Abstract
Auxin regulates a variety of physiological and developmental processes in plants. Although auxin acts as a suppressor of leaf senescence, its exact role in this respect has not been clearly defined, aside from circumstantial evidence. It was found here that ARF2 functions in the auxin-mediated control of Arabidopsis leaf longevity, as discovered by screening EMS mutant pools for a delayed leaf senescence phenotype. Two allelic mutations, ore14-1 and 14-2, caused a highly significant delay in all senescence parameters examined, including chlorophyll content, the photochemical efficiency of photosystem II, membrane ion leakage, and the expression of senescence-associated genes. A delay of senescence symptoms was also observed under various senescence-accelerating conditions, where detached leaves were treated with darkness, phytohormones, or oxidative stress. These results indicate that the gene defined by these mutations might be a key regulatory genetic component controlling functional leaf senescence. Map-based cloning of ORE14 revealed that it encodes ARF2, a member of the auxin response factor (ARF) protein family, which modulates early auxin-induced gene expression in plants. The ore14/arf2 mutation also conferred an increased sensitivity to exogenous auxin in hypocotyl growth inhibition, thereby demonstrating that ARF2 is a repressor of auxin signalling. Therefore, the ore14/arf2 lesion appears to cause reduced repression of auxin signalling with increased auxin sensitivity, leading to delayed senescence. Altogether, our data suggest that ARF2 positively regulates leaf senescence in Arabidopsis.
Keywords: Ageing, Arabidopsis, ARF2, auxin, leaf longevity, leaf senescence
Introduction
Senescence is the process of age-dependent programmed cell death that occurs at the cellular, tissue, organ, or organismal level, leading to the end of a lifespan (Noodén, 1988). As with many other biological processes, plant senescence is developmentally programmed and involves orderly, sequential changes in cellular physiology, biochemistry, and gene expression (Bleecker and Patterson, 1997; Nam, 1997; Buchanan-Wollaston et al., 2003; Lim et al., 2007a).
Senescence is the last stage in leaf development, during which cells undergo a major transition from carbon assimilation and other anabolic reactions to a catabolic pattern that results in cell dysfunction, structural disintegration and cell death (Hajouj et al., 2000). The most visible characteristics are colour changes, as in autumn foliage or the yellowing of leaves in monocarpic plants. This is due to a preferential breakdown of chlorophyll during chloroplast degradation and the synthesis of other pigments. Other metabolic changes include the hydrolysis of macromolecules such as lipids, proteins, and nucleic acids, which accumulated during the growth phase, followed by remobilization to young leaves and reproductive organs. Thus, although it is a deteriorative cellular process, leaf senescence critically contributes to the fitness of whole plants by ensuring their better survival and the optimal production of offspring (Noodén, 1988; Nam, 1997; Lim et al., 2007a).
Because of the rapid decline in photosynthetic capacity in leaves, senescence limits crop yields and plant biomass production. It can also cause post-harvest spoilage, such as leaf yellowing and nutrient loss in vegetable crops. Therefore, studying leaf senescence not only enhances our understanding of a fundamental biological process, but may also provide information on how to control senescence and improve the agricultural traits of crop plants. For example, delayed leaf senescence, with a concomitant preservation of the photosynthetic apparatus in a soybean mutant, increases its seed yield by 44% (Guiamét et al., 1990). Transgenic tobacco with a delayed leaf senescence phenotype also exhibits a 50% improvement in both seed yield and biomass (Gan and Amasino, 1995).
Leaf senescence, as an integral part of plant development, occurs in the last stage of leaf development. It is thus basically governed by developmental age. However, it is also influenced by various internal and environmental signals that are integrated into age information. Environmental factors may include pathogen infection, shading by neighbours, leaf detachment, drought, limited nutrients, extreme temperatures, and oxidative stress. Internal factors may include the formation of reproductive organs and changes in the levels of plant growth regulators (Weaver et al., 1998; Quirino et al., 1999, 2000; Buchanan-Wollaston et al., 2003; Lim et al., 2007a). Recent genomics and genetics studies in Arabidopsis have provided many insights into molecular events and their regulation during leaf senescence, thereby revealing the nature of regulatory factors and a highly complex molecular network (Woo et al., 2001; Gepstein et al., 2003; Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; Guo and Gan, 2006; Kim et al., 2006, 2009; van der Graaff et al., 2006; Lim et al., 2007b; Miao et al., 2008; Zhou et al., 2009). In particular, pathways involving the signalling factors sugar and phytohormones, such as cytokinin, ethylene, abscisic acid (ABA), jasmonic acid (JA), salicylic acid, and auxin, are of key importance in senescence (Buchanan-Wollaston et al., 2003, 2005). However, complex interactions among these signalling pathways during senescence have made it difficult to define the actual role and importance of genes within each pathway (He et al., 2001; van der Graaff et al., 2006). Therefore, to date, there is no clear understanding of the key controlling points within senescence signalling networks.
To enhance our knowledge of the molecular basis for leaf senescence, mutagenized Arabidopsis populations were screened with ethyl methane sulphonate (EMS) for delayed senescence mutants. This approach allowed us to identify various important positive elements of leaf senescence. Two allelic mutants with markedly delayed leaf senescence were investigated here. These were designated as oresara14 (ore14-1 and 14-2; oresara means ‘long-living’ in Korean). The ORE14 gene was identified by map-based cloning, and showed that it encodes auxin response factor 2 (ARF2), one of a family of transcription factors that bind to auxin-responsive elements (AuxREs) in the promoters of auxin-regulated genes (Ulmasov et al., 1997; Liscum and Reed, 2002; Guilfoyle and Hagen, 2007). Recent studies have shown that the arf2 mutation causes a pleiotropic phenotype, including the increased growth of aerial organs and seed size due to extra cell division, the inhibition of floral bud opening, and delays in flowering, leaf senescence, floral organ abscission, and silique ripening (Ellis et al., 2005; Okushima et al., 2005a; Schruff et al., 2006). These results suggest the involvement of ARF2 in regulating several developmental processes including leaf senescence, although the leaf senescence phenotype of the arf2 mutants was assessed only visually or was minimally analysed with respect to senescence symptoms or markers.
