Abstract
Metabolic phenotyping at cellular resolution may be considered one of the challenges in current plant physiology. A method is described which enables the cell type-specific metabolic analysis of epidermal cell types in Arabidopsis thaliana pavement, basal, and trichome cells. To achieve the required high spatial resolution, single cell sampling using microcapillaries was combined with routine gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) based metabolite profiling. The identification and relative quantification of 117 mostly primary metabolites has been demonstrated. The majority, namely 90 compounds, were accessible without analytical background correction. Analyses were performed using cell type-specific pools of 200 microsampled individual cells. Moreover, among these identified metabolites, 38 exhibited differential pool sizes in trichomes, basal or pavement cells. The application of an independent component analysis confirmed the cell type-specific metabolic phenotypes. Significant pool size changes between individual cells were detectable within several classes of metabolites, namely amino acids, fatty acids and alcohols, alkanes, lipids, N-compounds, organic acids and polyhydroxy acids, polyols, sugars, sugar conjugates and phenylpropanoids. It is demonstrated here that the combination of microsampling and GC-MS based metabolite profiling provides a method to investigate the cellular metabolism of fully differentiated plant cell types in vivo.
Keywords: Arabidopsis thaliana, basal cell, GC-TOF-MS, metabolites, pavement cell, single cell, trichome
Introduction
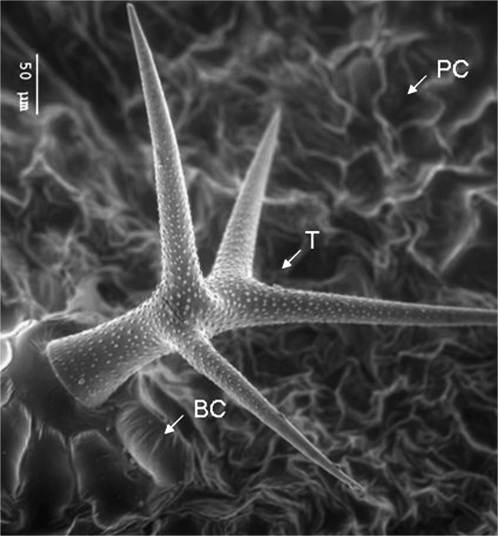
Epidermal cells represent the outer barrier of the plant to its environment. These cells perform important functions that are related to gas exchange, water homeostasis, defence and protection of the plant (Gutierrez-Alcala et al., 2000; Calo et al., 2006; Nawrath, 2006; Martin and Glover, 2007). The leaf epidermis of Arabidopsis thaliana consists of several specialized cell types: trichomes, stomata, pavement, and basal cells (Fig. 1).
Fig. 1.
Electron micrograph of epidermal cell types. Shown are all the leaf epidermal cell types analysed in this study, namely pavement cells (PC), trichomes (T), and its supporting basal cells (BC).
The most frequently occurring cell type in the epidermal cell layers of all plant organs is the pavement cell. A key function of this cell type is the protection of the tissue layers below. The pavement cells of dicotyledonous leaves are usually shaped like the interlocking pieces of a jigsaw puzzle which promote the mechanical strength necessary to support the large apoplastic cavities of the underlying mesophyll cells that promote the fast diffusion of carbon dioxide for photosynthesis. Furthermore, pavement cells are involved in the correct spacing of morphologically specialized cell types such as stomata and trichomes (Glover, 2000).
Trichomes of Arabidopsis thaliana comprise a single cell, each 300–500 μm in length with, usually, three to four branches. In contrast to trichomes from other plants species, Arabidopsis trichomes are non-glandular and appear to act as a simple mechanical barrier on the leaf surface (Esau, 1953; Glover, 2000). Basal cells surround the base of the trichomes and are likely to be involved in the support of the trichome structure (Marks, 1997). Due to their location on the leaf surface, basal and trichome cells provide a feasible and physiologically interesting model system for single cell analysis (Tomos and Sharrock 2001; Schellmann and Hülskamp, 2005).
Approaches to analyse gene functions are usually applied to samples derived from whole organisms or a defined organ which leads to a mixture of diverse cell types in the final sample. Because of this experimental limitation, the results need to be interpreted using an estimation of the cellular composition of the sample and its respective predominant cell type. Due to the resulting averaging effect, the information on defined cell types is diluted considering the most abundant cell type in the sample or even obscured in the case of rare cell types. Therefore, special techniques which are able to overcome such a limitation are required for highly spatially resolved investigations at the single cell level. Tissue- and cell-specific sampling methods such as protoplast preparation combined with cell sorting (Birnbaum et al., 2005), laser microdissection or microsampling have been developed and successfully applied to the plant system (Fricke et al., 1994; Tomos et al., 1994; Kehr, 2003; Leonardt et al., 2004). Transcript profiling has effectively contributed to novel insights into differentiated gene expression at the single cell level (Brandt et al., 1999, 2002; Tomos and Sharrock, 2001; Laval et al., 2002; Liu et al., 2004; Brandt, 2005). Furthermore, ground-breaking studies have started to address protein profiling and metabolic composition in single cells and restricted tissue samples (Arlt et al., 2001; Tomos and Sharrock, 2001; Wienkoop et al., 2004; Schad et al., 2005).
Along with the identification of gene and protein expression patterns, the metabolite status is of particular interest for characterizing the physiology of specialized cell types (Oliver et al., 2002; Fernie, 2003; Bino et al., 2004; Schad et al., 2005). While metabolic approaches have been widely applied to elucidate the metabolic phenotype of plant tissues at specific time points with medium to high temporal resolution and at time scales relevant for developmental processes, so far only little is known about the spatial resolution of metabolism in a living plant. Early pioneering studies demonstrated the asymmetric distribution of metabolic compounds on the subcellular (Farre et al., 2001) and/or tissue level, for example, enzymes involved in CO2 fixation (Outlaw and Fisher, 1975), cyanogenic glucosides (Saunders and Conn, 1977; Kojima et al., 1979; Thayer and Conn, 1981) or enzymes of the glucolytic pathway (Wurtele and Nikolau, 1986). Moreover, Slack et al. (1969) demonstrated the importance of the co-ordinated mode of action of enzymes distributed in chloroplasts of different cell types for the C4-dicarboxylic acid pathway of photosynthesis. However, comparative metabolic profiling of internal metabolites from specific single cell types, in contrast to tissue-specific profiling, has not yet been achieved. Up to now, investigations of metabolites from individual plant cells collected using microcapillaries have been restricted to a limited number of metabolites determined by capillary electrophoresis or enzymatic assays (Tomos et al., 1994; Kehr, 1998a, b; Arlt et al., 2001; Tomos and Sharrock, 2001). In contrast to previous studies, gas chromatography mass spectrometry (GC-TOF-MS) was used as a read-out that allows the simultaneous, non-targeted detection and identification of a multitude of compounds, predominantly primary metabolites from different classes and pathways (Fiehn et al., 2000; Roessner et al., 2000).
We report here on more than 90 different metabolites from a variety of classes associated with central cellular functions, which were identified without the need for analytical background correction in extracts from trichome, pavement, and basal cells of the Arabidopsis thaliana leaf epidermis.
Materials and methods
Plant material and sampling procedure
Arabidopsis thaliana plants (ecotype Columbia-0) were grown in a greenhouse at 60% humidity and a photoperiod of 16 h light (200 μmol m−2 s−1, 21 °C) and 8 h dark (17 °C). Single cell extracts were collected from 6-week-old plants at growth stage 3.9 (Boyes et al., 2001). Plant handling and the sampling procedure were always standardized under the same conditions and at the same time to ensure a similar comparable metabolic status of the plant. Single cell sampling was performed between 10.00 h and 11.00 h to avoid circadian modulation of the sampled metabolite pools.
Cell type-specific metabolite preparation
Single cell sampling was carried out as described by Brandt et al. (1999). Briefly, borosilicate glass capillaries (WPI, Berlin, Germany) were pulled on a List pipette puller (Darmstadt, Germany) with a tip aperture of 1–10 μm. Glass microcapillaries capillaries were mounted on a micromanipulator and rosette leaves of an intact plant were fixed under an Optiphot 2 microscope (Nikon, Duesseldorf, Germany). A microcapillary was inserted into a single pavement, basal, or trichome cell via an Eppendorf remote-controlled micromanipulator for sample collection (Hamburg, Germany). A single capillary was used to collect the content of a pavement, basal or trichome cell, respectively. For metabolic inactivation, the cell sap was immediately diluted into 200 μl extraction buffer containing water, methanol, and chloroform in a ratio of 1:2.5:1 by vol. Single cell sampling using microcapillaries was optimized to avoid metabolic activity after sampling and to accomplish the smallest possible time lag between sampling and metabolic inactivation, a routine procedure to avoid possible sampling artefacts of metabolite profiling experiments (Fiehn et al., 2000; Roessner et al., 2000).
