Abstract
Starving Dictyostelium amoebae emit pulses of the chemoattractant cAMP that are relayed from cell to cell as circular and spiral waves. We have recently modeled spiral wave formation in Dictyostelium. Our model suggests that a secreted protein inhibitor of an extracellular cAMP phosphodiesterase selects for spirals. Herein we test the essential features of this prediction by comparing wave propagation in wild type and inhibitor mutants. We find that mutants rarely form spirals. The territory size of mutant strains is approximately 50 times smaller than wild type, and the mature fruiting bodies are smaller but otherwise normal. These results identify a mechanism for selecting one wave symmetry over another in an excitable system and suggest that the phosphodiesterase inhibitor may be under selection because it helps regulate territory size.
When randomly located Dictyostelium cells begin to starve on a surface, a few will emit a single cAMP pulse that is relayed from cell to cell, spreading outward as a circular wave. At the same time, cells more chemotactically toward the cAMP source. Early during starvation, the system is said to be weakly excitable, because new circles emerge only to die out. As excitability increases with time, circular waves begin to propagate, and they occasionally interact with new pulses, breaking up and forming spirals (for review, see ref. 1).
These cAMP wave patterns can form in a field of randomly distributed cells because of the properties of the cAMP signaling system: When secreted cAMP levels rise above a certain threshold, cAMP binds to its receptor, stimulating internal cAMP production. Most of the cAMP is secreted, and external levels rise, which in turn stimulate increased production by binding to more receptors, creating a positive feedback loop. The system soon becomes adapted, however, and cAMP production halts. External cAMP levels then drop because of diffusion and the activity of a secreted cAMP-degrading enzyme (phosphodiesterase or PDE), and cells slowly reenter a regime where they become sensitive to cAMP once again. These properties are characteristic of excitable systems and are summarized in Fig. 1A.
Figure 1.
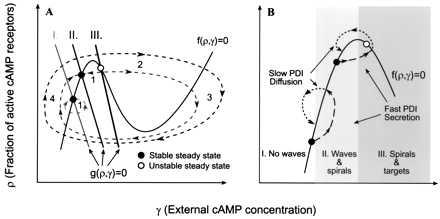
Idealized phase plane illustrating excitability in Dictyostelium. (A) The time-dependent variables ρ and γ describe the state of the system. The solid lines are nullclines where dρ/dt = 0 and dγ/dt = 0. I, II, and III are the g(ρ, γ) = 0 nullclines as the system changes with time. I corresponds to the nonexcitable or weakly excitable state, II corresponds to the excitable state, and III corresponds to the oscillatory state. The dotted lines represent the behavior of the system when it is perturbed from the steady state. After subthreshold stimuli, the system quickly relaxes to the previous steady state (region 1), but suprathreshold stimuli lead to rapid cAMP production (region 2), followed by a refractory period as cAMP levels drop (region 3), and then begin to recover (region 4). The outermost trajectory shows the behavior of the system when the steady state is unstable, region III. In this region, no stimulus is necessary and the system oscillates autonomously (based on ref. 19). (B) The system evolves with time as excitability increases. Waves cannot propagate in region I, waves and spirals can form in region II, and spirals and targets form in region III. Each phase lasts approximately 0–4, 4–6, and 6+ h, respectively. The regulation of excitability by PDI in two regions of the phase plane is illustrated. Circles correspond to the intersection of the g(ρ, γ) and f(ρ, γ) nullclines and indicate the local excitability. Global excitability of the system is an average of all cells, but there is a local variation in excitability for each cell, so the population has a spread around the average. Local PDI secretion rapidly shifts the intersection of the g(ρ, γ) and f(ρ, γ) nullclines in the direction of higher excitability. Diffusion slowly shifts the intersection back toward the previous value, which is not restored because PDI diffusion increases global excitability (1). PDI secretion in region I does not give rise to a cAMP pulse but does increase global excitability. In region II, PDI secretion may push local excitability into region III, forming a temporary oscillator and thus initiating cAMP pulses (based on A and ref. 1).