In our study, the in-depth senescence phenotype of the ore14/arf2 mutant was further analysed under various senescence-inducing conditions using many senescence parameters in order to unravel the physiological function of ARF2 in the plant leaf senescence network. Our results showed that ORE14/ARF2 plays an important role in the senescence process modulated by various senescence accelerating conditions, including leaf detachment in darkness, oxidative stress, and phytohormone treatment (ABA, ethylene, or MJ), as well as in age-dependent senescence. It is also demonstrated that ARF2 is a repressor of auxin signalling. Through the analysis of the ore14/arf2 mutant, it is suggested that the repression of auxin signalling by ARF2 might positively regulate the onset and progression of leaf senescence in Arabidopsis. Our results also illustrate that ARF2, among ARF family genes, might be a major player in controlling auxin-mediated leaf longevity.
Materials and methods
Plant materials and isolation of the ore14 mutant
The ore14 mutants in the Arabidopsis thaliana ‘Col’ wild-type background were identified by screening EMS-mutagenized M2 seeds for delayed leaf senescence, as determined by the delayed loss of chlorophyll during the dark-induced senescence of detached leaves (Oh et al., 1997). Isolated mutants were backcrossed twice with wild-type plants and used for all analyses. Plants for physiological experiments were grown in an environmentally controlled growth room (Korea Instrument, Seoul, Korea) with a 16/8 h light/dark cycle at 23 °C.
Assay of age-dependent leaf senescence
Age-dependent leaf senescence was assayed as described by Woo et al. (2001). Quantitative measurements were made of physiological and molecular parameters: chlorophyll content, photochemical efficiency, membrane ion leakage, and gene expression. Chlorophyll was extracted from individual leaves by boiling them in 95% ethanol at 80 °C. The concentration per fresh weight of leaf tissue was calculated as described by Lichtenthaler (1987). The photochemical efficiency of Photosystem II (PSII) was deduced from the characteristics of chlorophyll fluorescence (Oh et al., 1997) using a portable plant efficiency analyser (Hansatech Instruments, Norfolk, England). The ratio of maximum variable fluorescence to maximum fluorescence yield, which corresponds to the potential quantum yield of the PSII photochemical reactions, represented PSII photochemical reactions (Oh et al., 1997). Membrane ion leakage was determined by measuring electrolytes leaked from leaves (Woo et al., 2001). Two leaves were immersed for 3 h in 3 ml of 400 mM mannitol at 23 °C, with gentle shaking, after which initial conductivity was recorded. Total conductivity was determined after boiling for 10 min. Conductivity was expressed as the percentage of initial conductivity versus total conductivity.
Assay of artificially induced leaf senescence
For dark treatments, the third and fourth leaves at 12 d after their emergence (DAE) were detached and floated on 3 mM MES buffer solution (pH 5.7) and incubated at 23 °C in darkness. For hormone treatments, detached leaves were floated in the same buffer in the presence or absence of 50 μM abscisic acid (ABA; Sigma, USA), 100 μM methyl jasmonate (MJ; Sigma, USA), or 50 μM 1-aminocyclopropane-1-carboxylic acid (ACC; Sigma, USA). All hormone treatments were performed at 23 °C under continuous light. For oxidant treatments, detached leaves were floated on MES buffer in the presence or absence of 15 mM hydrogen peroxide. Chlorophyll content and photochemical efficiency were measured as described above.
RNA gel blot analyses
Total RNA was isolated from leaf tissue with Tri-Reagent (Molecular Research Center, USA). Total cellular RNA (10 μg) was size-fractionated by electrophoresis through a 1.2% formaldehyde-agarose gel and transferred onto a nylon membrane. Radiolabelled probes were prepared using a random labelling kit according to the manufacturer's instructions (Amersham, USA). After hybridization, the membranes were washed as previously described (Woo et al., 2001).
Mapping and sequencing
Homozygotes of ore14-1 in the ‘Col’ background were crossed with wild-type ‘Ler’ plants to create a mapping population, and 1391 plants with a delayed leaf senescence phenotype were selected from the F2 progeny. Genomic DNA from each plant was utilized for mapping the ORE14 locus relative to the loci of known Cleaved Amplified Polymorphic Sequence (CAPS) markers. New CAPS markers for the fine mapping of that locus were generated in the sequence of the BAC clones MTG10 and K22G18. The region near the mapped ORE14 locus was PCR-amplified and cloned into the pGEM T-easy vector (Promega, USA). Two independent PCR products were sequenced to identify the mutated sequence.
Inhibition of hypocotyl elongation in response to phytohormones
To measure hypocotyl lengths, Arabidopsis seedlings were grown for 4 d in continuous darkness on 0.1× Murashige and Skoog plates that contained various concentrations of NAA (α-naphthaleneacetic acid; Sigma, USA), BA (benzyladenine; Sigma, USA), or ACC (Sigma, USA). The length of hypocotyls was measured from individual plants (n >15), as previously described by Cary et al. (1995).
