed (Figure 1C). These findings suggest that larger and more heterogeneous Snf7 oligomers on endosomes (Supplementary Figure 1B) induced the formation of fewer, but larger and more heterogeneous, MVB vesicles. It seems that Vps24-dependent capping is lagging behind Snf7 oligomerization, which results in the assembly of larger Snf7 filaments. Yet, Vps24 capping is a prerequisite for MVB vesicle formation (vma4Δ vps24Δ, Table in Figure 1A). In addition, loss of ESCRT-II function, which is required for the assembly of ESCRT-III (vma4Δ vps25Δ, Figure 1A lower panel or vma4Δ vps36Δ, Table in Figure 1A), blocked MVB vesicle formation. No MVB vesicles were detected in 80% of all vacuoles (24 out 30 vacuoles). In 6 out of 30 vacuoles (20%), one abnormal vesicle was detected (Figure 1B; Table in Figure 1A). Taken together, these in vivo observations seem relevant for the mechanism of ESCRT-III-mediated MVB vesicle formation. During MVB sorting, ∼20 Snf7 molecules, with a length of 70 Å each (Muziol et al, 2006) would be sufficient to delineate a circular 50 nm-sorting domain that could result in the formation of MVB vesicle with a diameter of 25 nm. It follows that longer Snf7 filaments may delineate a larger MVB-sorting domain and hence generate larger MVB vesicles. Taken together, these data strongly suggest that the function of the ESCRT-III complex is controlled by the stoichiometry of its components. In particular, the length of Snf7 filaments seems to determine the size of MVB vesicles in vivo. Given the uniform size of MVB vesicles, the assembly of ESCRT-III on endosomes should be tightly controlled. This raises the important question, how is the assembly of a functional ESCRT-III complex regulated?
Figure 1.
ESCRT-III regulates the size of MVB vesicles. (A) Transmission electron microscopy of vma4Δ, vma4Δ (THD3)SNF7 and vma4Δ vps25Δ cells. Over-expression of Snf7 (vma4Δ (THD3)SNF7) resulted in the appearance of fewer, larger and more heterogeneous luminal vesicles inside the vacuole of vma4Δ mutant cells. vma4Δ vps25Δ mutants grew very slowly. Representative cells and magnifications are shown. Size bars (500 nm and 100 nm in the magnification). The table shows the range of the number of vesicles detected per vacuole/70 nm section in different ESCRT mutants. (n=30 vacuoles). (B) Average number of MVB vesicles/vacuole/70 nm section (n=30 vacuoles for each sample). vma4Δ mutants have on average 37±15 vesicles, vma4Δ vps25Δ have 0.3±0.5 vesicles and vma4Δ(THD3)SNF7 have 7±3 vesicles/vacuole/section. (C) Distribution of the vesicle diameter in 10 nm step (vma4Δ: n=300, vma4Δ(THD3)SNF7: n=300). In vma4Δ mutants, most vesicle diameters range from 20 to 45 nm (black line). In vma4Δ(THD3)SNF7 mutants, the smallest vesicle is 20 nm and the largest vesicle is 360 nm. In between, the vesicle diameters are evenly distributed.
ESCRT-II is required for Snf7 nucleation (oligomerization), but not Vps20 localization
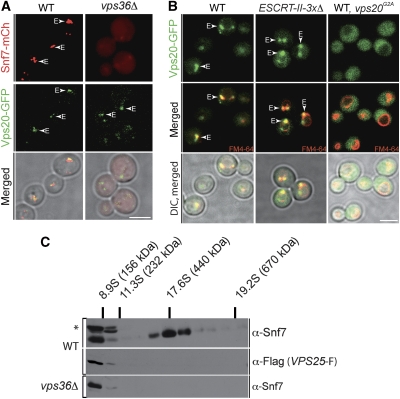
Earlier reports suggested that ESCRT-II is required for the assembly of ESCRT-III (Babst et al, 2002b). To begin to understand the underlying molecular mechanism, we first tested whether ESCRT-II was essential for the recruitment of different ESCRT-III subunits to endosomes. Therefore, we used C-terminal GFP fusions (or mCherry) of ESCRT-III molecules that were generated by chromosomal integration. The C-terminal GFP fusions exhibited a dominant negative phenotype and accumulated on aberrant endosomes (class E compartments), which allows easy identification of regulators of membrane recruitment (Teis et al, 2008). Deletion of Vps4 had no effect on the localization of Vps20-GFP or Snf7-mCherry (or Snf7-GFP) to the class E compartment (data not shown). Snf7-mCherry accumulated in large structures on endosomes. Vps20-GFP was found in the cytoplasm and on endosomes where it decorated the larger Snf7-mCherry structures (Figure 2A). Snf7-mCherry was mis-localized into the cytoplasm in vps36Δ (ESCRT-II) mutants (and vps22Δ, vps25Δ data not shown); yet in the same cells, the endosomal localization of Vps20-GFP was not affected (Figure 2A, second panel).
Figure 2.
ESCRT-II is required for Snf7 nucleation. (A) Live cell microscopy of cells harbouring chromosomal integrations for VPS20-GFP and SNF7-mCherry. Size bar (5 μm). Vps20-GFP and Snf7-mCherry co-localize on endosomes. In a vps36Δ mutant Vps20-GFP localizes to endosomes and Snf7-mCherry mis-localized into the cytoplasm. (B) Vps20-GFP localizes to endosomes independently of ESCRT-II (vps36Δ, vps22Δ, vps25Δ triple mutant). Vps20G2A-GFP did not localize to endosomes in WT cells. FM4-64 is shown in red. Size bar (5 μm). (C) Spheroplasts of WT cells harbouring a chromosomal integration of VPS25-Flag or vps36Δ mutants were cross-linked with 5 μm DSP. Solubilized membrane fractions (P13) were subjected to velocity sedimentation and analysed by SDS–PAGE and western blot with the indicated antibodies. *Background band.