After collection, complete extracts from an exact count of 200 single cells were dried by vacuum centrifugation (Concentrator 5301, Eppendorf, Hamburg, Germany) and stored dry at -80 °C until analysis.
Metabolite extraction from trichome preparations
An alternative sampling approach, leaf hair depilation (LHD), was specifically developed for trichome cells. Arabidopsis rosette leaves were fixed on a microscope slide by freeze-connecting it to a water droplet using liquid nitrogen. Single trichome cells were shaved off from the leaf surface using precooled fine-scaled forceps under a binocular microscope. The equivalent of 200 frozen trichome cells was immediately transferred into 200 μl of a water–chloroform–methanol solution. The extraction of whole trichome preparations was performed after shaking at 70 °C for 15 min and subsequently vortexing at room temperature. The complete liquid supernatant was separated by centrifugation at 16 000 g for 5 min and then dried by vacuum centrifugation (Concentrator 5301, Eppendorf, Hamburg, Germany). The amount of trichome cells was estimated from counts of trichome yield per leaf surface. Five replicates of representative rosette leaves, 9–14 of wild-type (WT) plants at growth stage 3.9 (Boyes et al., 2001) were chosen to determine the total trichome number (see Supplementary Fig. S1 at JXB online). The calculated average number of trichomes at stage 3.9, rosette leaf number 9–11, was 302±15, whereas for rosette leaves 12–14 an average of 189±23 was counted. Based on these observations, an estimated number of 200 trichomes per leaf were collected from rosette leaves 9–11 at growth stage 3.9 (Boyes et al., 2001).
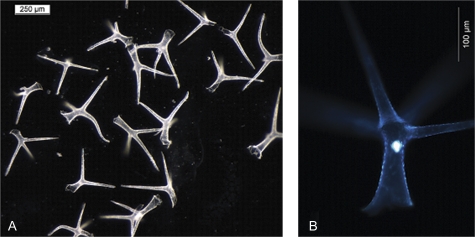
Structure and intactness of depilated trichome preparations were examined by light microscopy. It was confirmed that the trichome structure remained intact during LHD and contamination by other cell types was absent (Fig. 2A). Moreover, mature trichomes harvested by LHD were stained with 4′,6-diamidino-2-phenylindol (DAPI), a DNA staining dye. Staining of the nucleus was bright and consistent which confirmed the intactness of the cells (Fig. 2B).
Fig. 2.
(A, B) Arabidopsis thaliana trichomes sampled by leaf hair depilation (LHD). To confirm accuracy of the sampling and intactness of trichomes they were analysed under a binocular microscope. (A) Trichomes isolated by LHD (scale bar 250 μm) and (B) depicts an isolated trichome stained with DAPI. The trichome nucleus is visible as the bright spot in the centre of the image (scale bar 100 μm).
Control for laboratory contaminations
The analysis of single cell types requires high metabolite enrichment steps in order to achieve the best possible analytical sensitivity. The high enrichment steps, as compared to routine metabolite profiling of mixed tissues or full plant organ samples, bear the risk of accumulating so far overlooked or negligible laboratory contaminations. In order to detect and rule out such laboratory contaminations, so-called non-sample controls were performed. Profiles of non-sample controls were generated by in-parallel processing of pure sampling buffer without single cell extracts using the same containers, solvents, and equipment throughout the complete analytical procedure. The thus generated ‘empty’ profiles contain only mass spectral signals of laboratory contaminations which were used for the background subtraction of the GC-MS profile from single cell types.
GC-TOF (Time of Flight)-MS based steady-state metabolite profiling
Chemical derivatization (Fiehn et al., 2000; Roessner et al., 2000) and GC-TOF-MS metabolite profiling analysis (Wagner et al., 2003; Niehl et al., 2006) was performed essentially as described previously using the total dried metabolite preparation of each pool. The reagent volumes were reduced to 3 μl of 40 mg ml−1 methoxyamine hydrochloride in pyridine and 9 μl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA). Sample tubes were carefully manually agitated to allow full access of the reagents to the dried metabolite extracts. GC-TOF-MS profiling was performed using a 1 μl injection onto a FactorFour VF-5ms capillary column, 30 m length, 0.25 mm inner diameter, 0.25 μm film thickness with a 10 m EZ-guard precolumn (Varian BV, Middelburg, Netherlands), and an Agilent 6890N24 gas chromatograph with splitless injection and electronic pressure control (Agilent, Böblingen, Germany) mounted to a Pegasus III time-of-flight mass spectrometer (LECO Instrumente GmbH, Mönchengladbach, Germany). Chromatograms were manually assessed after mass spectral deconvolution (ChromaTOF software version 1.00, Pegasus driver 1.61, LECO, St Joseph, MI). Peak height of at least three mass fragments representing each analyte was normalized using the number of cells per sample and internal standardization by U-13C-sorbitol. Retention indices (RIs) were calculated from standard additions of a mixture of C12, C15, C19, C22, C32, C36 n-alkanes to each chromatogram (Strehmel et al., 2008). Tagfinder software was applied for the above chromatography data processing (Lüdemann et al., 2008).
Identification of metabolites within GC-TOF-MS metabolite profiles
Metabolites were identified using the NIST05 mass spectral search and comparison software (National Institute of Standards and Technology, Gaithersburg, MD, USA; http://www.nist.gov/srd/mslist.htm) and the mass spectral and retention time index (RI) collection (Schauer et al., 2005) of the Golm Metabolome Database (GMD; Kopka et al., 2005). Mass spectral matching was manually supervised and matches accepted with thresholds of match >650 (with maximum match equal to 1000) and RI deviation <1.0 %. Compounds, which could be matched to an as yet unidentified mass spectral and RI library entry, are named mass spectral tags (MSTs). Information on the mass spectral and retention index properties of MSTs may be retrieved form the web interface of GMD (Kopka et al., 2005) using the MIP-MP ID identifiers provided in Table 1 (see Supplementary Table S1 at JXB online).
Table 1.
Loading analysis of IC1 and IC2 and corresponding metabolites as shown in Fig. 4B
| Metabolite | MPI-MP ID | Loadings | IC | |
| A | A151002 | –0.0022 | IC1 | |
| B | DL-Glutamine | A178001 | –0.0018 | IC1 |
| C | Dehydroascorbic acid dimer | A185002 | 0.0013 | IC2 |
| D | DL-Isoleucine | A132002 | –0.0013 | IC1 |
| E | Xylitol | A171001 | –0.0013 | IC1 |
| F | DL-Asparagine | A168001 | 0.0013 | IC2 |
| G | DL-Tyrosine | A194002 | –0.0012 | IC1 |
| H | DL-Alanine, 3-cyano- | A138005 | –0.0011 | IC1 |
| I | DL-Phenylalanine | A157001 | –0.0010 | IC1 |
| J | A304001 | –0.0010 | IC1 | |
| K | DL-Threonine | A140001 | –0.0010 | IC1 |
| L | DL-Asparagine | A168001 | –0.0010 | IC1 |
| M | DL-Phenylalanine | A164001 | –0.0008 | IC2 |
| N | Ascorbic acid | A195002 | 0.0008 | IC1 |
| O | α,α′-Trehalose | A274002 | 0.0007 | IC1 |
| P | Citric acid | A182004 | 0.0007 | IC1 |
Metabolites, which influenced IC1 and IC2 above a threshold of 0.1 or below –0.1 are listed. A and J represent non-identified metabolites (cf. Supplementary Table S1 at JXB online).
Independent component analysis and statistical assessment of GC-TOF-MS data
Independent component analysis (ICA; Scholz et al., 2004) was applied to the metabolite fingerprints, i.e. all mass spectral information available prior to metabolite identification. Arbitrary ion currents of each observed mass fragment were normalized by the calculation of response ratios using the median of all samples as denominator and subsequent logarithmic transformation. Missing value substitution was as described earlier (Scholz et al., 2005). ICA is publicly available through the MetaGenAlyse web service (Daub et al., 2003; http://metagenealyse.mpimp-golm.mpg.de). Statistical testing was performed using Student's t test. Logarithmic transformation of response ratios approximated required Gaussian normal distribution of metabolite profiling data.