Results
Identification of Arabidopsis mutants with delayed leaf senescence
Arabidopsis, a representative monocarpic plant, is a favoured model for molecular and genetic studies of leaf senescence because its leaves readily undergo easily distinguishable developmental stages and show a well-defined and reproducible senescence programme (Hensel et al., 1993). Arabidopsis populations mutagenized with EMS for delayed leaf senescence mutants were screened, which allowed us to identify positive elements of senescence. Initial screening was carried out by visual evaluation of the degree of yellowing caused by chlorophyll loss when detached leaves were incubated in the dark. This technique is widely used for the consistent acceleration of leaf senescence (Oh et al., 1997). One of the mutants, ore14-1, which exhibited a delayed leaf senescence phenotype, was selected for further study. This mutation has a pleiotropic effect on vegetative and reproductive development, including the increased growth of aerial organs, inhibited floral bud opening, late flowering, and a delay in stem senescence (Fig. 1). These traits co-segregated with the delayed leaf senescence phenotype (data not shown). Genetic segregation analysis of the senescence phenotype was performed in F1 and F2 progeny derived from crosses between wild-type plants and the ore14-1 mutant. All F1 plants had normal phenotypes, including senescence symptoms, demonstrating that the ore14-1 mutation is recessive. In the F2 generation, 99 plants exhibiting wild-type phenotypes and 29 mutant plants were scored, indicative of 3:1 segregation (χ2=0.37, P >0.05). This showed that the mutation is inherited as a monogenic recessive trait.
Fig. 1.
Whole-plant phenotypes of Arabidopsis wild-type (‘Col’, left) and ore14-1 mutant (right) plants at 35, 46, and 65 d after germination (DAG).
Another mutant, ore14-2, with a phenotype similar to that of the ore14-1 mutant was also isolated. Genetic complementation tests revealed that these two mutations were in fact allelic (data not shown).
ORE14 is allelic to ARF2, which encodes an auxin response factor
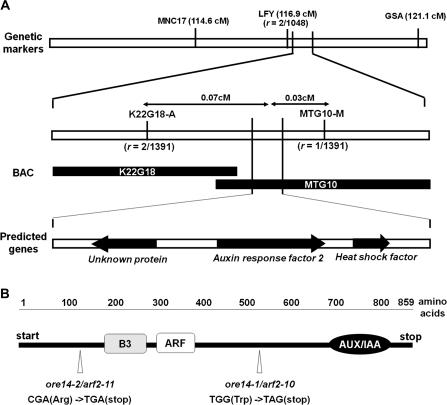
The gene responsible for the ore14-1 mutation was identified by map-based cloning (Fig. 2A). An initial genetic mapping, with CAPS markers, located ORE14 close to the LFY locus on Chromosome 5. CAPS markers were also generated using the genomic sequences of bacterial artificial chromosome clones (Fig. 2A). Among 1391 F2 progeny, one and two recombinant chromosomes were observed for the markers MTG10-M and K22G18-A, respectively. These mapping data located ORE14 at approximately 0.03 cM and 0.07 cM from these respective markers (Fig. 2A). The wild-type and mutant nucleotide sequences for the region around the map location of ORE14 were then compared. A single base change in the ore14-1 mutant—a G to A substitution—was identified at position 1673 from the translational start of the gene At5g62000 (Fig. 2B), which encodes auxin response factor 2 (ARF2). This mutation was expected to result in a premature stop codon at position 558 (Trp→stop) in the ARF2 protein. The same region was then sequenced from the ore14-2 mutant and it was found that a mutation here led to an early termination of translation at position 115 (Arg→stop) (Fig. 2B). Although the levels of ARF2 transcript of both mutants were comparable to that of wild-type plants (data not shown), these mutations probably cause a complete loss of function through premature translation termination. We simultaneously investigated a T-DNA insertion line in the At5g62000 gene, Salk_041472 line (arf2-5). The arf2-5 plants also exhibited phenotypes similar to those with the ore14 mutation, including delayed leaf senescence (data not shown). Several arf2 alleles were previously identified and designated as arf2-1 to arf2-9 (Ellis et al., 2005; Schruff et al., 2006); the ore14-1 and ore14-2 alleles are arf2-10 and arf2-11, respectively.
Fig. 2.
ORE14 is auxin response factor 2 (ARF2). (A) Map-based cloning of ORE14. The number of recombination events between CAPS markers and the ORE14 locus (r) is shown; BAC, bacterial artificial chromosome; cM, centimorgan. (B) Schematic representation of ORE14 with positions of the ore14-1 and 14-2 mutations; B3, DNA binding domain; ARF, auxin response region; AUX/IAA, domain involved in dimerization with other ARFs or Aux/IAA.
Auxin response factor 2 (ARF2) belongs to a family of transcription factors that bind to auxin-responsive elements (AuxREs) in the promoters of auxin-regulated genes (Ulmasov et al., 1997). In previous studies utilizing T-DNA insertion lines, the ARF2 gene was suggested to play a role in several developmental processes, such as cell division, organ growth, flowering, senescence, and abscission (Ellis et al., 2005; Okushima et al., 2005a; Schruff et al., 2006). However, the effect of ARF2 on leaf senescence has not been extensively examined. This led us to examine further the phenotype of the ore14/arf2 mutant with several senescence parameters under various senescence induction conditions.
The ore14/arf2 mutant exhibits extended leaf longevity during age-dependent senescence
The senescence symptoms of the ore14-1/arf2-10 mutant were first examined in detail during age-dependent in planta senescence. For an accurate assay, two factors were considered. First, in our experiment, leaf senescence was measured on a single leaf base with its age information. Measuring senescence parameters with a mixture of several leaves at a given plant age is not a valid analysis for leaf senescence, since the individual leaves of a plant have different ages. Only the third and fourth rosette leaves of a given plant were used here. The emergence time and growth rate of the fourth leaf was almost identical in wild-type plants and the ore14-1/arf2-10 mutant. Second, the senescence symptom was measured with various parameters that cover various aspects of senescence physiology.