Although the ESCRT-II subunit Vps25 interacted directly with Vps20, but not with Snf7 (Teo et al, 2004; Saksena et al, 2009), ESCRT-II was essential to recruit Snf7-GFP to endosomes. Yet, the localization of Vps20 to endosomes is independent of ESCRT-II. A triple deletion of all ESCRT-II subunits (vps22Δ, vps25Δ, vps36Δ) did not affect the targeting of Vps20-GFP to membranes co-labelled with FM4-64 (Figure 2B). In contrast, mutation of the conserved N-terminal myristoylation site G2A (Babst et al, 2002a; Yorikawa et al, 2005) resulted in mis-localization of Vps20G2A-GFP into the cytoplasm (Figure 2B) of WT cells, suggesting that the N-terminal myristoylation of Vps20 is crucial for endosomal targeting. Thus, in vivo the N-terminal myristoylation and not ESCRT-II first directs Vps20 to endosomes. On endosomes, Vps20 interacts with ESCRT-II. However, without ESCRT-II, Vps20 fails to recruit Snf7 to endosomes.
In vitro, Vps25 induces conformational re-arrangement(s) in Vps20 on liposomes (Saksena et al, 2009). Hence, the interaction with Vps25 seems to be required for the activation of Vps20 before the nucleation of Snf7 oligomers. To directly test this idea, we determined the molecular weight of cross-linked ESCRT-III in ESCRT-II mutant cells using velocity sedimentation centrifugation. In WT cells, expressing Vps25-Flag, endosomal Snf7 cross-linked into a 450-kDa complex and did not contain detectable amounts of Vps25-F (Figure 2C). In ESCRT-II mutants (vps36Δ (Figure 2C), vps22Δ and vps25Δ data not shown), Snf7 did not form the 450 kDa complex, suggesting that ESCRT-II is required for the nucleation of Snf7 oligomers. Taken together, these findings strongly suggest that transient interactions with ESCRT-II (Vps25) activate Vps20, which then nucleates Snf7 oligomerization and thus initiates ESCRT-III assembly.
Vps20 activation by Vps25 (ESCRT-II) is required for ESCRT-III assembly
To better understand the molecular mechanism of ESCRT-II-dependent Vps20 activation, we next used mutational analysis of Vps20 and Vps25. Expression of the long N-terminal α1/α2 hairpin (aa 1–116, Vps20H1,2) of Vps20 caused a strong MVB-sorting defect (class E compartment, data not shown). Yet, the N-terminal α1/α2 hairpin of Vps20 was sufficient to recruit Snf7-GFP to endosomes (Figure 3A). However, the deletion of the N-terminal helix α2 (Δ55–116 aa, Vps20ΔH2) resulted in cytoplasmic mis-localization of Snf7-GFP (Figure 3A). Vps20ΔH2 is properly expressed and localized to endosomes (data not shown), but cannot nucleate Snf7 oligomerization as shown by velocity sedimentation centrifugation (Figure 3B, lower panel). Recently, the molecular details of the Vps25 and Vps20 interface have been characterized (Im et al, 2009). Vps25 binds specifically to helix α1 of Vps20, suggesting that this interaction could induce Vps20 activation and subsequent Snf7 oligomerization. To directly test this idea in vivo, we mutated critical residues in Vps25 (T150K) that were shown to be required for the interaction with Vps20 (Im et al, 2009). Importantly, the Vps25T150K mutant failed to activate Vps20, which caused the mis-localization of Snf7-GFP into the cytoplasm (Figure 3A, lower panel).
Figure 3.
Vps20 activation by Vps25 is required for ESCRT-III assembly. (A) Live cell microscopy of vps20Δ, SNF7-GFP cells expressing either VPS20, vps20H1,2, vps20ΔH2 or vps25ΔSNF7-GFP cells expressing vps25T150K. FM4-64 is shown in red. Size bar (5 μm). (B) Spheroplasts of WT or vps20Δ mutants expressing the indicated Vps20 mutants were cross-linked with 5 μM DSP. Solubilized membrane fractions (P13) were subjected to velocity sedimentation and analysed by SDS–PAGE and western blot with the indicated antibodies. (C) Cartoon representation of Chmp3 hVps24 (PDB: 3FRT, (Bajorek et al, 2009)). Indicated are the loop region and helix α1,α2 and auto-inhibitory helix α5, and K61 of Vps20. The auto-inhibitory helix α5 was proposed to move away from the loop region, thereby releasing auto-inhibition. (D) The cytoplasm of cells expressing VPS20-Flag, VPS25-Flag or vps20Δ, vps20loop-HA or vps20Δ, vps25Δ, vps20loop-HA was subjected to size exclusion chromatography on an S200 column. Two fractions were pooled and analysed by SDS–PAGE and western blot with the indicated antibodies. (E) Fluorescence emission spectra of Vps2061−NBD (red trace) and Vps20loop61−NBD (blue trace). (F) In vitro oligomerization experiment. Liposomes were mixed with purified H6-Snf7 alone, or ESCRT-II (E-II), H6-Vps20G2A and H6-Snf7 or ESCRT-II (E-II), H6-Vps20loop and H6-Snf7 (20-fold excess) and incubated for 30 min. The proteins bound to liposomes were solubilized and subjected to velocity sedimentation and analysed by SDS–PAGE and western blot with the indicated antibodies.
Together, these findings emphasize that Vps25 binding to helix α1 of Vps20 is required for activation, whereas helix α2 of Vps20 is essential for the subsequent nucleation of Snf7 oligomers (Figure 3C). Helix α2 of different ESCRT-III proteins has been proposed to mediate the interaction between ESCRT-III molecules. Importantly, in each of the ESCRT-III proteins, helix α2 is auto-inhibited by helices α5/α6, which prevents premature assembly of ESCRT-III (Shim et al, 2006; Zamborlini et al, 2006; Bajorek et al, 2009; Saksena et al, 2009). The loop region that connects helix α1 with helix α2 has also been found to provide a crucial function. It holds the two helices α1/α2 in place and seems to position them for proper function. Furthermore, this α1/α2 loop region was proposed to mediate contacts between different ESCRT-III subunits (Muziol et al, 2006) and/or intra-molecular contacts with the auto-inhibitory domain of α5/α6 (Bajorek et al, 2009). We reasoned that any reduction in the length of the α1/α2 loop would compromise the ability of the loop to hold the two N-terminal helices in place and would generate an ESCRT-III molecule with dramatic alterations in its three-dimensional folding and structure. If the intra-molecular interactions between α1 and α2 were critical for maintaining auto-inhibition, then physical displacement of α1 and α2 (by reduction in the length of the α1/α2 loop) would generate a more-‘active' form of the ESCRT-III molecule and thereby result in impaired MVB-sorting function.