Studies of excitable media show that there is a fundamental difference between spirals and target-like patterns. Targets are formed when a cell or a group of cells send out cAMP signals periodically. These oscillating (or pacemaker) cells are necessary to sustain the target patterns. Spirals, in contrast, arise from a break in the symmetry of circular waves and, once formed, do not need further stimuli to persist. This is because the tip of a spiral rotates around a central core, and by the time the tip has completed one rotation, cells outside the core where the tip started have recovered and can be excited again. In a sense, the tip follows a receding wave of excitable cells and for this reason does not require a pacemaker. At the same time, cAMP levels in the core remain fairly constant, and indeed the core can even be devoid of cells.
The cAMP signaling system is complex and includes not only cAMP receptors, adenylyl cyclase, and many other downstream signaling molecules (2) but also two secreted diffusible proteins that modulate excitability, the aforementioned PDE, and its inhibitor, PDI (inhibitor of PDE). Because PDI is known to be one of the first molecules secreted during starvation (3) and because cells are caught in different stages of the cell cycle when they begin to starve (4), we postulated (1, 5) that randomly located cells secrete small pulses of PDI. The secretion of PDI has the effect of reducing the degradation of cAMP by inhibiting PDE, which leads to local increases in cAMP. This increases the local excitability of the cells past a critical point, triggering these cells to secrete cAMP pulses that propagate outwards as circular cAMP waves. PDI soon diffuses away, however, and this returns pulsing cells to a “ground state” that is excitable but nonpulsing (see Fig. 1B). As PDI diffuses, global excitability increases. Random local PDI secretion thus gives rise to circular waves, which, because of interaction with new pulses, break up and form spirals. Even though there may be many firing centers, only a few spirals can form, because their formation requires two independent events—a pulse of cAMP must first initiate a circular wave and a second uncorrelated pulse must interact with it (1). At this time, permanent oscillators have not formed, and spirals are the only signaling centers (region II, Fig. 1B). This allows them to form large aggregation territories by entraining neighboring cells.
Herein we test these ideas by comparing signaling in PDI+ and PDI− strains. We show that spirals tend to dominate in PDI+ strains but are virtually absent in PDI− strains. Territory sizes in PDI− strains are at least 50 times smaller, and the mature fruiting body is smaller but otherwise normal. It is possible, therefore, that secreted PDI is selected for because it helps guarantee the establishment of spiral waves. This in turn governs territory size, an important element of organismal fitness (6). We also compare our experimental results to a modified version of our earlier model, showing that random secretion and diffusion of PDI is in and of itself sufficient both to trigger cAMP pulses and select for spiral waves.
METHODS
Cell Growth and Imaging.
Cell populations were prepared and observed by dark-field microscopy at room temperature as described (7). Briefly, vegetative amoebae were grown on bacteria on Petri dishes. They were harvested, freed of bacteria by centrifugation, and allowed to settle on an agar surface at a density of 5.4 × 105 cells cm−2. To control for starvation time and environmental heterogeneity, mutant and wild-type cells were harvested while both were in the vegetative growth phase and plated on a single agar plate separated one from the other by a thin glass partition. Both strains could then be viewed simultaneously. Dark-field images were enhanced by subtracting successive frames, as described (7).
Dictyostelium Strains.
The PDI− strains 9–2, 9–6, 10–2, and 27 are deletions created by recombination in Dictyostelium discoideum DH1 (8). They grow on bacteria at the same rate as the DH1 parent.
Simulations.
These were carried out as described (1), with the following modification. In ref. 1, random spatial pulses of cAMP were used to initiate waves. In Fig. 4, both wave initiation and spiral selection are driven entirely by spatially random PDI pulses that are allowed to diffuse. The probability of PDI secretion = 0.5 × 10−6 per time step per cell. The diffusion constants for cAMP and PDI were assigned values of 4 × 10−6 cm2/sec and 8.3 × 10−7 cm2/sec, respectively. The model was solved by using a second-order Runge–Kutta method for time and an alternating direction implicit scheme for space. The grid resolution was 400 × 400 with Δx and Δy = 0.1 mm and Δt = 0.04 min. For details of the implementation and justification of these choices, see refs. 1 and 5.
Figure 4.
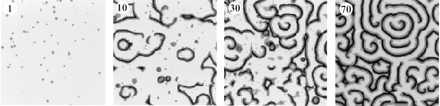
Computer simulation: random spatial secretion of PDI can account for the establishment and evolution of wave patterns. The transition from pulsing cells (1 min) to circles (10 min) to spirals (30 and 70 min) begins with the spatially random secretion of PDI. Increasing cAMP and PDI levels are shown in gray scale. The model is taken from ref. 1 where cAMP waves were triggered by random spatial secretion of cAMP rather than PDI.