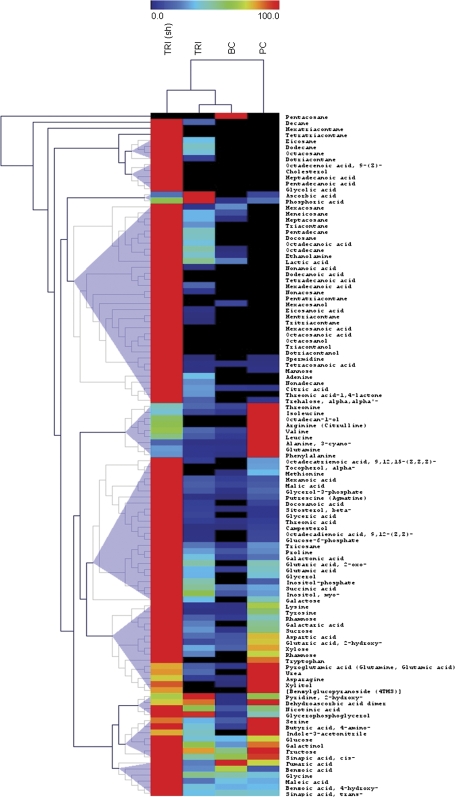
Graphical visualization of the metabolite heat map and hierarchical clustering (HCA) was performed using the software package TMEV (Saeed et al., 2003). HCA was based on the Euclidian distance measure applied to the maximum normalized averaged metabolite profiles (n=5–6). Tree generation and cluster formation was according to the complete linkage method (cf. Fig. 3).
Fig. 3.
Heat map visualisation of relative differences in metabolite pools between different epidermal cells. Data were maximum normalized (cell types with the highest pool size were set to 100%). Each cell type is visualized in a single column (average of n=5–6) and each metabolite is represented by a single row. Red indicates high abundance, whereas low relative metabolites are deep blue (cf. scale above heat map). Samples and metabolites were submitted to the hierarchical clustering using Euclidian distance (metabolite clusters with similar cellular profiles are indicated by triangle shading). Trichomes shaved by LHD [TRI (sh)] and micro sampled trichomes (TRI), basal cells (BC), and pavement cells (PC).
Estimation of metabolite pool sizes
To estimate the absolute concentrations of subsets of metabolites within single trichome cells, two standard calibration stocks were produced. Stock solution 1 contained 4 ng α-α trehalose (CAS 6138-23-4; Sigma T 9531; gmd-nr 2802); 4 ng saccharose (CAS 57-50-1; Supelco 4-7289; gmd-nr 523/1689); 4 ng myo-inositol (CAS 87-89-8; Sigma I 5125; gmd-nr 105), 4 ng citric acid (CAS 77-92-9; Sigma C 4540; gmd-nr 112/598); and 16 ng glutamic acid (CAS 56-86-0; Sigma G 1251; gmd-nr 688/687). Stock solution 2 contained 12 ng β-sitosterol (CAS 83-46-5; Sigma S 1270; gmd-nr 1711), 4 ng octacosanoic acid (CAS 506-48-9; Sigma O 4004; gmd-nr 978); 4 ng tetradecanoic acid (CAS 544-63-8; Sigma M 3128; gmd-nr 814); 4 ng cholesterol (CAS 57-88-5; Sigma C 8667; gmd-nr 760/761), and 8 ng tetratriacontane (CAS 14167-59-0; Fluka 88152; gmd-nr 527). These stock solutions were dissolved in a solvent solution composed of MeOH/CHCL3/H2O (2.5/1/1 by vol.), containing the internal standard ribitol. This ribitol concentration matched the one used in our trichome samples. From these calibration stocks the following dilution series were aliquoted: 1/1; 1/2; 1/4; 1/6; 1/8; 1/10; 1/12; 1/14; 1/16; 1/18; 1/20. Derivatization prior to GC-MS was performed as described earlier for the biological samples. The entire set of dilution series was measured by GC-MS and calibration curves were produced from the chromatograms. These calibration curves served to determine the absolute concentration of the selected metabolites in our trichome samples. Based on the molecular weight of the metabolites, upper concentration limits of selected metabolites were estimated which were within the picogram range in individual trichome cells (cf. Table 4). These apparent concentration limits were considered to represent rough approximations as most metabolite levels were near the analytical detection limit.
Table 4.
Estimation of the upper concentration limits of selected metabolite pool sizes in trichomes
| Metabolite | Upper concentration limit per single cell (pmol cell−1) |
| Sucrose | ∼ 0.7 |
| Trehalose | ∼ 0.7 |
| myo-Inositol | ∼ 1.2 |
| Citric acid | ∼ 1.1 |
| Glutamic acid | ∼ 5.5 |
| Sitosterol | ∼ 1.5 |
| Octacosanoic acid | ∼ 0.5 |
| Tetradecanoic acid | ∼ 2.1 |
| Cholesterol | ∼ 0.5 |
| Tetratriacontane | ∼ 1.1 |
Metabolite quantities of single trichomes harvested by LHD were estimated using standard calibration curves (n=10 pools of cells). Abundant metabolites which exhibited differential accumulation in trichome cells compared to epidermal pavement or basal cells were chosen.
Environmental Scanning Electron Microscopy (ESEM)
Arabidopsis leaves were fixed on the sample holder of the environmental scanning electron microscope Quanta 600 FEG (FEI Europe). Uncoated surfaces of leaves were investigated under ESEM vacuum mode conditions of the microscope at a low temperature of 5 °C and at a pressure of 865 Pa, where no drying processes of the samples takes place.
Results
GC-TOF-MS analysis applied to extracted pools of single cell samples yielded 117 identified metabolites among all tested cell types (Fig. 1; see Supplementary Table S1 at JXB online).
In addition, 23 metabolites were detected, which are currently not yet annotated or classified. All metabolite observations were carefully compared to non-sample experiments to control the complete sampling and analysis procedure. After qualitative and quantitative correction for laboratory contaminations, which considered sample concentration factors, 90 metabolites were found not to be present in non-sample control experiments and 27 further metabolites were accessible after quantitative background correction.
Estimation of replicate and global experimental variability
It was subsequently estimated if cell types could be distinguished by their respective metabolic phenotype. Visualization of differential cell-specific metabolite profiles was performed by hierarchical cluster analysis (HCA; Fig. 3) and independent component analysis (ICA; Fig. 4) (Scholz et al., 2004).
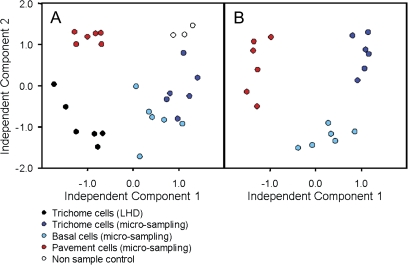
Fig. 4.
(A, B) Independent component analysis. Independent component one (IC1) is plotted against independent component two (IC2). (A) The complete dataset is visualized; whereas (B) shows only the samples obtained by microsampling. The metabolite profiling dataset was a fingerprint of all observed mass fragments, which was normalized and transformed to represent log10 of response ratios prior to independent component analysis (cf. methods section).
HCA (Fig. 3) provides a clear separation between the sampling methods, LHD and microsampling. Furthermore, several metabolites cluster in a characteristic manner associated with the analysed individual epidermal cell types; and the close relation between trichomes and basal cells, in contrast to the clear diversity from pavement cells (Fig. 3), became apparent using HCA.
Independent component analysis was chosen as it represents a non-supervised approach, which is not biased by knowledge of sample classification and choice of metabolite target. The so-called scores plot using independent component 1 (IC1) versus IC2 for visualization demonstrated distinct metabolite phenotypes of the different cell types as well as clear separation from non-sample controls and LHD trichome preparations (Fig. 4A). Figure 4B depicts the metabolic differentiation according to the microcapillary sampling method only. IC1 of this analysis clearly separated trichomes from pavement cells whereas IC2 represents metabolic parameters which separate basal cells from both trichome and pavement cells.
To investigate further which metabolites may account for the distinction of the cell types, sampling methods and also non-sample controls, the influence values, so-called loadings of IC1 and IC2 were plotted against the corresponding retention-time indices of the original metabolite data (see Supplementary Fig. S2 at JXB online). The loading for each component was calculated using the MetaGenAlyse software (Daub et al., 2003; http://metagenealyse.mpimp-golm.mpg.de). Extreme loading values indicate a strong contribution of this feature to the sample separation encoded into each respective IC. The metabolites which were predominant for the separation observed by IC1 and IC2, respectively, were glutamine, isoleucine, xylitol, tyrosine, 3-cyano alanine, phenylalanine, threonine, ascorbic acid, α,α-trehalose, and citric acid. The main contributions to the sample separation described by IC2 were dehydroascorbic acid, asparagine, and phenylalanine (see Supplementary Fig. S2 at JXB online; Table 1).