To directly test these hypotheses, we generated a Vps20 mutant (hereafter referred to as the Vps20loop mutant) in which we deleted residues 48–59 that lie within the α1/α2 loop (Figure 3C). As predicted, cells expressing the Vps20loop mutant exhibited a strong MVB-sorting defect (Figure 4B), but the localization of Snf7-GFP to endosomes was not affected (data not shown). Furthermore, the Vps20loop mutant induced the assembly of slightly larger Snf7 complexes (>500 kDa), as shown by velocity sedimentation centrifugation (Figure 3B). This suggested that the Vps20loop mutant represented a more-active Vps20 molecule that may overcome auto-inhibition more rapidly than the WT protein. Normally, Vps20 does not interact with ESCRT-II in the cytoplasm as shown by size exclusion chromatography. Vps20 elutes at around 44 kDa, whereas Vps25 elutes together with ESCRT-II at around 150 kDa (Figure 3D) and at a second peak around 30 kDa (data not shown). Interestingly, the Vps20loop mutant eluted at a molecular weight similar to that of ESCRT-II, which was entirely dependent on Vps25 (Figure 3D). It seemed that the deletion in the α1/α2 loop generated a more-‘active' form of Vps20, thereby resulting in the premature association of the Vps20 α1/α2 loop mutant with ESCRT-II in the cytoplasm. To directly show that the Vps20loop mutant has a different conformation in solution than the ‘auto-inhibited' WT Vps20, we labelled recombinant Vps20 and the Vps20loop mutant with the ‘environmentally sensitive' 7-nitrobenz-2-oxa-1,3-diazole (NBD) dye. The fact that Vps20 is a cysteine-free protein allowed us to site specifically label the WT and mutant form of Vps20 at a single site by introducing a cysteine residue, and then labelling the cysteine-containing protein with the thiol-reactive NBD. We chose to label Vps20 and the Vps20loop mutant at residue 61 because of its proximity to the α1/α2 loop and because we have earlier shown that residue 61 serves as a good reporter for the activation status of Vps20 (Saksena et al, 2009) (Figure 3C). NBD is environmentally sensitive, and its spectral properties are dramatically different in aqueous and non-aqueous environments (Johnson, 2005; Shepard et al, 1998). When NBD moves from an aqueous milieu (such as the surface of a soluble protein) to a hydrophobic environment (such as the non-polar interior of a membrane or protein), its emission intensity and fluorescence life time (τ) increase, whereas its wavelength of maximum emission (λem max) shifts to the blue (referred to as ‘blue shift'). Thus, monitoring the NBD emission intensity of two proteins labelled at the same site allows one to make conclusions regarding the environment of the NBD dye at that particular site within the two proteins (aqueous versus hydrophobic), and in doing so, allows one to determine whether the two proteins have different conformations around that particular site.
Figure 4.
The Vps25–Vps20 nucleation complex controls Snf7 oligomerization during MVB sorting. (A, B) Representative images of cells expressing Mup1-GFP (green). Cells were treated for 60 min with methionine and FM4-64 (red). In snf7Δ vps20Δ or vps20Δ vps20PW cells, Mup1-GFP accumulated on the E-compartment (arrowheads) and was detected at the PM (arrows). In vps24Δ, vps20Δ vps20loop and snf7Δ snx41Δ cells Mup1-GFP was detected only on the class E compartment. Size bar (5 μm). (C) Quantification of the fluorescence ratio (FR) of Mup1-GFP at the PM and on the endosomes in different Vps20 mutants. High FR indicates the accumulation of Mup1-GFP on the E-compartment; n>25 cells for each mutant. (D) Canavanine sensitivity assay with WT cells and the indicated mutants. vps20Δ and snf7Δ were sensitive to 0.6 μg/ml canavanine. vps24Δ, vps2Δ and snf7Δ snx41Δ were more resistant to 0.6 μg/ml canavanine.
As shown in Figure 3E, the NBD emission intensity of Vps2061−NBD (indicated by the red trace) was five-fold higher than the emission intensity of Vps20loop61−NBD (blue trace) mutant labelled with NBD at the same site. Furthermore, the λem max of the Vps20loop61−NBD mutant was blue shifted compared with Vps2061−NBD. This indicated that the NBD dye at residue 61 in the Vps20loop mutant is in a more aqueous environment compared with the NBD dye at residue 61 in wt Vps20 and thus supports the concept that mutations in the α1/α2 loop region generate a mutant Vps20 molecule that might be more active.
To address the effect of the Vps20loop mutant on Snf7 oligomerization more directly, we used in vitro ESCRT-III assembly reactions that we have earlier characterized (Saksena et al, 2009). Conditions were chosen under which purified Snf7 did not efficiently oligomerize on liposomes in the absence of its nucleator Vps20, as revealed by velocity sedimentation centrifugation of the liposome-bound proteins (Figure 3F). Upon addition of recombinant ESCRT-II and Vps20, Snf7 (present in 20-fold excess) formed higher molecular weight oligomers on liposomes. Importantly, the Vps20loop mutant induced Snf7 oligomerization more efficiently, which was readily detected because the Vps24 cap and the Vps4 complex were omitted to avoid disassembly in these assay conditions (Figure 3F).
Taken together, these findings imply that the first step in the ordered assembly of ESCRT-III is mediated by the transient binding of Vps25 to Vps20. Binding of Vps25 to Helix α1 of Vps20 could induce a conformational change in the loop region between α1/α2 that would displace the auto-inhibitory loop α5/α6. Consistent with this, the mutant Vps20PW(P183W, P189W and P192W), which fails to displace the auto-inhibitory loop, cannot be activated and hence does not nucleate Snf7 oligomerization (Saksena et al, 2009). Thus, Vps25 would facilitate the release of auto-inhibition and convert Vps20 into an active nucleator, a step required to induce Snf7 oligomerization on endosomes. Our findings suggest a mechanism that helps to explain how ESCRT-II provides necessary spatial and temporal regulation for the assembly of ESCRT-III in close proximity to cargo molecules.