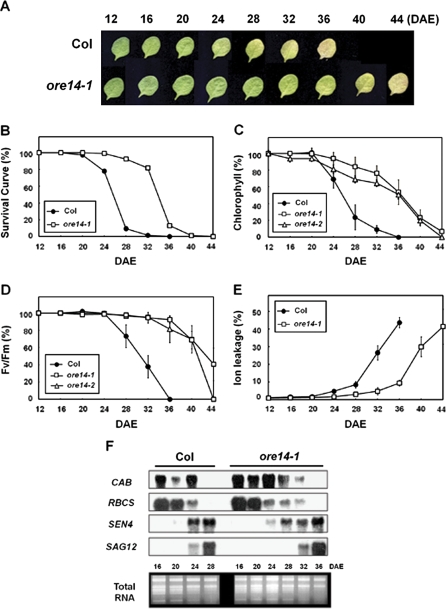
Leaf yellowing is a convenient visible indicator, mainly reflecting the chloroplast senescence of mesophyll cells. Age-dependent leaf yellowing using the fourth rosette leaf of mutant and wild-type plants were examined first. Wild-type leaves started to turn yellow at the leaf age of 20 d. Leaf age here means the age of the leaf under assay, counting from its visible emergence. After 20 d, yellowing gradually spread to the whole leaf. Wild-type leaves finally showed signs of necrosis at the age of 36 d (Fig. 3A). By contrast, ore14-1/arf2-10 mutant leaves started to turn yellow at the age of 28 d, i.e. 8 d later than wild-type leaves. They progressed more slowly, so that complete yellowing was not seen in the mutant until 44 d after emergence (DAE). Even then, however, the integrity of the mutant leaf shape was still maintained (Fig. 3A). When the point was determined at which the survival of the entire leaf population per plant was 50%, it was found that the leaf longevity of the ore14-1/arf2-10 mutant was extended by 35%, from 26 DAE to 35 DAE (Fig. 3B).
Fig. 3.
Extended leaf longevity in ore14/arf2 mutants. (A) Age-dependent senescence phenotype of the third and fourth rosette leaves of wild-type (‘Col’) and ore14-1/arf2-10 mutant plants at different ages. The photographs show representative leaves at each time point. (B) Survival curve. Values indicate the percentage of leaves alive on a given day after emergence (DAE). n=100. (C–E) Chlorophyll content (C), photochemical efficiency (Fv/Fm) of PSII (D), and membrane ion leakage (E) were measured from the third and fourth leaves starting at 12 DAE, when leaves had just reached full growth. Error bars indicate standard deviation (SD, n=30). (F) Age-dependent changes in gene expression. Total cellular RNAs isolated at the indicated ages were probed with CAB, RBCS, SEN4, and SAG12.
Symptoms of age-dependent senescence were then quantitatively investigated by measuring changes in several parameters, such as chlorophyll content, photochemical efficiency, membrane ion leakage, and the expression of senescence-associated genes. Chlorophyll amounts in wild-type leaves started to decrease rapidly after 20 DAE. By the age of 28 d, the chlorophyll content was reduced to 27% of that of 12 DAE leaves (mature green stage). By contrast, losses in the ore14-1/arf2-10 mutant were slower, with 52% of its chlorophyll remaining at 36 DAE (Fig. 3C). The change in the photochemical efficiency (Fv/Fm) of PSII was also monitored as an index of functional leaf senescence (Oh et al., 1997). As shown in Fig. 3D, the photochemical efficiency of wild-type leaves declined to 38% by the age of 32 d, whereas that of ore14-1/arf2-10 leaves remained almost unchanged until the age of 32 d and declined only to 39%, even at the age of 44 d. This showed that the functional integrity of PSII was maintained much longer in mutant leaves. Similar observations were made for these two parameters with the ore14-2/arf2-11 mutant (Fig. 3C, D). Senescence also involves the disruption of plasma membrane integrity as the final step in cell death. This can be conveniently quantified by monitoring ion leakage. It was found that leaf senescence was delayed in the ore14-1/arf2-10 mutant, as shown by slower increases in membrane ion leakage of the leaves with aging (Fig. 3E).
Moreover, in the ore14-1/arf2-10 mutant, a photosynthesis-related chlorophyll a/b-binding protein gene (CAB) and a ribulose biphosphate carboxylase small subunit gene (RBCS) were expressed at higher levels at the later stages, while the induction of two senescence-associated genes, SEN4 and SAG12, was delayed (Fig. 3F). These results indicated that the ore14-1/arf2-10 mutation considerably extends leaf longevity and delays leaf senescence, controlling various senescence-associated physiological and molecular symptoms. Thus, these results suggest that ORE14/ARF2 plays an important role in regulating the leaf senescence process.
Delays in artificially induced senescence
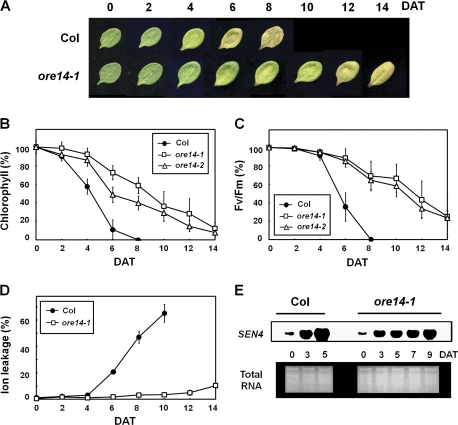
Darkness is one of the most potent external stimuli that accelerate leaf senescence. The responses were compared of the ore14-1/arf2-10 mutant and wild-type plants during dark-induced senescence. To minimize developmental effects, fully grown third and fourth leaves were detached at 12 DAE (mature green stage). Those from the wild-type plants lost 90% of their chlorophyll after 6 d of dark incubation. However, for the mutant, the chlorophyll content declined much more slowly; even after 6 d, over 70% was retained (Fig. 4A, B). Measurement of the photochemical efficiency showed that, after 8 d of incubation, ore14-1/arf2-10 mutant leaves still maintained over 65% of their initial PSII activity, while wild-type leaves completely lost this activity (Fig. 4C). A relatively slower increase in membrane ion leakage in the ore14-1/arf2-10 mutant was also observed (Fig. 4D). Consistent with these physiological data, the induction level of SEN4, a molecular marker for dark-induced senescence, was much lower in the mutant during dark incubation (Fig. 4E).