When analysing the dataset of all the metabolites identified in this study it became apparent that the most complete set was detectable in trichomes collected by leaf hair depilation (LHD). A comparison of metabolites abundant in microsampled trichome cell sap with metabolites detected in complete trichomes (LHD) revealed several compounds with similar abundance (see Supplementary Fig. S3 at JXB online) as was expected, comparing equivalent samples collected either with or without apoplast and cuticula. Moreover, in addition to metabolites having similar abundance in trichomes, using both alternative sampling methods, notable differences were found (Table 2; Fig. 3).
Table 2.
Metabolite distribution in trichomes obtained by LHD in comparison to micro sampled trichome cell sap (n=6 pools of cells)
| Metabolite | Trichomes (LHD) (Normalized response) |
Trichomes (microsampling) (Normalized response) |
Comparison |
|||
| Name | AVG | Count (%) | AVG | Count (%) | P | Ratio |
| Alkane | ||||||
| Nonacosane | 0.0214 | 100 | 0.0066 | 100 | 0.009 | 3.231 |
| Hentriacontane | 0.1232 | 100 | 0.0106 | 100 | 0.002 | 11.609 |
| Tritriacontane | 0.0245 | 100 | 0.0013 | 100 | 0.003 | 18.616 |
| Pentatriacontane | 0.0027 | 100 | 0.0004 | 100 | 0.006 | 6.661 |
| Fatty acids | ||||||
| Hexanoic acid | 0.0020 | 100 | 0.0003 | 50 | 0.035 | 6.942 |
| Nonanoic acid | 0.0009 | 83 | 0.0003 | 100 | 0.002 | 2.773 |
| Tetradecanoic acid | 0.0177 | 83 | 0.0080 | 100 | 0.046 | 2.226 |
| Docosanoic acid | 0.0016 | 83 | 0.0006 | 100 | 0.022 | 2.683 |
| Tetracosanoic acid | 0.0060 | 83 | 0.0009 | 100 | 0.004 | 6.836 |
| Hexacosanoic acid | 0.0086 | 83 | 0.0003 | 83 | 0.001 | 26.068 |
| Octacosanoic acid | 0.0021 | 83 | n.d.a | 0 | Absent | |
| Fatty alcohols | ||||||
| Octadecan-1-ol | 0.0016 | 100 | 0.0009 | 100 | 0.025 | 1.915 |
| Hexacosanol | 0.0003 | 100 | n.d. | 0 | Absent | |
| Octacosanol | 0.0012 | 100 | n.d. | 0 | Absent | |
| Triacontanol | 0.0013 | 100 | n.d. | 0 | Absent | |
| Dotriacontanol | 0.0015 | 100 | n.d. | 0 | Absent | |
| Lipids | ||||||
| Cholesterol | 0.0026 | 100 | 0.0006 | 100 | 0.000 | 4.018 |
| Campesterol | 0.0051 | 100 | 0.0001 | 17 | 73.026 | |
| β-Sitosterol | 0.0181 | 100 | 0.0011 | 100 | 0.000 | 16.714 |
| α-Tocopherol | 0.0003 | 83 | n.d. | 0 | Absent | |
| Amino acids | ||||||
| 3-Cyano-alanine | 0.0030 | 100 | 0.0004 | 67 | 0.020 | 7.211 |
| Aspartic acid | 0.0128 | 100 | 0.0026 | 100 | 0.036 | 4.920 |
| Asparagine | 0.0414 | 83 | 0.0029 | 83 | 0.009 | 14.188 |
| Glutamic acid | 0.0185 | 83 | 0.0064 | 100 | 0.008 | 2.919 |
| Valine | 0.0044 | 100 | 0.0017 | 100 | 0.040 | 2.646 |
| Isoleucine | 0.0192 | 83 | 0.0046 | 100 | 0.018 | 4.197 |
| Acids | ||||||
| Fumaric acid | 0.0252 | 83 | 0.0064 | 83 | 0.004 | 3.940 |
| Maleic acid | 0.0156 | 83 | 0.0047 | 67 | 0.038 | 3.338 |
| Malic acid | 0.0346 | 83 | 0.0057 | 83 | 0.033 | 6.107 |
| 2-Oxo-glutaric acid | 0.0006 | 50 | 0.0003 | 83 | 0.022 | 2.170 |
| 2-Hydroxy-glutaric acid | 0.0016 | 100 | 0.0003 | 100 | 0.019 | 5.118 |
| Polyhydroxy acids | ||||||
| Glyceric acid | 0.0116 | 83 | 0.0011 | 83 | 0.012 | 10.282 |
| Threonic acid | 0.0075 | 83 | 0.0004 | 83 | 0.000 | 19.744 |
| Galactonic acid | 0.0046 | 83 | 0.0017 | 100 | 0.020 | 2.718 |
| Polyols | ||||||
| Xylitol | 0.0119 | 100 | 0.0005 | 83 | 0.000 | 25.883 |
| myo-Inositol | 0.0032 | 100 | 0.0019 | 100 | 0.039 | 1.652 |
| Sugars | ||||||
| Xylose | 0.0017 | 100 | 0.0005 | 67 | 0.003 | 3.195 |
| Rhamnose | 0.0026 | 100 | 0.0005 | 83 | 0.012 | 5.347 |
| Glucose | 0.0025 | 83 | 0.0009 | 50 | 0.032 | 2.642 |
| Sucrose | 0.0039 | 100 | 0.0016 | 100 | 0.039 | 2.444 |
| α,α'-Trehalose | 0.0120 | 100 | 0.0019 | 100 | 0.042 | 6.314 |
| Phenylpropanoids | ||||||
| cis-Sinapic acid | 0.0011 | 83 | 0.0005 | 100 | 0.000 | 2.076 |
| trans-Sinapic acid | 0.0063 | 83 | 0.0022 | 100 | 0.001 | 2.849 |
| MSTsb | ||||||
| 0.0179 | 100 | 0.0014 | 100 | 0.000 | 12.349 | |
| 0.0012 | 100 | n.d. | 0 | Absent | ||
| 0.0015 | 83 | n.d. | 0 | Absent | ||
| 0.0103 | 100 | 0.0017 | 83 | 0.027 | 5.949 | |
| 0.0113 | 83 | 0.0022 | 100 | 0.007 | 5.054 | |
| 0.0015 | 100 | 0.0006 | 100 | 0.005 | 2.627 | |
| 0.0032 | 83 | 0.0008 | 100 | 0.003 | 4.029 | |
| 0.0024 | 83 | 0.0006 | 100 | 0.001 | 3.838 | |
| 0.0055 | 100 | n.d. | 0 | Absent | ||
| 0.0016 | 100 | 0.0005 | 83 | 0.001 | 3.456 | |
| 0.0007 | 100 | 0.0003 | 100 | 0.008 | 2.393 | |
| 0.0005 | 100 | 0.0001 | 100 | 0.001 | 3.591 | |
| 0.0023 | 100 | n.d. | 0 | Absent | ||
| 0.0043 | 100 | 0.0006 | 100 | 0.005 | 6.679 | |
n.d. not detectable.
MSTs, mass spectral tags.
Metabolites such as the fatty acid octacosanoic acid and the fatty alcohols hexacosanol, octacosanol, triacontanol, and dotriacontanol were only detected in trichomes harvested by LHD (Fig. 3; Table 2). Furthermore, the lipid component α-tocopherol could not be identified in trichome cell sap in contrast to complete trichomes (Table 2). To get an overview about the differences in the metabolic content between trichome samples obtained by LHD in contrast to microcapillary sampled trichomes, Student's t test (Table 2) was applied. Comparing the different sampling methods it becomes clear that a larger number of metabolites was accessed with the LHD sampling. Also, this sampling method allows the detection of higher apparent quantities of metabolites (Table 2; Fig. 3).
A detailed investigation of the metabolite occurrence within samples obtained by microcapillary-based collection revealed various metabolites that could be detected in specific microsampled cell types only. In particular, when comparing pavement cells to microsampled trichome and basal cells it became obvious that several metabolites were found above detection limits exclusively in pavement cells, namely arginine, tryptophan, α-tocopherol, and benzylglucopyranoside. Threonic acid-1,4-lactone was only detectable in trichome cell sap, whereas no exclusive metabolic marker could so far be identified in basal cells. Comparisons of the whole datasets indicated that 27 annotated metabolites in this study showed significant differences in terms of abundance in the investigated cell types.
Application of Student's t test provided additional information on significant differences in the metabolic content of cell types collected by microcapillaries (Table 3; Figs 3, 5). In particular, amino acids such as serine, 3-cyano-alanine, asparagine, 4-aminobutyric acid, threonine, pyroglutamic acid, glutamic acid, valine, isoleucine, leucine, and lysine exhibited highly elevated levels in pavement cells in contrast to trichome and basal cells (Figs 3, 5). Furthermore, the same tendencies in amino acid abundance were observed when comparing trichome and pavement cells with basal cells. Nearly all the above-mentioned metabolites displayed significantly increased levels in trichomes and pavement cells in comparison to basal cells.