The Vps25–Vps20 nucleation complex controls Snf7 oligomerization during MVB sorting
To test the functional significance of the Vps25-dependent activation of Vps20, we used two different in vivo assays that directly measure the ESCRT-dependent vacuolar degradation of two different cell surface receptors, the methionine transporter Mup1 and the arginine transporter Can1.
First, we used quantitative live cell microscopy to examine the effect of the two different Vps20 mutants on the membrane trafficking of the methionine permease, Mup1. Mup1-GFP localized to the plasma membrane and was efficiently degraded through the MVB pathway, 60 min after addition of methionine (Teis et al, 2008). We have shown that loss of Snf7 oligomerization (e.g. vps20Δ or snf7Δ) (Figure 4A and B) resulted in recycling of Mup1 from aberrant endosomes back to the plasma membrane. If Snf7 oligomerization is not capped (e.g. vps24Δ), Mup1 is trapped on endosomes (Teis et al, 2008) (Figure 4A). However, in snf7Δ vps24Δ double mutants, Mup1-GFP was no longer trapped at the class E compartment, suggesting that the formation of Snf7 filaments is essential for cargo trapping (Supplementary Figure 2A and B). Importantly, loss of Snx41, a sorting nexin required for recycling cargo to the plasma membrane (Hettema et al, 2003), blocked the recycling of Mup1-GFP from the class E compartment in an snf7Δ mutant (Figure 4A). As the analysis of Mup1-GFP (as well as CPS and Can1 (see Supplementary data and Supplementary Figures 2–4) traffic can distinguish different functional states of ESCRT-III assembly/disassembly, we next compared two functionally different Vps20 mutants, Vps20loop and Vps20PW(P183W, P189W and P192W). Vps20loop resulted in enhanced Snf7 oligomerization (Figure 3B and F). In contrast, Vps20PW restricted conformational re-arrangements of the auto-inhibitory loop in Vps20 (helices α5/α6) and locked the closed conformation, which did not nucleate Snf7 oligomerization (Saksena et al, 2009). Interestingly, in cells expressing the Vps20loop mutant, Mup1-GFP was trapped on endosomes and very little Mup1-GFP recycled back to the cell surface. In contrast, in cells expressing the Vps20PWmutant, Mup1 was not trapped on endosomes and recycled back to the plasma membrane (Figure 4B). Quantification of the fluorescence intensities (IF) on the class E compartment and at the plasma membrane allowed the calculation of a fluorescence ratio (FR) (IF at the class E compartment divided by the IF at the plasma membrane). A high FR represents predominant localization of Mup1-GFP at the class E compartment. The Vps20PW mutant has a low FR, which is comparable with vps20Δ or snf7Δ mutants (Figures 4C and 6B). In contrast, the Vps20loop mutant has a high FR, comparable with vps24Δ mutants (Figures 4C and 6B). These findings showed that the Vps20PW mutant, which failed to nucleate Snf7 oligomerization, permitted cargo recycling from endosomes. In contrast, the Vps20loop mutant, which hyper-activated Snf7 nucleation, trapped more cargo on endosomes.
Figure 6.
Two Snf7 filaments are required to trap/encircle cargo. (A) Representative images of cells expressing Mup1-GFP (green). Cells were treated for 60 min with methionine and FM4-64 (red). In all cells, Mup1-GFP accumulates on the E-compartment (arrowheads) and is detected at the PM (arrows). Size bar (5 μm). (B) Fluorescence ratio (FR) of Mup1-GFP at the PM and on the endosomes in different class E mutants; n>25 for each mutant. (C) Canavanine sensitivity assay with WT cells and the indicated mutants. Similar to vps20Δ and snf7Δ, all ESCRT-II mutants (vps36Δ, vps22Δ and vps25Δ) are sensitive to 0.6 μg/ml canavanine. ‘One-armed' and ‘armless' ESCRT-II mutants (Vps22D214A or Vps25R83D) are sensitive to 0.6 μg/ml canavanine. (D) Model for ESCRT-II-mediated assembly of ESCRT-III. Both Vps25 molecules of ESCRT-II would transiently interact with two Vps20 molecules. This interaction induces conformational re-arrangements that simultaneously activate both Vps20 molecules (Step 1). Activated Vps20 then nucleates the oligomerization of two Snf7 filaments (Step 2) that are either branched (I) or run in parallel (II). Capping by Vps24 and Vps2 terminates Snf7 oligomerization (Step 3). Thereby two ESCRT-III filaments could generate an MVB-sorting domain to sequester cargo and form MVB vesicles.
In a complementary approach, a simple plate assay was used to determine the sensitivity of yeast to the toxic arginine analogue canavanine, which enters the cell through the arginine transporter Can1. Yeast strains impaired in endocytosis and degradation of Can1 take up excess canavanine and are, therefore, more canavanine sensitive as compared with WT cells (Lin et al, 2008). All ESCRT mutants are more canavanine sensitive as compared with WT cells and do not grow on 1 μg/ml canavanine (Supplementary Figure 1C). Interestingly, ESCRT mutants can be grouped into two distinct classes with respect to canavanine sensitivity. Class I mutants interfere with ESCRT-III assembly (ESCRT-II mutants as well as vps20Δ or snf7Δ mutants). All class I mutants were hyper-sensitive to canavanine and did not grow on 0.6 μg/ml canavanine (Figures 4D and 6C). Class II included mutants that block ESCRT-III disassembly (vps24Δ, vps2Δ and vps4Δ (data not shown)). In contrast to class I mutants, class II were less sensitive to canavanine and grew comparable with WT cells on 0.6 μg/ml canavanine (Figure 4D). Importantly, snf7Δ vps24Δ double mutants were hyper-sensitive class I mutants and did not grow on 0.6 μg/ml canavanine (Supplementary Figure 3C). These findings are most consistent with the idea that Snf7 oligomers trap MVB cargo on endosomes. When Snf7 oligomers cannot be formed (such as in ESCRT-II or vps20Δ or snf7Δ, snf7Δ vps24Δ double mutant cells), Can1 will not be degraded and recycles back to the plasma membrane. In mutants that do not cap or disassemble Snf7, the Snf7 oligomers will trap the majority of Can1 on endosomes and less Can1 will recycle back to the plasma membrane. Hence, less canavanine will enter the cells. Therefore, class II mutants are less sensitive to canavanine when compared with class I mutants. Consistently, the snf7Δ snx41Δ double mutant is more resistant to canavanine as compared with snf7Δ mutants, because the recycling from the class E compartment to the plasma membrane was blocked. These results were complemented by quantification of the Can1-GFP fluorescence intensities on the class E compartment and the plasma membrane (Supplementary Figure 3A and B). In vps24Δ mutants, more Can1-GFP accumulated at the class E compartment as compared with snf7Δ and snf7Δ vps24Δ double mutants (Supplementary Figure 3A and B). Both Vps20 mutants, Vps20loop and Vps20 PW, caused an MVB-sorting defect (Figure 4B) and are, therefore, sensitive to 1 μg/ml canavanine (Supplementary Figure 1C). However, the Vps20loop mutant behaved as a class II mutant and was more resistant to lower canavanine concentration (Figure 4D). The Vps20PW mutant was a hyper-sensitive class I mutant (Figure 4D). This finding correlated well with the proposal that the Vps20loop mutant was more active and favoured Snf7 oligomerization (see also Figure 3) and hence trapped more Can1 on endosomes. In contrast, the Vps20PW mutant did not nucleate Snf7 oligomerization and more Can1 recycled back to the plasma membrane.