Fig. 4.
Delayed leaf senescence of ore14/arf2 mutants during dark-induced senescence. (A) Phenotypes of wild-type and ore14 mutant leaves. The third and fourth rosette leaves were detached at the age of 12 d and incubated in darkness; DAT, days after treatment. (B–D) Chlorophyll content (B), photochemical efficiency (Fv/Fm) of PSII (C), and membrane ion leakage (D) were examined every 2 d after dark treatment. Error bars indicate SD (n=24). (E) Expression of SEN4. Total cellular RNAs were isolated from wild-type and ore14-1/arf2-10 mutant leaves at the days indicated.
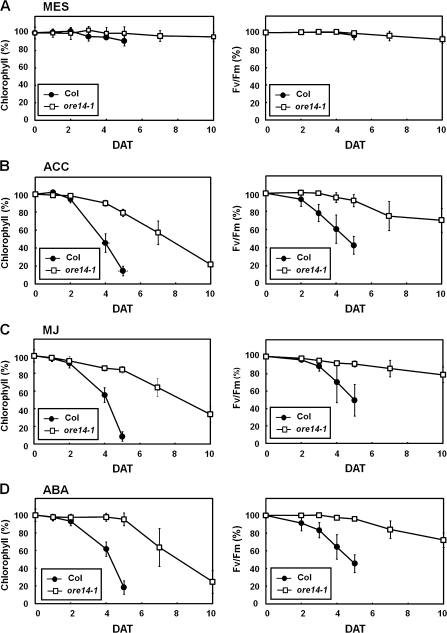
It was then tested whether the ore14-1/arf2-10 mutation affects phytohormone-induced leaf senescence. Three hormones in particular—ABA, MJ, and ethylene— strongly hasten leaf senescence (He et al., 2001; Woo et al., 2001; Lim et al., 2007b). Before examining hormone-induced leaf senescence symptoms, the effect of light alone was evaluated first as a control experiment. When detached leaves were exposed to light in the presence of MES buffer alone, no significant changes in chlorophyll content and PSII activity in wild-type or mutant leaves were observed (Fig. 5A). To determine whether ORE14/ARF2 is involved in the senescence process induced by senescence-accelerating phytohormones, the senescence symptoms of the ore14-1/arf2-10 mutant were examined after treatment with these hormones. When detached leaves were treated with MJ, ABA, or ethylene, a rapid decrease in chlorophyll was observed in wild-type leaves and, 5 d after incubation, the chlorophyll of wild-type leaves was reduced to less than 20%. However, ore14-1/arf2-10 mutant leaves retained more than 80% of chlorophyll following treatment with these hormones at 5 d after incubation (Fig. 5B–D). A similar pattern was observed when PSII activity was measured. These data clearly showed a delay of leaf senescence in the ore14-1/arf2-10 mutant during plant hormone-accelerated senescence. These results imply that ORE14/ARF2 might be a regulator in controlling the senescence process modulated by these hormones, as well as in age-dependent and dark-induced senescence.
Fig. 5.
Delay of leaf senescence in the ore14-1/arf2-10 mutant during senescence accelerated by phytohormones. The third and fourth rosette leaves were detached at the age of 12 d and incubated under continuous light in MES buffer alone (A), with 50 μM ACC (B), 100 μM MJ (C), or 50 μM ABA (D). Chlorophyll content and photochemical efficiency are presented as average percent values ±SD, relative to those of leaves incubated in light; n=24. DAT, days after treatment.
Resistance of the ore14-1/arf2-10 mutant to oxidative stress
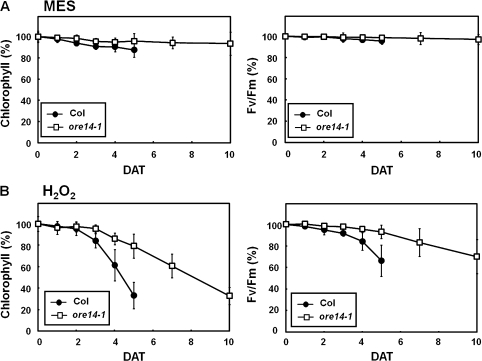
Reactive oxygen species play a critical role in mediating oxidative damage and accelerating senescence in a variety of organisms (Reilly et al., 2004). Potential life spans are positively correlated with plant tolerance to oxidative stress. The oxidative stress tolerance of the ore14-1/arf2-10 mutant was therefore assessed. For this experiment, the procedure that was used for assaying phytohormone-induced senescence symptoms was adapted and hydrogen peroxide was used as an oxidant. Under such exposure, both chlorophyll content and PSII activity declined rapidly in the wild type. By contrast, the ore14-1/arf2-10 mutant retained over 80% of its chlorophyll and 95% of the original PSII activity, even at 5 d after treatment (Fig. 6A, B). Thus, the ore14-1/arf2-10 mutation conferred greater resistance to oxidative stress, one of the major determinants of senescence. These results further indicate that ORE14/ARF2 might be a central regulator of leaf senescence. The data also support our previous finding that oxidative stress tolerance is linked to the control of leaf longevity (Woo et al., 2004).
Fig. 6.
Delay of leaf senescence in the ore14-1 mutant during senescence accelerated by oxidative stress. The third and fourth rosette leaves were detached at the age of 12 d and incubated under continuous light in MES buffer alone (A) and with 15 mM hydrogen peroxide (B). Chlorophyll content and photochemical efficiency are presented as average per cent values ±SD, relative to those of leaves incubated in light (n=24). DAT, days after treatment.