RESULTS
Time-lapse frames comparing signaling in PDI+ and PDI− strains are shown in Fig. 2. Scattered PDI+ cells (left half of each video frame) begin to pulse 4–5 h after starvation under our conditions, sending out single circular waves of cAMP, which then die down (Fig. 2A). This early stage corresponds to region I of Fig. 1. Later, firing centers appear at random, with circular waves appearing and disappearing, evolving with time into a complex pattern of spirals and fragments of circles, but there are no permanent oscillators (Fig. 2 B and C). This period corresponds to region II of the phase plane, Fig. 1. With time (Fig. 2, compare C with E and F), the wavelength shortens and a single rapidly rotating spiral dominates a territory of many mm2. By viewing the nonsubtracted images, one can see that the cell density is essentially homogenous for the first 300 min. These findings are consistent with those of others (9–11) and recent results from our laboratories (7).
Figure 2.
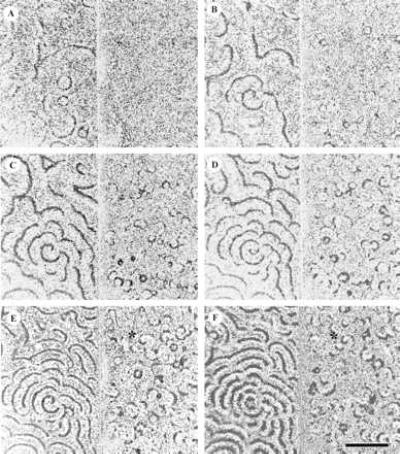
Evolution of targets and spirals in a parental and a PDI deletion of Dictyostelium. (A–F) The parental strain is on the left and the mutant is on the right in each video frame. Dark-field intensity in gray-scale corresponds to cAMP concentration. Vegetative Dictyostelium discoideum DH1 and PDI− mutant 27 were plated side by side on agar surfaces, separated by a thin glass plate at 5.4 × 105 cells cm−2. Video images were gathered by dark-field illumination. Mutant 27 carries an extensive PDI deletion (8). Similar results were obtained with three other independently isolated deletions, 10–2, 9–2, and 9–6. (A–F) Images were taken 271, 321, 344, 368, 396, and 422 min after harvesting and plating the cells. The asterisk marks an apparent small spiral (see text). (Bar = 5 mm.)
Compare this behavior to a PDI− mutant strain. Pulsing centers spring up at random, as before, but they appear a few hours later and are more numerous (Fig. 2 B and C). Rather than emitting just a few pulses before dying down, as observed in the parental strain, they oscillate periodically and rarely form spirals. When they do (asterisks), they are soon disrupted by nearby oscillators. In terms of the phase plane sketched in Fig. 1, they may be thought of as having entered the oscillatory regime III. Aggregation streams and fruiting bodies form in the mutant but are much smaller and more numerous than those in the parent (Fig. 3). Aggregation streams form and fruiting body formation occurs at approximately the same time as wild type. Of the four PDI mutants tested in these experiments, only 10–2 formed a few persistent spirals, possibly because it retains some PDI activity.
Figure 3.
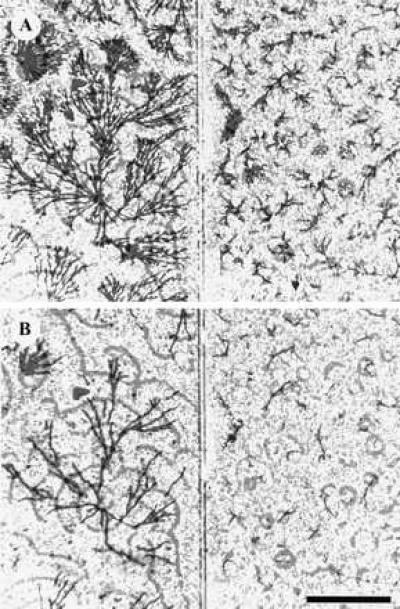
Aggregation territories in parental and mutant strains. (Left) Parent. (Right) PDI deletion 27. (A) t = 494 min after harvesting and plating. (B) t = 619 min after harvesting and plating. A reference wave pattern in gray scale (367 min) has been superimposed on aggregating streams of amoebae, emphasizing the relationship between the size of the wave and the size of the resulting territory several hours later. (Bar = 5 mm.)