Taken together, these findings suggested that the ESCRT-II-mediated activation of Vps20 is a tightly regulated multi-step process that is required to control the nucleation of Snf7 oligomers.
A single Vps25 molecule is sufficient to activate Vps20
Our results strongly indicate that ESCRT-II, in particular the interaction of Vps25 with Vps20, controls the first step in the assembly of ESCRT-III. Interestingly, each ESCRT-II complex contains two Vps25 molecules (arms) in equivalent positions, which generate an unusual Y-shape (Hierro et al, 2004; Teo et al, 2004). Therefore, we next addressed how this characteristic structural feature of ESCRT-II contributes to the assembly of ESCRT-III. Well-characterized point mutations specifically disrupt the formation of the paired Vps25 arms and MVB sorting (Hierro et al, 2004). The Vps25R83D mutant cannot bind to either Vps36 or Vps22, and hence resulted in the formation of an ‘armless' ESCRT-II complex (Vps22, Vps36 without Vps25) (Figure 5A) (Hierro et al, 2004). Mutation of Vps36D548R specifically disrupted the binding of Vps25 to Vps36, but not the binding of Vp25 to Vps22. Therefore, the Vps36D548R mutant generated a Vps22–Vps25 ‘one-armed' ESCRT-II mutant complex (Figure 5A) (Hierro et al, 2004). Similarly, the Vps22D214A mutant cannot bind to Vps25 and resulted in the formation of a Vps25–Vps36 ‘one-armed' ESCRT-II complex (Figure 5A) (Hierro et al, 2004). First, we examined the recruitment of Snf7-GFP to endosomes in cells expressing the ‘armless' or the ‘one-armed' ESCRT-II complexes. An ‘armless' ESCRT-II (Vps25R83D) no longer recruited Snf7-GFP to endosomes (class E compartments) (Figure 5A), most likely because endosomal Vps20 cannot be activated. Surprisingly, ‘one-armed' ESCRT-II complexes (Vps36D548R or Vps22D214A) were sufficient to activate Vps20 and recruit Snf7-GFP to endosomes (class E compartments) (Figure 5A). Next, we addressed the oligomerization status of Snf7 in cells expressing the mutant ESCRT-II complexes. Velocity sedimentation centrifugation showed that ‘armless' ESCRT-II could not have triggered Snf7 oligomerization (Figure 5B). In contrast, ‘one-armed' ESCRT-II complexes (Vps36D548R or Vps22D214A) were sufficient to induce the nucleation of a 450-kDa Snf7 filament (Figure 5B). Surprisingly, a ‘one-armed' ESCRT-II complex, containing a single Vps25 molecule, is sufficient to activate Vps20. Moreover, it seems that Snf7 oligomers, which are nucleated from ‘one-armed' ESCRT-II complexes, are still disassembled by Vps4, because larger Snf7 oligomers were not detected (Figure 5B). Yet, Snf7 oligomerization on endosomes is not sufficient for MVB sorting under physiological conditions, which suggests that an intact ESCRT-II complex in some ways is required to produce functional ESCRT-III assemblies.
Figure 5.
A single Vps25 molecule is sufficient activate Vps20. (A) The cartoons depict the Y-shaped structure of ESCRT-II. ESCRT-II consists of one Vps36 (light blue), one Vps22 (dark blue) and two Vps25 molecules/arms (red). The Vps25R83D mutation results in an ‘armless' ESCRT-II complex. Vps36D548R or Vps22D214A mutations generate a ‘one-armed' ESCRT-II complex. Live cell microscopy of Snf7-GFP in the different ESCRT-II mutants. In ESCRT-II mutants, Snf7-GFP no longer localize to the class E compartment. Snf7-GFP eventually aggregated in cytoplasmic dots that are not FM4-64 positive. ‘One armed' ESCRT-II complexes were sufficient for the recruitment of Snf7-GFP to the class E compartment. FM4-64 is shown in red. Size bar (5 μm). (B) Spheroplasts of cells expressing ‘armless' or ‘one-armed' ESCRT-II were cross-linked with 5 μM DSP. Solubilized membrane fractions (P13) were subjected to velocity sedimentation and analysed by SDS–PAGE and western blot with the indicated antibodies.
ESCRT-II might nucleate at least two Snf7 filaments to sort MVB cargo
It seems that ESCRT-II functions as a scaffold, in which a single Vps25 molecule is sufficient to induce nucleation of the ESCRT-III complex. However, it was clear from the earlier experiments that ESCRT-II with only one Vps25 arm is not sufficient for MVB sorting (Hierro et al, 2004). Therefore, we next asked how two Vps25 arms contribute to the functional assembly of ESCRT-III during MVB sorting using quantitative live cell microscopy of Mup1-GFP and the canavanine sensitivity assay (Figure 6).