ARF2 is a repressor of auxin signalling
The ARF family of transcription factors regulates many responses to auxin. These proteins bind to auxin response elements in the promoters of auxin-regulated genes and activate or repress transcription (Ulmasov et al., 1997). Most ARFs have three domains: an N-terminal DNA-binding domain, a C-terminal dimerization domain, and a middle region (MR) that activates or represses transcription (Ulmasov et al., 1999). ARFs containing glutamine-rich MRs, for example, ARF5 and ARF19, function as activators of auxin-responsive gene expression in vivo (Nagpal et al., 2005; Okushima et al., 2005b; Wilmoth et al., 2005) and in transiently transfected protoplasts (Ulmasov et al., 1999). By contrast, ARFs containing proline- and/or serine-rich MRs, such as ARF1 and ARF2, have been suggested to repress auxin-responsive gene expression in protoplast transient assays (Tiwari et al., 2003, 2004).
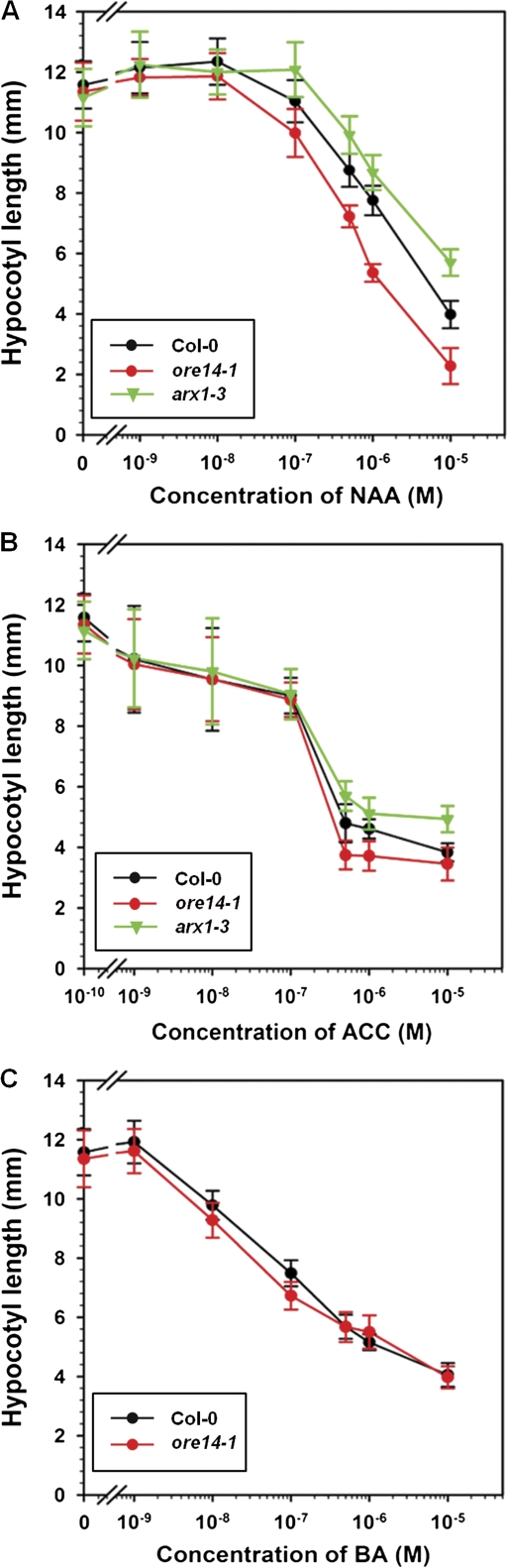
To test whether ARF2 represses auxin signalling in vivo, the growth responses of ore14-1/arf2-10 hypocotyls to exogenous phytohormones was examined. In wild-type seedlings, exogenous NAA (a synthetic auxin) inhibits hypocotyl growth. This response was enhanced in the ore14-1/arf2-10 mutant (Fig. 7A). The hypocotyl growth inhibition responses of the ore14-1/arf2-10 mutant to cytokinin and ethylene was also examined. Exogenous applications of BA (a synthetic cytokinin) or ACC (an immediate biosynthetic precursor of ethylene) inhibited wild-type hypocotyl growth. However, in contrast to the case with NAA, ore14-1/arf2-10 hypocotyls showed a nearly normal inhibition response to BA or ACC (Fig. 7B, C), although they were slightly more sensitive at >1 μM ACC. These results indicate that the ore14-1/arf2-10 mutation does not cause defects in the known signal transduction pathways that underlie the perception and response to ethylene or cytokinin. They also suggest that ore14-1/arf2-10 preferentially alters the auxin response, at least with regard to hypocotyl growth. Together, these data demonstrate that ARF2 is a true repressor of auxin signalling, supporting previous results from protoplast transient assays (Tiwari et al., 2003).
Fig. 7.
Hypocotyl growth inhibition responses to exogenous phytohormones. Seedlings of wild type (‘Col’) and the ore14-1 mutant were grown for 4 d in various concentrations of NAA (A), ACC (B), or BA (C). The auxin resistant 1-3 (axr1-3) mutant was included as an internal control. At least 20 seedlings were sampled for each treatment. Bars indicate standard errors.