Table 3.
Comparative analysis of metabolites detected in microcapillary sampled trichome cell sap (TRI), pavement cells (PC) and basal cells (BC)
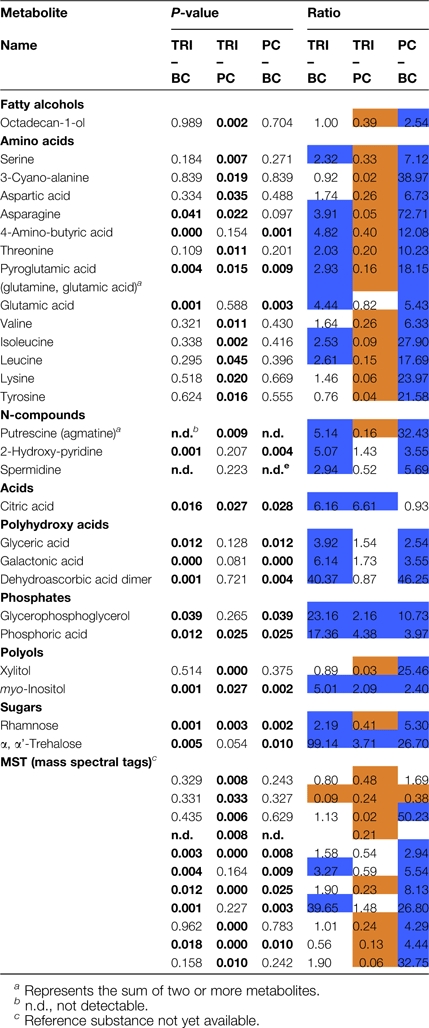 |
Fig. 5.
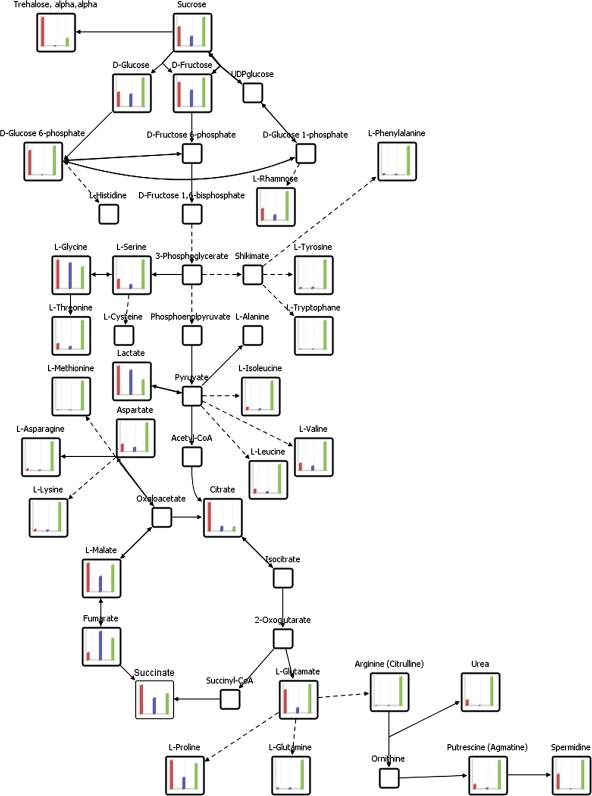
Comparative visualization of central metabolism as determined by metabolic profiling of trichomes, basal and pavement cells collected by micro sampling. The pathway connectivity and bar diagram visualisation of relative metabolite pool sizes (cf. Fig. 3) was performed using the software package VANTED (Junker et al., 2006). Empty squares indicate relevant central metabolites which were not detectable by GC-MS based profiling of single cell types. From left to right: trichomes, red; basal cells, blue; pavement cells, green.
Higher levels of N-compounds like putrescine, agmatine, spermidine, and 2-hydroxy-pyridine were detected in trichomes and in pavement cells when compared to basal cells. However, only for putrescine was a significant elevation of 6.25-fold in pavement cells shown in contrast to trichome cell sap (Table 3; Fig. 5). Furthermore, the fatty alcohol octadecan-1-ol was significantly more abundant in pavement cells when compared to trichomes and basal cells, whereas no difference for this metabolite was found between trichome and basal cells (Table 3). Similarly, in comparison to basal cells, polyhydroxy acids also showed significantly enhanced quantities in trichomes and pavement cells for which up to 46-fold higher levels could be verified for a dehydroascorbic acid. Highly elevated citric acid levels were determined in trichomes compared to basal and pavement cells. The citric acid content of microsampled trichomes was 6.16-fold and 6.61-fold elevated compared to basal cells and pavement cell respectively (Table 3; Fig. 5). Phosphates, like glycerophosphoglycerol and phosphoric acid (Table 3), were also found to a higher degree in trichomes when compared to pavement and basal cells as well as in pavement cells compared to basal cells. The polyols, xylitol and myo-inositol, however were present in different quantities in the cell types analysed. It is noteworthy that xylitol was present at similar levels in both trichomes and basal cells but displayed a 33-fold higher occurrence in pavement cells compared to trichomes and a 25-fold increased level compared to basal cells. By contrast, myo-inositol was found to exhibit 5-fold higher levels in trichomes than in basal cells and 2-fold higher levels than in pavement cells. A 2.4-fold elevation was observed when comparing the contents of myo-inositol in pavement cells to basal cells.
Different amounts of the sugar α,α-trehalose were found among the three cell types analysed. Trichomes and pavement cells accumulate enhanced amounts of this sugar when compared to basal cells. In trichomes, a 3.71-fold higher α,α-trehalose level could be measured compared to pavement cells (Table 3, Fig. 5) while the opposite trend was found for rhamnose. Again rhamnose and α,α-trehalose were less abundant in basal cells compared to the other cell types analysed.
With respect to as yet unknown metabolites, one seems particularly interesting as it exhibits an 11-fold increase in basal cells when compared to trichomes and 2.6-fold elevated levels in basal cells compared to pavement cells (Table 3). Identification of this metabolite promises to provide a metabolic marker for this cell type.
A summary of metabolic trends observed within the three epidermal cell types is schematized in a condensed pathway diagram (Fig. 5). Components of central metabolic pathways such as the TCA cycle, amino acid biosynthesis, carbohydrate metabolism, and nitrogen metabolism are linked in a functional context. Together, this visualization of metabolic pools underlines biochemical pathway specification between pavement, basal, and trichome cells of Arabidopsis thaliana.
Limitations to estimation of metabolite pool sizes
Metabolite profiling approaches typically allow the multi-parallel and non-targeted screening of metabolite pools relative to a defined reference sample. These screens can be performed using an internal standard for the correction of technical errors or, alternatively, by performing normalization to the sum of all the observed metabolites. This latter case is typically less preferred as the normalization by sum of all observed metabolites provides information on metabolite composition rather than the preferred information on the relative change of metabolite pool sizes. Typically, the exact quantification is deemed unnecessary for the discovery of metabolic processes defining genotypic or environmental acclimation effects (Fiehn et al., 2000; Roessner et al., 2000). However, the intention here was to investigate the limitations of exact quantification by routine external calibration curves. These curves were used to standardize the quantitative GC-TOF-MS response of relevant differentially accumulated metabolites from the previous investigations, such as sucrose, α,α-trehalose, citric acid, and myo-inositol (Table 4). Initial attempts to inject stable isotope-labelled internal standards into each cell and subsequent extraction into the same or a second capillary proved not to be feasible. Furthermore, infusion of an internal standard similar to the application of fluorescent dyes required extended diffusion times and, therefore, was judged prone to wounding and delayed metabolic inactivation artefacts. In conclusion, currently no method is available to quantify volume recovery from microsampled cells. Moreover, the qualitative visual inspection of cell ghosts after sampling did not yield indications of residual internal structures, such as nuclei or chloroplasts, or of variable behaviour between cell types. We therefore estimated the identical recoveries or volume losses of all three different cell types by analysing the volume of sample in the capillary after sampling in three repetitions for each cell type, changing the capillary for each repetition. Thus, the internal standard was administered after microsampling to monitor any residual recovery losses. As attempts to elucidate cellular metabolite concentrations would require estimations of both cell volume and recovery during sampling, it was felt justified to suggest the traditional profiling as described above as an advisable routine approach.