All ESCRT-II mutants (vps22Δ, vps25Δ and vps36Δ) phenocopied snf7Δ and vps20Δ mutants; more Mup1-GFP recycled back to the plasma membrane (Figure 6A and B). Similarly, in mutants expressing the ‘armless' (Vps25R83D) or ‘one-armed' ESCRT-II complexes (Vps22D214A, Vps36D548R), Mup1-GFP was no longer sorted to the lumen of the vacuole (Figure 6A and B). In the ‘one-armed' ESCRT-II mutant cells, Mup1-GFP accumulated at the class E compartment and was detected at the plasma membrane, despite the nucleation of Snf7 oligomers. Although these Snf7 oligomers are non-functional for sorting MVB cargo, they do seem to be substrates for Vps4-mediated disassembly (Figure 5B) and, therefore, cannot sequester Mup1-GFP on the endosomes. Using the Can1 plate assay showed that similar to snf7Δ and vps20Δ, all ESCRT-II mutants are class I mutants and hyper-sensitive to canavanine when compared with class II mutants (e.g.vps24Δ) (Figure 6C). ‘Armless' ESCRT-II (Vps25R83D) was a class I mutant and canavanine hyper-sensitive. Interestingly, the mutants with ‘one-armed' ESCRT-II complexes (Vps22D14A) (and Vps36D548R, data not shown) were also phenotypic class I mutants (Figure 6C), although they supported the nucleation of Snf7 filaments. Yet, Snf7 filaments that are generated from a ‘one-armed' ESCRT-II complex phenocopied loss of Snf7 filaments in the growth assay. The Vps22Q216A mutant (only two amino-acid residues away from D214) was earlier shown not to affect binding of Vps25 and fully rescued the loss of Vps22 (Figure 6C). These findings indicated that ESCRT-II required both Vps25 molecules to generate a functional ESCRT-III complex. Taken together, our data indicate that both Vps25 arms in ESCRT seem to provide certain geometry during the nucleation of ESCRT-III. This might result in the assembly of at least two Snf7 oligomers (Figure 6D) that function together during MVB sorting. The first model proposes that at least two Snf7 filaments would be branched by ESCRT-II (Figure 6D, I) to define an MVB-sorting domain. Alternatively, two Snf7 filaments could be nucleated in parallel and run side-by-side on the membrane to define an MVB-sorting domain (Figure 6D, II). Maybe one Snf7 filament would sequester MVB cargo at the neck of the growing vesicle, whereas the second filament assists in membrane deformation.
Discussion
Recent findings suggested that the ESCRT-III complex bends and drives the fission of membranes in ESCRT-dependent processes, including MVB vesicle formation and the budding and release of HIV (Saksena et al, 2009; Wollert et al, 2009). The assembly of the ESCRT-III machinery on membranes is tightly regulated. In the cytoplasm, its individual subunits are kept inactive by auto-inhibition (Zamborlini et al, 2006; Shim et al, 2007). On endosomes, sequential conformational changes in the four-core ESCRT-III subunits result in the ordered assembly of the active ESCRT-III complex. Now, ESCRT-III assembly on endosomes can best be described in three distinct steps. The first step in ESCRT-III formation is the Vps25-dependent activation of the inactive Vps20 monomer on endosomes. In the second step, the activated Vps20 nucleates the homo-oligomerization of Snf7. Capping by Vps24 and Vps2 is the third step, which terminates Snf7 oligomerization (Teis et al, 2008; Saksena et al, 2009). Vps2 mediates the recruitment of the Vps4 AAA–ATPase, which then initiates the disassembly of ESCRT-III (Obita et al, 2007; Stuchell-Brereton et al, 2007). Current data indicate that Snf7 homo-oligomers form spiral-like filaments that deform membranes (Hanson et al, 2008). In vitro, individual ESCRT-III subunits assembled into filaments (Vps24), rings and sheets (Snf7) and helical tubes (hVps24, hVps2) that were disassembled by Vps4 (Ghazi-Tabatabai et al, 2008; Lata et al, 2008). Interestingly, ESCRT-III seems to generate membrane vesicles with different diameters depending on the biological context. The diameter of MVB vesicles is ∼25 nm in yeast (Nickerson et al, 2006) and ∼50 nm in HeLa cells (Doyotte et al, 2005). HIV-induced membrane stalks are between 50 and 100 nm in diameter and complete virions are on average 124 nm (Briggs et al, 2006) in diameter. The membrane tubule that connects the daughter cells just before abscission has diameter of 200 nm (Mullins and Biesele, 1977). Thus, it seems likely that the control of ESCRT-III assembly, in particular the regulation of Snf7 oligomerization, could be one way to regulate the size of these intermediates in membrane fission. Our results indicated that the regulated assembly and disassembly of ESCRT-III complexes is required for MVB vesicle formation. ESCRT-III with an excess of Snf7 subunits is capable of generating MVB vesicles that are larger and no longer of uniform size. This finding is in good agreement with the earlier findings. Over-expressed hSnf7 assembled into large polymeric ring-like arrays that formed protruding buds and tubular evaginations at the plasma membrane of mammalian cells (Hanson et al, 2008). Loss of DID2, which has a function in coordinating Vps4-dependent ESCRT-III disassembly (Nickerson et al, 2006) and RNAi-mediated depletion of hVps24 resulted in the formation of slightly larger MVB vesicles (Bache et al, 2006). Reconstitution experiments indicated that ESCRT-III induces deformation of liposomes and can generate luminal vesicles that bud into the lumen of giant unilamellar vesicles (GUVs) (Saksena et al, 2009; Wollert et al, 2009). Interestingly, the luminal vesicles generated from GUVs had a variable diameter and were much larger than physiological MVB vesicles. Taken together, these findings emphasize a mechanistic function of ESCRT-III, in particular Snf7 homo-oligomers, during membrane deformation and vesicle scission. Therefore, it will be important to understand how the assembly of the ESCRT-III complex, in particular the nucleation of Snf7 filaments, is controlled.