The number of oscillators that form in PDI− fields is approximately 50 times larger than the number of spirals in wild-type fields (48 compared with a single large spiral with 5 spiral fragments on the edge of the field for the experiment in Fig. 2). This difference in number is reflected in territory (Fig. 2), aggregate (Fig. 3), and fruiting body size (data not shown).
Strong oscillators form in PDI mutants (region III, Fig. 1) and rapidly extinguish each other by collision. This is apparent from an analysis of the digital images used to gather the data for Fig. 2. Seventy-five oscillating centers could be followed in this experiment. All of them gave rise to waves that propagated strongly at velocities similar to wild type, going extinct by collision with neighboring targets. In addition, the average wavelength of these targets (1.89 ± 0.67 mm, n = 18) was shorter and less uniform than the wavelength of fully developed spirals in the wild type (2.44 ± 0.23 mm, n = 8), whereas the propagation velocity was approximately wild type, strong evidence that the mutant is if anything more excitable than wild type (12, 13). Its failure to form large territories is not, therefore, because of lower excitability or the inability to propagate waves.
In our earlier model of spiral evolution, waves were triggered by random cAMP pulses, because this mirrors random PDI pulses. In the modified version shown in Fig. 4, this is accomplished by random pulses of secreted PDI that diffuse with time. Small changes in the local concentration indirectly elevate cAMP levels, which lead to cAMP pulses (Fig. 4, 1 min). These initiate waves of cAMP (Fig. 4, 10 min), and they interact with other waves to produce spirals (Fig. 4, 30 min), which become more frequent with time (Fig. 4, 70 min). These images illustrate the transition through phase space outlined in Fig. 1 and demonstrate by simulation that random spatial and temporal PDI pulses are sufficient for the establishment of spiral waves.
DISCUSSION
Our chief finding is that cells mutant in the structural gene for PDI fail to develop spiral cAMP waves. Aggregation territories are consequently much smaller, and this suggests a role for PDI in the evolution of territory size. These results support a prediction of a recently published model for spiral wave generation (1), and numerical results with a modified version of this model demonstrate that spatially and temporally random pulses of PDI are sufficient for spiral formation.
PDI and PDE are secreted in a wide variety of Dictyostelium species (3). Mutant PDE strains of Dictyostelium discoideum fail to aggregate and thus cannot form fruiting bodies (14). This is the expected behavior, because the primary role for PDE is the degradation of cAMP so that the signaling system can be reset and new waves can propagate. The inhibition of PDE by secreted PDI might then be seen as a mechanism to modify the excitability of the signaling system. This is unlikely, however, because PDI synthesis is strongly repressed by cAMP (15), and, as shown herein, PDI− cells develop oscillators and propagate waves at the expected frequencies. (The reasons for this may be understood with reference to Fig. 1. Even in the absence of PDI, cells proceed to region III of the phase plane where they emit pulses of cAMP independently of extracellular perturbations.) Furthermore, when well-developed spiral waves are abolished by a mist of 10−3 M cAMP, only circular waves and target patterns resume when the system recovers (K.J.L., E.C.C., and R.E.G., unpublished results). This observation also argues that PDI plays a major role in spiral selection, because at the time the signaling system is reset by cAMP, PDI synthesis has long ceased. Thus, it is unlikely that PDI plays a continuing role in signal propagation, and this is consistent with the view that its primary function is to regulate pattern selection.
At low cell densities, target patterns predominate in wild-type cells (7). We interpret this observation in the following way. At low cell densities, the excitability of a population in region I (Fig. 1) is shifted to the left relative to cells at higher densities. With time, both low- and high-density populations move into region II, where waves can propagate and spirals potentially form. However, in the low-density populations, the frequency of pulses per unit area is not high enough to achieve the conditions necessary to break the symmetry of expanding circles, and spirals fail to form. Nonetheless, cells do continue to develop, finally moving into region III, and becoming oscillators centered on targets.