In the following, it was tested if pools of 100–1000 LHD sampled trichomes (n=10) may be amenable to the exact quantification of metabolites. Replicate preparations of replicate cell numbers and external calibration series of the selected metabolites (Table 4) were used. Due to the high standard deviation of LHD trichome samples, single replicates frequently tend to fall below the detection limits and, because of the requirement to estimate the number of trichome cells after depilation, it was decided to estimate the upper concentration limits based on 800–1000 cells per pool. According to the lower concentration limit of our quantitative calibration samples, metabolites may not amount to more than 0.5–5.5 pmol per trichome cell, for example 0.5 pmol cell−1 of octacosanoic acid or 5.5 pmol cell−1 of glutamic acid. In conclusion, it was extrapolated that exact quantification will ideally require pools of 2 000–5 000 cells, at least 10-fold more than was required for the profiling of relative pool sizes following the conventional metabolite profiling concept. By contrast, metabolite profiling of relative pool sizes was already feasible using microsamples of 200 single cells as described within the Materials and methods section.
Discussion
To investigate the metabolic status of specific Arabidopsis thaliana epidermal cell types, pools of single cell extracts collected from trichome, basal, and pavement cells via microcapillaries (Kehr, 2001, 2003; Tomos and Sharrock, 2001) were analysed. Hitherto, this is the only direct method which allows sample collection with the highest spatial resolution for subsequent analysis. Alternative methods, like laser microdissection and fluorescence activated cell sorting, enable the collection of homogeneous cell populations in a large-scale range; however, these methods have to deal with the inevitable tissue preparation procedures before collecting and analysing cell materials (Kehr, 2003). Due to these limitations, single cell sampling, using microcapillaries, still provides the only applicable method to analyse cell extracts from living plants in an unbiased approach.
GC-TOF-MS was successfully applied to pooled single cell extracts, which, in contrast to other metabolite analytical methods, enabled the identification of 117 compounds within single epidermal cell types. Previously, several approaches have been developed for the investigation of solute concentrations on the single cell level. These comprised ion-selective microelectrodes (Felle, 1993), energy dispersive X-ray analysis (Williams et al., 1993) of frozen tissue sections, or extracted vacuolar sap and microfluometric enzymatic assays (Fricke et al., 1995). However, these methods are restricted to the analysis of a small number of ions. Parallel detection of four anions or five cations became available by the application of capillary electrophoresis (Bazzanella et al., 1998). In contrast to the GC/TOF-MS approach described in the present work, the capillary electrophoresis approach enabled subsamples of the content that was collected from one single cell to be analysed (Bazanella et al., 1998). To this end, our present study demonstrates that metabolite analyses at the single cell level requires merging of individually collected single cell extracts, when a high number of metabolites is targeted.
The routine GC-TOF-MS metabolite profiling technology is reported to have fair analytical reproducibility, 2–12% relative standard deviation (RSD), compared to a typical biological variability of 17–56% RSD observed when analysing whole rosettes of Arabidopsis plants (Fiehn et al., 2000) As the challenge of analysing single cell types may introduce a higher degree of variability, the overall precision of our GC-TOF-MS measurements of different cell types was assessed by calculating the RSDs of each cell type and mode of preparation, respectively. Using a pooling strategy, namely by combining multiple single cell samples from different plants, the expected increase of experimental error compared to bulk tissue analysis was negated (Fiehn et al., 2000; Roessner et al., 2000). Comparing previous reports on the biological variation of such bulk samples, it is possible to demonstrate that many of our single cell analyses were within or close to the previously reported range of 17–56% RSD indicating that it is possible to achieve a reproducibility comparable to other published studies. It was also possible to demonstrate that, in our hands, microcapillary-based sampling yielded more precise results in terms of reproducibility of obtained metabolite measurements compared to trichome samples collected by LHD.
To visualize differences of the particular cell types, as well as from the diverse sampling methods used in this approach, independent component analysis (ICA) and hierarchical clustering analysis (HCA) were employed. Applying independent component analysis to our metabolite dataset proved that the experimental error, as estimated by replicate analysis of the same cell type, was substantially smaller than the differentiation of metabolic phenotypes between cell types. Thus it is clearly demonstrated that microsampling linked to GC-TOF-MS profiling represents a valid and feasible method to explore the cellular physiology of epidermal tissue (Fig. 4A).
Moreover, ICA demonstrated that the metabolite profiles of trichome, basal, and pavement cells are specific and can be distinguished by their particular metabolite composition (Fig. 4B). Independent components one and two have a great influence on the separation of the cell types analysed. The loading values for each component were calculated and thus depict the influence of each mass on each of the components (see Supplementary Fig. S2 at JXB online). Furthermore, our analysis revealed the expected and clearly demonstrated insight that cell types have differentiated metabolic properties, which can now be characterized further and utilized for enhanced insight into the cell specificity of expressed genes.
The clear separation of cell types, but also sampling methods for trichomes, which was obtained by ICA, could be confirmed by using HCA. Both means of data mining clearly demonstrated the characteristic differences in the metabolic phenotype of specialized leaf epidermal cells, trichome isolation methods, and the close relationship between trichomes and its adjacent basal cells (Fig. 3).
Metabolite analysis of trichomes isolated by LHD
The heat map (Fig. 3) demonstrates that trichome samples which were collected by LHD contain nearly all metabolites that could be measured in this study and also the highest metabolic pool sizes were observed in these samples. Complete trichomes as harvested by LHD are comprised of cell sap plus membrane and cell wall structures. Thus compound classes such as alkanes, fatty acids, fatty alcohols, and lipids, which are associated with the cell membrane and cell wall, were detected only in LHD extracts. (Fig. 3; Table 2). Besides differences in metabolite concentrations caused by the harvesting protocols, identical metabolite concentrations were revealed for several substances. This congruence in metabolite levels confirmed the validity of our sampling protocols.
Metabolite analysis of trichomes, basal, and pavement cells collected using microsampling
In general, pavement and trichome cells exhibit a significantly higher abundance of several metabolites than basal cells (Fig. 5). Basal cells seem to have an overall lower metabolite content than the other cell types analysed. Only a few metabolites were detectable and accumulated to greater pool sizes in this specific cell type (Fig. 3; Table 3). Among these metabolites are glycine, lactate, and fumarate (Fig. 5). Taking into account the spatial localization and also the close relation observed between trichomes and basal cells using HCA, it is conceivable that a basal cell itself might not be highly metabolically active but rather serves as a transfer cell to and from trichomes (Fig. 1).
Overall, a clear differentiation between trichomes, basal, and pavement cells could be observed (Figs 3, 4). Visualization of the primary metabolism comprising TCA, amino acid biosynthesis, and sugar metabolism demonstrated in most cases the highest activity in pavement cells. However, diverse compounds, for example organic acids, were highly abundant in trichomes.
Metabolites with significant higher pool sizes in pavement cells
Amino acids observed in this study were abundant in all cell types. However, it can be noted that pavement cells seemed to accumulate amino acids to a higher extent and nearly all the amino acids identified were evidently more highly abundant in pavement cells than in trichome or basal cells (Table 3). Therefore, a conclusion might be that pavement cells contain enhanced or activated amino acid biosynthesis pathways or that they import amino acids from other tissues. For example, tryptophan and the sum of arginine and citrulline represented by a common chemical derivative were exclusively found in this cell type (Fig. 5).
Amines were mainly associated with pavement cells. The polyamine spermidine was detectable in all pavement cell measurements. By contrast, the same compound is found above the detection limit in only 50% of the trichome cell pools and is even less frequent, namely only in one-third of the basal cell measurements (see Supplementary Table S1 at JXB online). The calculated ratio showed a 1.9-fold increased occurrence in pavement cells compared to trichome cells (see Supplementary Table S1 at JXB online). The diamine putrescine was 6.25-fold significantly higher in pavement cells in comparison to trichome cell sap (Table 3). In addition, a chemical derivative representing the sum of arginine and citrulline, the precursor of the two synthesis pathways of putrescine, was only measurable in pavement cells (Fig. 5). In the plant kingdom, polyamines like spermidine are ubiquitously found together with their diamine precursor putrescine. Moreover, they have been suggested to be involved in biotic and abiotic stress responses (Torrigiani et al., 1997; Walters, 2000, 2003).
Rhamnose was detectable at a significantly 2.4-fold increased amount in pavement cells in comparison to trichome cells and was present in 5.3-fold higher levels in pavement compared to basal cells (Table 3). Aside from being present in the pectic polysaccharides rhamnogalacturonan I and II (Diet et al., 2006), rhamnose is also present in a variety of secondary metabolites including flavonoids, anthocyanins or triterpenoids (Barber and Neufeld, 1961; Bar-Peled et al., 1991; Watt et al., 2004). In this context, it is conceivable that rhamnose accumulates in pavement cells due to its association with secondary metabolites which are known to accumulate in pavement cells.