Discussion
ORE14/ARF2 affects a broad spectrum of leaf senescence symptoms
In a screen for Arabidopsis mutants exhibiting delayed leaf senescence, two ore14 mutations were identified, each of which contains a lesion in ARF2. These mutations also confer pleiotropic developmental phenotypes, including delays in flowering and floral organ abscission, and inhibition of floral bud opening in early-formed flowers. These phenotypes have also been reported for other arf2 mutant alleles (Ellis et al., 2005; Okushima et al., 2005a). The arf2/mnt1 mutation can also cause the increased growth of aerial organs and more cell divisions (Schruff et al., 2006). The pleiotropic effects of arf2/mnt1/ore14 mutations may hamper assaying the effect of the gene on senescence. In particular, when minimally analysed with respect to senescence symptoms or markers, for example, the use of a single senescence symptom such as yellowing or loss of chlorophyll, interpretation may be misleading. In order to show that the ARF2 mutation delays functional senescence, the ore14 mutations were analysed in detail by examining several anabolic and catabolic parameters. The fact that these mutations influenced all the parameters indicated that the affected plants are functional ‘stay-green’ mutants, and that they are not just delayed in their loss of chlorophyll. This implies that ORE14/ARF2 might be a key regulatory factor involved in leaf senescence rather than just one of the components that executes the process (Smart, 1994; Oh et al., 1997). It was also found that ORE14/ARF2 is required for the proper progression of leaf senescence induced by darkness, phytohormones, and oxidative stress, as well as by age. These results suggest that ORE14/ARF2 functions in a step common to all these processes. The data also support the idea that these senescence-inducing factors, developmental age, dark, hormones, and oxidative stress, all share a common senescence pathway.
The apparently pleiotropic effects of the arf2 mutation, such as reduced sterility or late flowering, may be associated with delayed leaf senescence, but are unlikely to be its cause. This is because, unlike many other monocarpic species, senescence of individual Arabidopsis leaves is not closely linked with the development of reproductive structures (Hensel et al., 1993; Lim et al., 2007a). Both ethylene and cytokinin have a major effect on leaf senescence. Ethylene-insensitive plants, such as etr1 and ein2 mutants, have a delayed leaf senescence phenotype (Grbic and Bleecker, 1995). Simularly, ore12-1, a gain-of-function mutation in AHK3 (a cytokinin receptor), confers extended leaf longevity (Kim et al., 2006). To determine whether our ore14/arf2 phenotype is independent of the ethylene and cytokinin signal transduction pathways, hypocotyl elongation was monitored in response to these hormones. No significant differences were found between mutant and wild-type hypocotyls (Fig. 7), although the mutant exhibited a slightly enhanced sensitivity at high ACC concentrations (1–10 μM). This enhanced sensitivity might be due to an indirect effect of altered auxin signalling. Nevertheless, it is inferred that the delayed leaf senescence phenotype of ore14/arf2 plants is not due to alterations in cytokinin or ethylene sensitivity. A similar report was made by Ellis et al. (2005), who based their findings on the fact that the ein2 and arf2 mutations act additively in delaying leaf senescence symptoms, and that the relative deferment in chlorophyll loss caused by exogenous cytokinin is similar in both wild-type and arf2 plants. It was also observed that the responses of the ore14/arf2 seedlings to ABA and MJ were similar to those of wild-type seedlings (data not shown). These results, in turn, indicate that the delayed senescence symptoms of the ore14/arf2 mutant during phytohormones-induced senescence are not due to defects in the perception of these hormones.
Since leaf senescence is intimately related to previous developmental stages, such as initiation, growth, and maturation, it is possible that genes controlling all these processes, including cell division, could affect age-dependent senescence. This has been demonstrated in leaves from the bop1-1 mutant, which has enhanced meristematic activity (Ha et al., 2003). Likewise, organs in arf2/mnt1/ore14 mutants, including the leaves, are larger than those of wild-type plants, due to extra cell divisions and expansion (Schruff et al., 2006). Thus, it is possible that extended leaf longevity in ore14 mutants might be related to increased cell division activity. However, because the delayed senescence phenotype of ore14/arf2 was also observed under artificially induced senescence conditions, we believe that this possibility is very low.
Role of ARF2 in auxin signalling
ARFs with a glutamine-rich MR might function as activators of auxin-responsive gene expression in transiently transfected protoplasts, while ARFs with proline- and/or serine-rich MRs, such as ARF1 or ARF2, repress the transcription of reporter genes under the control of synthetic AuxREs (Tiwari et al., 2003). However, when compared with the global gene expression profiling of liquid-cultured arf2 seedlings or the inhibition of root growth by exogenous auxin, no significant effect of the mutation was observed (Okushima et al., 2005a). The fact that the arf2 mutation does not affect auxin-induced or -repressed genes indicates that ARF2 does not participate in auxin signalling in a particular developmental stage or organ. However, in contrast to earlier reports, it was observed that the ore14/arf2 mutant exhibited enhanced sensitivity to auxin, as assessed by the inhibition of hypocotyl growth. This implies that ARF2 functions as a transcription repressor of auxin-responsive gene expression, as previously proposed by Tiwari et al. (2003). The results seen here along with other data also suggest that ARF2-mediated auxin signalling is temporally and spatially regulated.
Other studies have shown that ARF2 links brassinosteroid (BR) and auxin biosynthetic pathways (Vert et al., 2008). Thus, the effect of the ore14/arf2 mutation on auxin responses might be due to an altered BR response. However, in the study of Vert et al. (2008) no difference in response to exogenous BR was detected between the wild type and the mutant, indicating that the ore14/arf2 mutation does not cause defects in BR perception and response. Conceivably, an increased sensitivity to auxin conferred by the arf2 mutation might not be due to an altered BR response.
ARF2 is a major player in the auxin-mediated control of leaf longevity
The role of auxin in leaf senescence has been elusive, particularly due to its involvement in various aspects of plant development. However, evidence that auxin has a role in suppressing leaf senescence has accumulated. Shoji et al. (1951) reported that auxin levels decline with leaf age. The exogenous application of auxin to leaves represses transcription of some senescence-associated genes (Noh and Amasino, 1999). Together, these results imply that decreased auxin levels along with leaf age might induce the initiation and/or progression of leaf senescence, indicating that auxin is a negatively acting factor of leaf senescence. Genetic mutations that alter auxin signalling also support the involvement of auxin in controlling leaf senescence (Ellis et al., 2005).
