Abstract
The roles of starch branching enzyme (SBE, EC 2.4.1.18) IIa and SBE IIb in defining the structure of amylose and amylopectin in barley (Hordeum vulgare) endosperm were examined. Barley lines with low expression of SBE IIa or SBE IIb, and with the low expression of both isoforms were generated through RNA-mediated silencing technology. These lines enabled the study of the role of each of these isoforms in determining the amylose content, the distribution of chain lengths, and the frequency of branching in both amylose and amylopectin. In lines where both SBE IIa and SBE IIb expression were reduced by >80%, a high amylose phenotype (>70%) was observed, while a reduction in the expression of either of these isoforms alone had minor impact on amylose content. The structure and properties of the high amylose starch resulting from the concomitant reduction in the expression of both isoforms of SBE II in barley were found to approximate changes seen in amylose extender mutants of maize, which result from lesions eliminating expression of the SBE IIb gene. Amylopectin chain length distribution analysis indicated that both SBE IIa and SBE IIb isoforms play distinct roles in determining the fine structure of amylopectin. A significant reduction in the frequency of branches in amylopectin was noticed only when both SBE IIa and SBE IIb were reduced, whereas there was a significant increase in the branching frequency of amylose when SBE IIb alone was reduced. Functional interactions between SBE isoforms are suggested, and a possible inhibitory role of SBE IIb on other SBE isoforms is discussed.
Introduction
Starch is composed of a single monomer type glucose, polymerized into large molecules through a combination of both α(1→4) and α(1→6) linkages. The polydisperse molecules of starch are generally classified as belonging to two component fractions, known as amylose and amylopectin, on the basis of their degree of polymerization (DP) and the ratio of α(1→6) to α(1→4) linkages. Typically, amylose molecules constitute 20–30% of the mass of starch, have a DP of between 500 and 5000, and contain less than 1% α(1→6) linkages. By contrast, amylopectin contributes 70–80% of the dry weight of starch, is a much larger molecule with DP ranging from 5000 to 50 000 and contains 4–5% α(1→6) linkages. There is considerable interest in generating starches in a range of species with altered amylose to amylopectin ratios for use in industrial processes (Fergason, 1994) and as a source of resistant starch for human health (Topping and Clifton, 2001).
Amylose is synthesized within the starch granule where the enzyme granule-bound starch synthase (GBSS) is essential for amylose synthesis to occur. The involvement of other starch synthetic enzymes in contributing to amylose synthesis cannot be ruled out, and the presence α(1→6) linkages in amylose, although in a lesser proportion than in amylopectin, indicates a role for branching enzymes. However, there is no genetic evidence that any other gene is essential for amylose synthesis. By contrast, amylopectin synthesis is more complex. A range of genes is required for amylopectin synthesis, including genes encoding starch synthases, branching enzymes, and debranching enzyme (Myers et al., 2000; Ball and Morell, 2003). The biochemical evidence suggests that these enzymes act at the surface of the granule, building up amylopectin molecules that can be incorporated into the structure of the growing starch granule.
Two classes of starch branching enzyme are known in dicotyledonous plants, such as pea and potato, starch branching enzyme I (also known as SBE B) and starch branching enzyme II (also known as SBE A). Arabidopsis contains three genes classified as SBE homologues, two SBE II-type genes (Accession numbers At2g36390 and At5g03650) and a third gene that is only distantly related to either of the known SBE classes. The activities of the two SBEs belonging to the SBE II class (SBE 2 and SBE 3) are redundant and either of these isoforms is sufficient for normal starch synthesis. The third SBE (SBE 1) has no apparent function in starch metabolism (Dumez et al., 2006). In pea embryos, a lesion in SBE II results in a reduction in starch synthesis yielding the rugosous wrinkled seed phenotype described by Mendel (Bhattacharyya et al., 1993) and an amylose content of >60%. In potato, down-regulation of SBE II leads to an increased amylose phenotype (amylose content approximately 35%; Jobling et al., 1999), whereas when SBE I was down-regulated no significant difference in the amylose content or amylopectin chain length profile was observed (Safford et al., 1998). Suppression of both SBE II and SBE I in potato yielded a starch with an amylose content of 70% or higher (Schwall et al., 2000).
In cereal species, such as rice, maize, barley, and wheat, three classes of starch branching enzymes (SBE I, SBE IIa, and SBE IIb) have been reported. Reduction or elimination of SBE I activity in wild-type backgrounds does not result in measurable differences in amylose content in rice endosperm (Satoh et al., 2003) and maize leaf or endosperm (Blauth et al., 2002). In wheat, apparent gene duplication events have increased the number of SBE I genes in each genome (Rahman et al., 1999). The elimination of greater than 97% of the activity by combining mutations in the highest expressed forms of the SBE I genes from the A, B, and D genomes had no measurable impact on starch structure or functionality (Regina et al., 2004). In maize endosperm, lesions in the SBE IIb gene result in the amylose extender (ae) phenotype with amylose contents from 50–80% (Garwood et al., 1976) whereas lesions in SBE IIa lead to unaltered endosperm starch, but leaf starch with elevated amylose (Blauth et al., 2001). In rice, mutations eliminating the expression of SBE IIb lead to increased amylose content (up to 35% in an indica background, and up to 25-30% in a japonica background; Mizuno et al., 1993; Nishi et al., 2001). In wheat, it has been demonstrated that suppression of the expression of SBE IIa is required to yield a very high amylose content (>70%) (Regina et al., 2006).