The time-lapse records of Fig. 2 reveal that the ability of the PDI nulls to enter the oscillatory regime is unimpaired. The oscillator frequency appears normal, and the extinction of successive waves occurs by collision, both properties of a functional excitable system that has fully entered the oscillatory regime. Thus the mutants make the transition through the phase plane at about the same time as their parent, they oscillate at the expected frequency, and they in general behave in a fashion very similar to wild-type cells recovering from a cAMP mist.
Why has this complex network evolved, in which the secretion of an inhibitor appears to promote spirals? We believe the explanation lies in the evolution of territory size and the properties of spiral waves in excitable media. Because the size of the mature slime mold fruiting body is roughly proportional to the size of the territory from which it forms (16, 17), any feature that tunes signaling to increase the range over which an attractant can be relayed may also increase evolutionary fitness. This is clearly so when large cell masses are important for spore dispersal, locomotion, and predator avoidance (18).
There will also be selection for spirals over circles and targets for reasons that are rooted in three properties of spiral waves in excitable media. (i) As we have noted, spirals rotate about a core of adapted cells and do not require oscillators. This means that early in starvation they can form and persist before the system reaches a level of excitability where permanent oscillators begin to appear (Fig. 1). In terms of the “developmental path” idea of Goldbeter and Segel (19), which has recently been used to demonstrate how spirals can be selected (20), spirals are found before oscillators on the developmental path (nullclines I → II → III, Fig. 1A). This gives them an early competitive edge, and once formed, they entrain oscillators that form later (this work and ref. 7). (ii) Because spirals are caused by two independent events—a propagating wave must first be established by a pulsing cell and then interact with a second pulse—they will be infrequent relative to oscillators if we assume both arise by the same basic mechanism. They will therefore be centered on larger territories (Fig. 3). This is because there is only a finite time between the emergence of the first signaling centers and the establishment of isolated aggregation territories, at which time no new centers emerge. Therefore, the number of territories that form on a field depends strongly on the frequency of formation of new signaling centers as well as on competition between centers (iii). Finally, that the spatial frequency of Dictyostelium spirals is higher than targets is an empirical fact (our data and ref. 10), and this is a generic property of excitable media under a wide variety of conditions (12). Thus when a spiral collides with a target wave front, at each collision the wave with the highest spatial frequency moves closer to the center emitting the wave of lower frequency, finally extinguishing it. This effect is enhanced if the oscillator at the center of the target is also irregular, as it appears to be in Dictyostelium (our data and ref. 10).
Recently, Levine et al. (21) have used our approach to initiate spirals in a cellular automaton model of aggregating Dictyostelium. They find that once spirals have been initiated, their size can be regulated by genetic feedback, by which they mean the regulation of genes by cAMP in such a way that excitability increases with time. Although their model does not attempt to capture the kinetic and biochemical reality of Dictyostelium signaling, it supplies a general way to realize the developmental path of Segel and Goldbeter (19) and shows how spiral size can be adjusted with developmental time. It is also possible to initiate spirals if there are large-scale inhomogeneities already present in the cell population (5, 22).
We have focused on the role PDI plays in pattern selection. There are, however, many other components of the cAMP relay system, and one can ask if by tuning them, spirals might be favored or disfavored. We might imagine, for example, that mutations in cAMP receptors or PDE itself could equally well mimic the results we have presented herein. We suggest, however, that by altering various components of the signaling system to change affinities, rate constants, and the like, the main effect, short of destroying wave propagation altogether, will be to alter excitability in such a way as to shift the spiral vs. wave frequency along the cell density axis as defined in earlier work from this laboratory (7). That is, the basic relationship will continue to hold, and in different genetic backgrounds, PDI mutations will have the same effect as shown herein, although the effect will be seen at different cell densities.
We have emphasized that the secretion of PDI acts as an extracellular perturbing force that indirectly selects for spirals that can form and persist in the absence of an oscillator. This feature of Dictyostelium, which we believe allows it to synthesize and secrete a protein that plays a determining role in pattern selection, is in striking contrast to nonliving systems. In the Belousov–Zhabotinskii reaction, for example, the kinds of patterns that will form are triggered by perturbations whose origins are usually unknown and external to the system. Although it is true that Dictyostelium also finally depends on random perturbations, just as nonliving systems do, it appears to have evolved a strategy that places the perturbations needed for pattern selection under genetic control in such a way that one geometry dominates. Whether or not these particular geometric properties of spirals in excitable media are under selection in other biological settings (23, 24) is an open question.