What is the major player in the auxin-mediated control of leaf senescence? ARFs regulate auxin-mediated transcriptional activation and repression, and each ARF protein is thought to play a central role in various auxin-mediated developmental processes. A few lines of our data, together with previous results, indicate that ARF2 might be a major player in the auxin-mediated control of leaf senescence. First, ARF2 affects many aspects of senescence, whether induced by age, darkness, hormones, or oxidative stress. Second, phenotypic analyses of T-DNA insertion lines for ARF family genes have revealed that ARF2 plays a major role in controlling leaf senescence and that ARF1 acts only in a partially redundant manner (Ellis et al., 2005). Third, microarray analysis has shown that ARF2 transcripts increase in senescing leaves when induced by developmental ageing or darkness (Buchanan-Wollaston et al., 2005; Ellis et al., 2005). The ARF7 and ARF19 genes are also induced in senescing leaves (Lin and Wu, 2004). However, mutations in these genes do not alter the leaf senescence phenotype, although they enhance the delay in senescence conferred by the arf2 mutation (Ellis et al., 2005).
How then is leaf senescence delayed in the arf2/mnt1/ore14 mutant? With the theory that auxin functions as a suppressor of leaf senescence and that ARF2 is a repressor of auxin signalling, it is conceivable that the reduced functioning of ARF2 in the mutant can cause less repression of auxin signalling with increased auxin sensitivity, leading to delayed senescence. Our data, as well as previous results, suggest that auxin has a role in suppressing leaf senescence, but suppression is decreased with leaf ageing, possibly through the increased activity of ARF2. Further studies on the identification of ARF2 downstream target genes or interacting proteins will help to dissect the senescence pathways involved in the ARF2-mediated control of leaf longevity.
Acknowledgments
We thank KH Suh, YS Park, and BH Kim for excellent technical assistance. We also thank Priscilla Licht and Patrick Hughes (Bioedit® Ltd) for critical proofreading of the manuscript. This work was supported by grants from the Crop Functional Genomics Research Program (to HGN; CG1311) and the MOST/KOSEF to the National Core Research Center for Systems Bio-Dynamics (R15-2004-033). The work by POL was supported by the Priority Research Centers Program (2009-0094060) and the Basic Research Program (2009-0067967) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology. This work was also supported by TDPAF (107098-3).
References
- Bleecker AB, Patterson SE. Last exit: senescence, abscission, and meristem arrest in Arabidopsis. The Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiology. 1995;107:1075–1082. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M. Large-scale identification of leaf senescence-associated genes. The Plant Journal. 2003;36:629–642. doi: 10.1046/j.1365-313x.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Grbic V, Bleecker AB. Ethylene regulates the timing of leaf senescence in Arabidopsis. The Plant Journal. 1995;8:595–602. [Google Scholar]
- Guiamét JJ, Teeri JA, Noodén LD. Effects of nuclear and cytoplasmic genes altering chlorophyll loss on gas exchange during monocarpic senescence in soybean. Plant and Cell Physiology. 1990;31:1123–1130. [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Current Opinion in Plant Biology. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- Ha CM, Kim GT, Kim BC, Jun JH, Soh MS, Ueno Y, Machida Y, Tsukaya H, Nam HG. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiology. 2000;124:1305–1314. doi: 10.1104/pp.124.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang W, Swain JD, Green AL, Jack TP, Gan S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiology. 2001;126:707–716. doi: 10.1104/pp.126.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel L, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. The Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. The Plant Cell. 2008;20:307–319. doi: 10.1105/tpc.107.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology. 1987;18:350–382. [Google Scholar]
- Lim PO, Kim Y, Breeze E, et al. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. The Plant Journal. 2007b;52:1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annual Review of Plant Biology. 2007a;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH. Molecular events in senescing Arabidopsis leaves. The Plant Journal. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology. 2002;49:387–400. [PubMed] [Google Scholar]
- Miao Y, Smykowski A, Zentgraf U. A novel upstream regulator of WRKY53 transcription during leaf senescence in. Arabidopsis thaliana. Plant Biology. 2008;10(Supplement 1):110–120. doi: 10.1111/j.1438-8677.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Nam HG. Molecular genetic analysis of leaf senescence. Current Opinion in Biotechnology. 1997;8:200–207. doi: 10.1016/s0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino R. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molecular Biology. 1999;41:181–194. doi: 10.1023/a:1006342412688. [DOI] [PubMed] [Google Scholar]
- Noodén LD. The phenomenon of senescence and aging. In: Noodén LD, Leopold AC, editors. Senescence and aging in plants. San Diego, CA: Academic Press; 1988. pp. 2–50. [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. The Plant Journal. 1997;12:527–535. doi: 10.1046/j.1365-313x.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR2 (ARF2): a pleiotropic developmental regulator. The Plant Journal. 2005a;43:29–46. doi: 10.1111/j.1365-313X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell. 2005b;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. Molecular aspects of leaf senescence. Trends in Plant Science. 2000;5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Molecular Biology. 1999;40:267–278. doi: 10.1023/a:1006199932265. [DOI] [PubMed] [Google Scholar]
- Reilly K, Gomez-Vasquez R, Buschmann H, Tohme J, Beeching JR. Oxidative stress responses during cassava post-harvest physiological deterioration. Plant Molecular Biology. 2004;56:625–641. doi: 10.1007/s11103-005-2271-6. [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Shoji K, Addicott FT, Swets WA. Auxin in response to leaf blade abscission. Plant Physiology. 1951;26:189–191. doi: 10.1104/pp.26.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CM. Gene expression during leaf senescence. New Phytologist. 1994;126:419–448. doi: 10.1111/j.1469-8137.1994.tb04243.x. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. The Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathway by auxin response factor 2. Proceedings of the National Academy of Sciences, USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Molecular Biology. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Nam HG, Lim PO. The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant and Cell Physiology. 2004;45:923–932. doi: 10.1093/pcp/pch110. [DOI] [PubMed] [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiology. 2009;150:167–177. doi: 10.1104/pp.108.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]