ESCRT-II controls the spatial/temporal activation of ESCRT-III assembly
Genetic and biochemical experiments showed that the ESCRT-II complex is required for the formation of ESCRT-III during MVB sorting (Babst et al, 2002b). The ESCRT-II subunit Vps25 binds specifically to helix α1 of Vps20 (and not to Snf7) and induces conformational re-arrangements in Vps20 on liposomes in vitro (Teo et al, 2004; Saksena et al, 2009). These findings suggested that the interaction of Vps25 with Vps20 could be the first step in the assembly of ESCRT-III. Alternatively, Vps20 was found to bind to the C-terminal domain of Vps28 (Pineda-Molina et al, 2006). How this interaction could contribute to MVB sorting is currently not understood. Interestingly, we found that ESCRT-II is not required for the recruitment of Vps20 to endosomes. N-terminal myristoylation of Vps20 and Chmp6 (hVps20) seems to be the primary targeting signal to endosomes (Babst et al, 2002a; Yorikawa et al, 2005). However, the interaction of the ESCRT-II subunit Vps25 with helix α1 of Vps20 is required for the recruitment and the nucleation of Snf7 oligomers. Although the localization of Vps20 to endosomes is not affected by loss of ESCRT-II, Vps20 fails to nucleate Snf7, suggesting that Vps25 activates Vps20. Our data suggest that Vps25 could facilitate the release of auto-inhibition by binding to helix α1 of Vps20 (Im et al, 2009) and thereby displaces the auto-inhibitory loops (α5/6) from α2, which subsequently triggers Snf7 nucleation. This idea was supported by mutations in the loop region between α1 and α2 of Vps20 (Vps20loop) and mutations in the auto-inhibitory loop (α5–α6) (Vps20PW) (Saksena et al, 2009). On membranes, the Vps20loop mutant more rapidly released auto-inhibition and more efficiently nucleated Snf7 oligomerization in vitro and in vivo. However, it is important to note that Vps20loop could not bypass the requirement for ESCRT-II, suggesting that the Vps20loop mutants behaved similar to a more-active Vps20 molecule. In contrast, the Vps20PW mutant cannot release auto-inhibition (Saksena et al, 2009) and behaved similar to a loss of function mutant, which was unable to nucleate Snf7 oligomers. Hence, our data indicate a two-step activation process for Vps20. First, Vps20 is targeted to endosomes, which requires N-myristoylation. Yet, endosomal targeting is not sufficient to recruit or nucleate Snf7 polymers, but may enhance the efficiency of the interaction between Vps20 and Vps25 (ESCRT-II). Next, Vps25 induces the release of Vps20 auto-inhibition by binding to α1 in Vps20, which probably displaces the auto-inhibitory region (α5/α6). This finally results in full activation of Vps20, which then triggers Snf7 nucleation through helix α2 of Vps20. This activation mechanism of Vps20 seems conserved in higher eukaryotes, in which the C-terminal half of hVps25 (EAP20) bound more tightly to the N-terminal half (helix α1/α2) of hVps20 (Chmp6) (Langelier et al, 2006). As ESCRT-II binds to PI(3)P (phosphatidyl-inositol-3-phosphate) and to ubiquitinated cargo, this two-step activation mechanism of Vps20 ensures spatial and temporal control of ESCRT-III formation in close proximity to MVB cargo.
ESCRT-II could assemble at least two Snf7 filaments during MVB sorting
Similar to ESCRT-0 (Vps27/Hse1) and ESCRT-I (Vps23, Vps37, Vps28 and Mvb12), ESCRT-II is recruited to endosomes as a preformed complex consisting of Vps36, Vps22 and two Vps25 molecules (Babst et al, 2002b; Hierro et al, 2004; Teo et al, 2004). One Vps25 molecule binds to Vps36 and the second Vps25 binds to Vps22. The two Vps25 arms create the characteristic Y-shape of the ESCRT-II complex. Specific mutations in either Vps22 or Vps36 disrupt the binding of one Vps25 molecule and result in the assembly of an ESCRT-II complex that contains only one Vps25 molecule. These ‘one-armed' ESCRT-II complexes were no longer functional, which showed that both Vps25 arms are each required for MVB sorting (Hierro et al, 2004). Unexpectedly, we found that one-armed ESCRT-II is nonetheless sufficient to activate Vps20 and subsequently nucleate Snf7 oligomerization on endosomes. Although one Vps25 molecule is sufficient for Vps20 activation, both Vps25 molecules are required for MVB sorting. Snf7 oligomers that are nucleated from a ‘one-armed' ESCRT-II are not functional during MVB sorting and accumulated abnormal endosomal compartments. Probably, a single Snf7 filament is not sufficient to generate a functional ESCRT-III lattice. Therefore, it is reasonable to speculate that ESCRT-II functions as a scaffold for Vps20 activation. Both Vps25 arms would provide geometry during the nucleation process that might result in the formation of two Snf7 filaments. Maybe the one filament would sequester MVB cargo at the neck of the growing vesicle, whereas the second filament assists in membrane deformation (Figure 6D). On the basis of the available ESCRT-II structures, it is difficult to speculate on the orientation of the two Snf7 filaments. Both Vps25 arms seem to be flexible (Hierro et al, 2004; Teo et al, 2004; Im and Hurley, 2008) and might undergo conformational changes when ESCRT-II binds to membranes. Interestingly, ESCRT-II as well as hVps20 (Chmp6) are dispensable for HIV budding and cytokinesis, whereas hSnf7 (Chmp4) is required for both (Martin-Serrano et al, 2003; Langelier et al, 2006; Carlton and Martin-Serrano, 2007; Morita et al, 2007), suggesting a different activation mechanism for ESCRT-III. During HIV budding and cytokinesis, the BRO domain of ALIX/AIP could provide an alternative activation scaffold for the formation of ESCRT-III. ALIX/AIP binds directly to hSnf7 (Chmp4) and is recruited to the site of HIV budding by p6-gag and to the mid-body by the centrosome protein 55 (Cep55) (Carlton and Martin-Serrano, 2007; Morita et al, 2007). This alternative, ALIX-dependent, ESCRT-III nucleation process could bypass the requirement for ESCRT-II and Vps20. As viral budding and cytokinesis do not require cargo sorting, ESCRT-III nucleated from ALIX may just be required for membrane fission. However, in the MVB pathway, ESCRT-II would provide the added function of cargo sorting and membrane deformation by regulating the nucleation of a second Snf7 filament. It seems possible that different nucleation mechanisms induce and regulate the assembly of ‘specialized' ESCRT-III machineries during MVB sorting, cell division and HIV budding. The ESCRT-II-dependent activation of Vps20 on endosomes may be dedicated to the assembly of an ESCRT-III complex that consists of at least two Snf7 oligomers that together execute the last steps of MVB sorting, cargo sequestration and MVB vesicle formation.