Acknowledgments
We thank Bob Austin for cheerfully sharing his laboratory with us, and R. Sucgang for thoughtful criticism. This work was supported by grants from the National Science Foundation (E.C.C. and R.E.G.), National Institutes of Health (J.F. and R.H.K.), and the Alfred P. Sloan Foundation (R.E.G.).
ABBREVIATIONS
- PDE
cAMP phosphodiesterase
- PDI
inhibitor of PDE
References
- 1.Pálsson E, Cox E C. Proc Natl Acad Sci USA. 1996;93:1151–1155. doi: 10.1073/pnas.93.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firtel R A. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- 3.Gerisch G, Malchow D, Riedel V, Muller E, Every M. Nat New Biol. 1972;235:90–92. doi: 10.1038/newbio235090a0. [DOI] [PubMed] [Google Scholar]
- 4.McDonald S A, Durston A J. J Cell Sci. 1984;66:195–204. doi: 10.1242/jcs.66.1.195. [DOI] [PubMed] [Google Scholar]
- 5.Pálsson E, Cox E C. In: Dictyostelium—A Model System for Cell and Developmental Biology. Maeda Y, Inouye K, Takeuchi I, editors. Tokyo: Universal Academy; 1997. pp. 411–423. [Google Scholar]
- 6.Bonner J T. Am Naturalist. 1982;119:530–552. [Google Scholar]
- 7.Lee K, Cox E C, Goldstein R. Phys Rev Lett. 1996;76:1174–1177. doi: 10.1103/PhysRevLett.76.1174. [DOI] [PubMed] [Google Scholar]
- 8.Wu L, Franke J, Blanton R L, Podgorski G J, Kessin R H. Dev Biol. 1995;167:1–8. doi: 10.1006/dbio.1995.1001. [DOI] [PubMed] [Google Scholar]
- 9.Alcantara F, Monk M. J Gen Microbiol. 1974;85:321–334. doi: 10.1099/00221287-85-2-321. [DOI] [PubMed] [Google Scholar]
- 10.Gross J D, Peacey M J, Trevan D J. J Cell Sci. 1976;22:645–656. doi: 10.1242/jcs.22.3.645. [DOI] [PubMed] [Google Scholar]
- 11.Höfer T, Sherratt J A, Maini P K. Physica D. 1995;85:425–444. [Google Scholar]
- 12.Keener J P, Tyson J J. Physica D. 1986;21:307–324. [Google Scholar]
- 13.Keener J P. Siam J Appl Math. 1986;46:1039–1056. [Google Scholar]
- 14.Barra J, Barrand P, Blondelet M H, Brachet P. Mol Gen Genet. 1980;177:607–613. doi: 10.1007/BF00272671. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Franke J. Gene. 1990;91:51–56. doi: 10.1016/0378-1119(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 16.Bonner J T, Dodd M R. Biol Bull. 1962;122:13–24. [Google Scholar]
- 17.Waddell D R. J Embryol Exp Morph. 1982;70:75–98. [PubMed] [Google Scholar]
- 18.Kessin R H, Gundersen G G, Zaydfudim V, Grimson M, Blanton R L. Proc Natl Acad Sci USA. 1996;93:4857–4861. doi: 10.1073/pnas.93.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldbeter A, Segel L A. Differentiation. 1980;17:127–135. doi: 10.1111/j.1432-0436.1980.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 20.Lauzeral J, Halloy J, Goldbeter A. Proc Natl Acad Sci USA. 1997;94:9153–9158. doi: 10.1073/pnas.94.17.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine H, Aranson I, Tsimring L, Truong T V. Proc Natl Acad Sci USA. 1996;93:6382–6386. doi: 10.1073/pnas.93.13.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dallon J C, Othmer H G. Phil Trans R Soc Lond B Biol Sci. 1997;352:391–417. doi: 10.1098/rstb.1997.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechleiter J, Girard S, Peralta E, Clapham D. Science. 1991;252:123–126. doi: 10.1126/science.2011747. [DOI] [PubMed] [Google Scholar]
- 24.Glass, L. (1996) Phys. Today August, 40–45.