Metabolites with significant higher pool sizes in trichomes
Citric acid revealed significantly enhanced levels in trichome cells when compared to pavement (6.6-fold) and basal cells (6.2-fold) (Table 3; Fig. 5). Citric acid is involved in diverse pathways such as the TCA, the glyoxylate cycle, and acetyl-CoA biosynthesis. Moreover, organic acids such as citric acid function as metal chelators in detoxification processes (Callahan et al., 2006) thus providing a direct link to citric acid elevation in trichomes. Trichomes can accumulate heavy metals and act as sinks for toxic molecules (Gutiérrez-Alcalá et al., 2000; Domínguez-Solís et al., 2001) which thereby might explain trichome-enriched citric acid contents.
Trehalose, was significantly more abundant in trichome cells (Table 3; Fig. 5) when compared to pavement (3.71-fold) and basal cells (99-fold). Besides its role as a storage carbohydrate and transport sugar, trehalose has been associated with the stress protection machinery (Wiemken, 1990; Crowe et al., 1998). The precursor trehalose-6-phosphate seems to play an important role as a signalling molecule and appears to regulate sugar metabolism (Eastmond et al., 2003). Recently, it was demonstrated that the trehalose-6-phosphate synthase/phosphatase (AtTPS6), beyond a role in development, also regulates plant architecture, the shape of pavement cells, and the branching of trichomes (Chary et al., 2008).
Estimation of metabolite pool sizes
Quantification of selected compounds was performed to estimate the concentration of metabolites that are highly abundant in complete trichomes or in trichome cell sap in comparison to pavement cell sap. Up to now, only a few studies have addressed the quantification of metabolite concentrations in single cells (Fricke et al., 1994; Kehr et al., 1999; Tomos and Sharrock, 2001). Most of these studies focused on the quantification of sugars and amino acids and used methods such as fluorescent microscope photometry or capillary electrophoresis. In this study, GC-TOF-MS analysis of complete trichomes and quantified selected compounds was combined using calibration curves (Table 4). Thus this approach provides another possibility of metabolite quantification at the single cell level. The concentrations of sucrose and trehalose which was found in complete trichomes are comparable with the concentrations for hexose equivalents in epidermal cells detected by Kehr et al. (1999) using microfluorometry. Therefore, our GC-MS approach is consistent with previous studies that were based on a lower throughput scale.
Conclusion
This study demonstrates that single cell sampling techniques can be successfully combined with GC-TOF-MS profiling analyses to obtain highly spatially resolved insights into the composition of intercellular metabolite pools of specific epidermal cells. Indeed, metabolite profiling at the level of single cell types is now feasible for applications in gene function analysis. Compared to previous studies, it was possible to demonstrate clearly the metabolic differentiation among specialized epidermal cell types. The measured metabolite distributions provide the basis for subsequent generation and testing of hypotheses with respect to biological processes especially relevant in those cell types. Furthermore marker metabolites could be identified which can be used for cell-specific analysis.
Thus, the next step was taken on the clearly laborious path towards full transcript, protein, and metabolite profiling at the single in vivo cell level. In the long run, single cell analysis and technological development may lead to enhanced understanding of cell to cell communication and spatial aspects of plant physiology. We are convinced that cell-specific metabolic differentiation can now be explored at the molecular and functional genomics levels.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Metabolites identified in this study.
Supplementary Fig. S1. Average number of trichomes of 40-d-old Arabidopsis Columbia-0 wild-type leaves
Supplementary Fig. S2. Independent scores of IC1 and IC2 and corresponding retention time indices
Supplementary Fig. S3. Metabolites showing similar mean normalized responses in complete trichomes harvested by LHD and trichome cell sap collected by microsampling (n=6 pools of cells)
Supplementary Material
Acknowledgments
This study was supported by BMBF in the context of the German plant genomics program GABI (project No. 0312277D). We thank Gareth Catchpole and Leonard Krall for critical reading of the manuscript.
References
- Arlt K, Brandt S, Kehr J. Amino acid analysis in five pooled single plant cell samples using capillary electrophoresis coupled to laser-induced fluorescence detection. Journal of Chromatography A. 2001;926:319–325. doi: 10.1016/s0021-9673(01)01052-4. [DOI] [PubMed] [Google Scholar]
- Barber GA, Neufeld EF. Rhamnosyl transfer from TDPL-rhamnose catalyzed by a plant enzyme. Biochemical and Biophysical Research Communications. 1961;6:44–48. doi: 10.1016/0006-291x(61)90182-6. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Lewinsohn E, Fluhr R, Gressel J. UDP-rhamnose:flavanone-7-O-glucoside-2″- O-rhamnosyltransferase. Purification and characterization of an enzyme catalyzing the production of bitter compounds in citrus. Journal of Biological Chemistry. 1991;266:20953–20959. [PubMed] [Google Scholar]
- Bazzanella A, Lochmann H, Tomos AD, Bächmann K. Determination of inorganic cations and anions in single plant cells by capillary zone electrophoresis. Journal of Chromatography A. 1998;809:231–239. doi: 10.1016/s0021-9673(01)00627-6. [DOI] [PubMed] [Google Scholar]
- Bino RJ, Hall RD, Fiehn O, et al. Potential of metabolomics as a functional genomics tool. Trends in Plant Science. 2004;9:418–425. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiting in plants via cell sorting of protoplasts from fluorescent reporter lines. Nature Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. The Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt S, Kehr J, Walz C, Imlau A, Willmitzer L, Fisahn J. Technical advance: a rapid method for detection of plant gene transcripts from single epidermal, mesophyll and companion cells of intact leaves. The Plant Journal. 1999;20:245–250. doi: 10.1046/j.1365-313x.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- Brandt S, Kloska S, Altmann T, Kehr J. Using array hybridization to monitor gene expression at the single cell level. Journal of Experimental Botany. 2002;53:2315–2323. doi: 10.1093/jxb/erf093. [DOI] [PubMed] [Google Scholar]
- Brandt SP. Microgenomics: gene expression analysis at the tissue-specific and single-cell levels. Journal of Experimental Botany. 2005;56:495–505. doi: 10.1093/jxb/eri066. [DOI] [PubMed] [Google Scholar]
- Callahan DL, Baker AJ, Kolev SD, Wedd AG. Metal ion ligands in hyperaccumulating plants. Journal of Biological and Inorganic Chemistry. 2006;11:2–12. doi: 10.1007/s00775-005-0056-7. [DOI] [PubMed] [Google Scholar]
- Calo L, Garcia I, Gotor C, Romero LC. Leaf hairs influence phytopathogenic fungus infection and confer an increased resistance when expressing a Trichoderma α-1,3-glucanase. Journal of Experimental Botany. 2006;57:3911–3920. doi: 10.1093/jxb/erl155. [DOI] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiology. 2008;146:97–107. doi: 10.1104/pp.107.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM. The role of vitrification in anhydrobiosis. Annual Review of Physiology. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- Daub CO, Kloska S, Selbig J. MetaGeneAlyse: analysis of integrated transcriptional and metabolite data. Bioinformatics. 2003;19:2332–2333. doi: 10.1093/bioinformatics/btg321. [DOI] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C. The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-l-rhamnose synthase. The Plant Cell. 2006;18:1630–1641. doi: 10.1105/tpc.105.038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis JR, Gutierrez-Alcala G, Vega JM, Romero LC, Gotor C. The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. Journal of Biological Chemistry. 2001;276:9297–9302. doi: 10.1074/jbc.M009574200. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Li Y, Graham IA. Is trehalose-6-phosphate a regulator of sugar metabolism in plants? Journal of Experimental Botany. 2003;54:533–537. doi: 10.1093/jxb/erg039. [DOI] [PubMed] [Google Scholar]
- Esau K. Plant anatomy. New York: John Wiley & Sons, Inc; 1953. [Google Scholar]
- Farre EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L. Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiology. 2001;127:685–700. doi: 10.1104/pp.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH. Ion-selective microelectrodes their use and importance in modern plant cell biology. Botanica Acta. 1993;106:5–12. [Google Scholar]