Materials and methods
Materials
The mouse monoclonal antibody specific for Flag was purchased from Sigma. Rabbit polyclonal antibodies specific for Snf7 were described earlier (Babst et al, 1998, 2000). DSP was from Pierce and FM4-64 from Invitrogen.
Yeast strains and DNA manipulation
Saccharomyces cerevisiae strains, Media and DNA manipulations are described in Supplementary data. vma4Δ vps25Δ and vma4Δ vps36Δ mutants were grown in YNB (pH=4.5).
In vivo cross-linking and velocity sedimentation on 10–40% glycerol gradients
These were performed as described earlier (Teis et al, 2008); 30 OD600 equivalents of yeast were spheroplasted, lysed in PBS and subjected to subcellular fractionation (Babst et al, 1997). Membrane fractions (P13) and cytoplasm (S100) were solubilized in PBS, 0.2% Tween 20. Linear glycerol gradients (10–40%)+0.2% Tween20 were prepared. Solubilized protein samples were loaded and spun for 210 min at 100 000 g. A total of 1 ml fractions were collected from the top and TCA precipitated.
Gel filtration
Gel filtration was performed as described earlier (Babst et al, 2000).
Fluorescence spectroscopy
Steady state fluorescence spectroscopy measurements were performed as described earlier (Saksena et al, 2009).
Microscopy and image analysis
Microscopy was performed as described earlier (Teis et al, 2008). The fluorescence intensity (IF) of Mup1-GFP was measured using a 19 × 19 Pixel ROI on an E-compartment (IFE) or at the plasma membrane (IFPM) and subtracted against background (19 × 19 Pixel ROI outside cells). FR was calculated FR=IFE/IFPM. Yeast cells were labelled with FM4-64 for 10 min, washed twice and incubated for 45 min before imaging (Vida and Emr, 1995).
In vitro assembly reaction
Liposomes were generated using Folch fraction I and extruded using 100 nm filters. ESCRT-II, H6-VPS20 and H6-SNF7 were purified as described earlier (Saksena et al, 2009). Equimolar ESCRT-II and Vps20 ratios were used. Snf7 was added in 20-fold excess and incubated with liposomes for 30 min at 22°C. Liposomes were collected by 30 min centrifugation at 55 0000 r.p.m. The pellet was re-suspended in solubilization buffer (PBS+0.2% (v/v) Tween 20) and layered on top of a 10–40% linear glycerol gradient.
Transmission electron microscopy and image analysis
Cells were grown in YNB (pH=4.5). A yeast pellet of 50 ODs was re-suspended in fix buffer (3% glutaraldehyde, 0.1 M sodium cacodylate, 5 mM calcium chloride, 5 mM magnesium chloride and 2.5% sucrose, pH 7.4) and fixed for 1 h at room temperature. For detailed information, refer to Supplementary data.
Supplementary Material
Acknowledgments
We thank Drs Jason A MacGurn and Chris J Stefan for comments on the manuscript, Dr Dan W Baird for transforming yeast strains and Manuel Alonso Y Adell for generating the Vps25T150K mutation. We are indebted to Prof. Arthur E Johnson for his assistance with the fluorescence spectroscopy experiments and helpful comments. DT was supported by an HFSP (LT00634/2006-L) fellowship and FWF (Y 444-B12). SS was funded by a fellowship of the American Heart Association (AHA 0826060D). This work was partially funded by a research award from Cornell University to SDE.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI (2006) Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol 13: 1029–1030 [DOI] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD (2002a) Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD (2002b) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G, Estepa EJ, Emr SD (2000) Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1: 248–258 [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17: 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache KG, Stuffers S, Malerod L, Slagsvold T, Raiborg C, Lechardeur D, Walchli S, Lukacs GL, Brech A, Stenmark H (2006) The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell 17: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI (2009) Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol 16: 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD (2006) The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14: 15–20 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J (2007) Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316: 1908–1912 [DOI] [PubMed] [Google Scholar]
- Doyotte A, Russell MR, Hopkins CR, Woodman PG (2005) Depletion of TSG101 forms a mammalian ‘Class E' compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci 118(Pt 14): 3003–3017 [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL (2008) Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE (2008) Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HR (2003) Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J 22: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, Hurley JH (2004) Structure of the ESCRT-II endosomal trafficking complex. Nature 431: 221–225 [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S (2006) Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol 13: 1031–1032 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35: 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Hurley JH (2008) Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell 14: 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, Hurley JH (2009) Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev Cell 17: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE (2005) Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic 6: 1078–1092 [DOI] [PubMed] [Google Scholar]
- Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI (2006) Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol 80: 9465–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W (2008) Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD (2003) Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA 100: 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Klionsky DJ (1994) Differential effects of compartment deacidification on the targeting of membrane and soluble proteins to the vacuole in yeast. J Cell Sci 107(Pt 10): 2813–2824 [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI (2007) Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26: 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ (1977) Terminal phase of cytokinesis in D-98s cells. J Cell Biol 73: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W (2006) Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell 10: 821–830 [DOI] [PubMed] [Google Scholar]
- Nickerson DP, West M, Odorizzi G (2006) Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol 175: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449: 735–739 [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W (2006) The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic 7: 1007–1016 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452 [DOI] [PubMed] [Google Scholar]
- Richter C, West M, Odorizzi G (2007) Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J 26: 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Emr SD (2009) ESCRTs and human disease. Biochem Soc Trans 37(Pt 1): 167–172 [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD (2009) Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK (1998) Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry 37: 14563–14574 [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Hayden MS, Lee KY, Trombetta ES, Pypaert M, Nara A, Yoshimori T, Wilm B, Erdjument-Bromage H, Tempst P, Hogan BL, Mellman I, Ghosh S (2006) CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J Cell Biol 172: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI (2007) Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8: 1068–1079 [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H (2005) Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem 280: 19600–19606 [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449: 740–744 [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD (2008) Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell 15: 578–589 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2006) ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125: 99–111 [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL (2004) ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell 7: 559–569 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S (2007) The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 8: 355–368 [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH (2009) Membrane scission by the ESCRT-III complex. Nature 458: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD (1998) Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J 17: 4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M (2005) Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J 387(Pt 1): 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H (2006) Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA 103: 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.