- Fernie AR. Metabolome characterization in plant system analysis. Functional Plant Biology. 2003;30:111–120. doi: 10.1071/FP02163. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fricke W, Hinde P, Leigh RA, Tomos AD. Vacuolar solutes in the upper epidermis of barley leaves. Intercellular differences follow patterns. Planta. 1995;196:40–49. [Google Scholar]
- Fricke W, Pritchard J, Leigh RA, Tomos AD. Cells of the upper and lower epidermis of barley (Hordeum vulgare L.) leaves exhibit distinct patterns of vacuolar solutes. Plant Physiology. 1994;104:1201–1208. doi: 10.1104/pp.104.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover BJ. Differentiation in plant epidermal cells. Journal of Experimental Botany. 2000;51:497–505. doi: 10.1093/jexbot/51.344.497. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Alcala G, Gotor C, Meyer AJ, Fricker M, Vega JM, Romero LC. Glutathione biosynthesis in Arabidopsis trichome cells. Proceedings of the National Academy of Sciences, USA. 2000;97:11108–11113. doi: 10.1073/pnas.190334497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J. Determination of gamma-aminobutyric acid in microdialysis samples by microbore column liquid chromatography and fluorescence detection. Journal of Chromatography B Biomedical Sciences and Applications. 1998a;708:49–54. doi: 10.1016/s0378-4347(97)00657-9. [DOI] [PubMed] [Google Scholar]
- Kehr J. Determination of glutamate and aspartate in microdialysis samples by reversed-phase column liquid chromatography with fluorescence and electrochemical detection. Journal of Chromatography B Biomedical Sciences and Applications. 1998b;708:27–38. doi: 10.1016/s0378-4347(97)00677-4. [DOI] [PubMed] [Google Scholar]
- Kehr J. High resolution spatial analysis of plant systems. Current Opinion in Plant Biology. 2001;4:197–201. doi: 10.1016/s1369-5266(00)00161-8. [DOI] [PubMed] [Google Scholar]
- Kehr J. Single cell technology. Current Opinion in Plant Biology. 2003;6:617–621. doi: 10.1016/j.pbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Kehr J, Wagner C, Willmitzer L, Fisahn J. Effect of modified carbon allocation on turgor, osmolality, sugar and potassium content, and membrane potential in the epidermis of transgenic potato (Solanum tuberosum L.) plants. Journal of Experimental Botany. 1999;50:565–571. [Google Scholar]
- Kojima M, Poulton JE, Thayer SS, Conn EE. Tissue distributions of dhurrin and of enzymes involved in its metabolism in leaves of Sorghum bicolor. Plant Physiology. 1979;63:1022–1028. doi: 10.1104/pp.63.6.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krüger S, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Laval V, Koroleva OA, Murphy E, Lu C, Milner JJ, Hooks MA, Tomos AD. Distribution of actin gene isoforms in the Arabidopsis leaf measured in microsamples from intact individual cells. Planta. 2002;215:287–292. doi: 10.1007/s00425-001-0732-y. [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. The Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ma L, Zhang JF, Lu YT. Determination of single-cell gene expression in Arabidopsis by capillary electrophoresis with laser induced fluorescence detection. Journal of Chromatography B Analytical Technology and Biomedical Life Sciences. 2004;808:241–247. doi: 10.1016/j.jchromb.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Lüdemann A, Strassburg K, Erban A, Kopka J. TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC–MS)-based metabolite profiling experiments. Bioinformatics. 2008;24:732–737. doi: 10.1093/bioinformatics/btn023. [DOI] [PubMed] [Google Scholar]
- Ma JF. Role of organic acids in detoxification of aluminum in higher plants. Plant and Cell Physiology. 2000;41:383–390. doi: 10.1093/pcp/41.4.383. [DOI] [PubMed] [Google Scholar]
- Marks MD. Molecular genetic analysis of trichome development in Arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:137–163. doi: 10.1146/annurev.arplant.48.1.137. [DOI] [PubMed] [Google Scholar]
- Martin C, Glover BJ. Functional aspects of cell patterning in aerial epidermis. Current Opinion in Plant Biology. 2007;10:70–82. doi: 10.1016/j.pbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Nawrath C. Unraveling the complex network of cuticular structure and function. Current Opinion in Plant Biology. 2006;9:281–287. doi: 10.1016/j.pbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Niehl A, Lacomme C, Erban A, Kopka J, Krämer U, Fisahn J. Systemic Potato virus X infection induces defence gene expression and accumulation of beta-phenylethylamine-alkaloids in potato. Functional Plant Biology. 2006;33:593–604. doi: 10.1071/FP06049. [DOI] [PubMed] [Google Scholar]
- Oliver DJ, Nikolau B, Wurtele ES. Functional genomics: high-throughput mRNA, protein, and metabolite analyses. Metabolical Engineering. 2002;4:98–106. doi: 10.1006/mben.2001.0212. [DOI] [PubMed] [Google Scholar]
- Outlaw WH, Fisher DB. Compartmentation in Vicia faba leaves. I. Kinetics of C in the tissues following pulse labeling. Plant Physiology. 1975;55:699–703. doi: 10.1104/pp.55.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. The Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saunders JA, Conn EE. Subcellular localization of the cyanogenic glucoside of Sorghum by autoradiography. Plant Physiology. 1977;59:647–652. doi: 10.1104/pp.59.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad M, Mungur R, Fiehn O, Kehr J. Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods. 2005;1:2. doi: 10.1186/1746-4811-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. International Journal of Developmental Biology. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J. Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics. 2004;20:2447–2454. doi: 10.1093/bioinformatics/bth270. [DOI] [PubMed] [Google Scholar]
- Scholz M, Kaplan F, Guy CL, Kopka J, Selbig J. Non-linear PCA: a missing data approach. Bioinformatics. 2005;21:3887–3895. doi: 10.1093/bioinformatics/bti634. [DOI] [PubMed] [Google Scholar]
- Slack CR, Hatch MD, Goodchild DJ. Distribution of enzymes in mesophyll and parenchyma-sheath chloroplasts of maize leaves in relation to the C4-dicarboxylic acid pathway of photosynthesis. Biochemical Journal. 1969;114:489–498. doi: 10.1042/bj1140489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. Retention index thresholds for compound matching in GC–MS metabolite profiling. Journal of Chromatography B Analytical Technology and Biomedical Life Sciences. 2008;871:182–190. doi: 10.1016/j.jchromb.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Thayer SS, Conn EE. Subcellular localization of dhurrin β-glucosidase and hydroxynitrile lyase in the mesophyll cells of Sorghum leaf blades. Plant Physiology. 1981;67:617–622. doi: 10.1104/pp.67.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomos AD, Sharrock RA. Cell sampling and analysis (SiCSA): metabolites measured at single cell resolution. Journal of Experimental Botany. 2001;52:623–630. [PubMed] [Google Scholar]
- Tomos AD, Hinde P, Richardson P, Pritchard J, Fricke W. Microsampling and measurements of solutes in single cells. In: Harris N, Oparka KJ, editors. Plant cell biology a practical approach. Oxford: IRL Press; 1994. [Google Scholar]
- Torrigiani P, Rabiti AL, Bortolotti C, Betti L, Marani F, Canova A, Bagni N. Polyamine synthesis and accumulation in the hypersensitive response to TMV in Nicotiana tabacum. New Phytologist. 1997;135:467–473. [Google Scholar]
- Wagner C, Sefkow M, Kopka J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry. 2003;62:887–900. doi: 10.1016/s0031-9422(02)00703-3. [DOI] [PubMed] [Google Scholar]
- Walters DR. Polyamines in plant–microbe interactions and plant disease. Physiological and Molecular Plant Pathology. 2000;57:137–146. [Google Scholar]
- Walters DR. Polyamines and plant disease. Phytochemistry. 2003;64:97–107. doi: 10.1016/s0031-9422(03)00329-7. [DOI] [PubMed] [Google Scholar]
- Watt G, Leoff C, Harper AD, Bar-Peled M. A bifunctional 3,5-epimerase/4-keto reductase for nucleotide-rhamnose synthesis in Arabidopsis. Plant Physiology. 2004;134:1337–1346. doi: 10.1104/pp.103.037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Van Leeuwenhoek. 1990;58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Zoeller D, Ebert B, Simon-Rosin U, Fisahn J, Glinski M, Weckwerth W. Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry. 2004;65:1641–1649. doi: 10.1016/j.phytochem.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Williams ML, Thomas BJ, Farrar JF, Pollock CJ. Visualizing the distribution of elements within barley leaves by energy dispersive X-ray image maps (EDX maps) New Phytologist. 1993;125:367–372. doi: 10.1111/j.1469-8137.1993.tb03888.x. [DOI] [PubMed] [Google Scholar]
- Wurtele ES, Nikolau BJ. Enzymes of glucose oxidation in leaf tissues: the distribution of the enzymes of glycolysis and the oxidative pentose phosphate pathway between epidermal and mesophyll tissues of C3-plants and epidermal, mesophyll, and bundle sheath tissues of C4-plants. Plant Physiology. 1986;82:503–510. doi: 10.1104/pp.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.