In barley, both SBE IIa and SBE IIb are expressed in the developing endosperm where the expressed proteins are partitioned between the starch granule matrix and the amyloplast stroma (Morell et al., 1997; Sun et al., 1997; Rahman et al., 2001). The relative expression levels of SBE IIa and SBE IIb barley are very different from the expression levels of the respective enzymes in maize. In maize endosperm, the level of SBE IIb protein present in the amyloplast stroma is about 50 times higher than SBE IIa (Gao et al., 1997). However, in barley, there are approximately equal levels of SBE IIa and SBE IIb proteins expressed in the endosperm (Sun et al., 1998).
No mutants in barley with lesions in either SBE IIa or SBE IIb have been described. In this study, RNAi technology was used to examine the roles of the SBE IIa and SBE IIb genes in controlling the content and branching of amylose and amylopectin in developing endosperm of barley. The study revealed that SBE IIa and SBE IIb enzymes play specific roles in determining the chain length distribution and branching pattern of amylose and amylopectin molecules. It was also established that suppression of both isoforms of SBE II was required to elevate the amylose content in barley to a level >65%.
Materials and methods
Generation of hairpin RNA constructs of SBE IIa (hp-SBE IIa) and SBE IIb (hp-SBE IIb)
hp-SBE IIa and hp-SBE IIb constructs for barley transformation were generated by cutting out the respective expression cassettes from pDV03-IIa and pDV03-IIb (described in Regina et al., 2006) that included a high molecular weight glutenin promoter and nos 3′ terminator using the restriction enzymes ApaI and NotI and ligating the respective inserts into the ApaI/NotI site of a binary vector, pWBVec8 (Wang et al., 1998) that contained a hygromycin (hph) resistance gene driven by the cauliflower mosaic virus (CaMV) 35S promoter.
Transformation of barley
hp-SBE IIa and hp-SBE IIb constructs generated as above were used to transform the barley variety Golden Promise (GP) through Agrobacterium (strain AGL1)-mediated transformation following the protocol of Tingay et al. (1997).
Analysis of transgenic plants
Southern blot hybridization was carried out on DNA from the hp-SBE IIa and hp-SBE IIb barley transgenic lines. DNA from barley plants was digested with ApaI that was known to cut only once within the T-DNA insert. Digested DNA was electrophoresed on 0.8% agarose gels and blotted on to Hybond N+ nylon membranes (Amersham Pharmacia Biotech UK Ltd). The probe for hp-SBE IIb lines was generated from the intron 3 region of wSBE II-DB1 (Regina et al., 2005) by PCR amplification using the primers ARA19F (5′-CACCCATTGTAATTGGGTACACTG-3′) and ARA23R (5′-CTGCGCATAAATCC AAACTTCTCG-3′). The probe for the hp-SBE IIa lines was generated from the intron 3 region of wSBE II-DA1 (GenBank Accession number AF338431) by PCR amplification using the primers AR2aKpnIF (5′-GGTACCGCAGAAAATATACGAGATTGACCC-3′) and AR2aSacIR (5′-GAGCTCCCACCTTCATGTTGGTCAATAGC-3′). The probe DNA (100–200 ng) was radioactively labelled using the Megaprime DNA labelling system (Amersham Pharmacia Biotech UK Ltd). The hybridization was carried out in 25% (v/v) formamide, 5× SSC, 0.1% SDS, 10× Denhardt's solution, 100 μg ml−1 salmon sperm DNA at 42 °C for 16 h, followed by washing in 2× SSC, 0.1% SDS at 65 °C for 3×1 h.
Expression analyses of SBEIIa and SBEIIb in developing barley endosperm
Protein expression
Endosperm soluble proteins from barley plants were extracted from developing endosperms (18 d after anthesis) by homogenizing at 4 °C using 50 mM potassium phosphate buffer, pH 7.5 (0.1 ml per endosperm), containing 5 mM EDTA, 5 mM DTT, and 1 mM pefabloc. The extracts were centrifuged at 14 000 g for 15 min and supernatant was used as crude soluble proteins. Protein concentration in the supernatant was estimated and 16 μg per sample was loaded onto an 8% non-denaturing polyacrylamide gel. Immunoblotting was carried out as described by Kosar-Hashemi et al. (2006). The primary antibodies used were wheat SBE IIa (anti-WBE IIa; Rahman et al., 2001) and wheat SBE IIb (anti-WBE IIb; Regina et al., 2005). Immunoreactive proteins were revealed using an Amersham ECL detection system according to the manufacturer's instructions. Densitometry analysis of immunoblots was done using TotalLab software package (Nonlinear Dynamics Ltd, Newcastle, UK).
Activity staining of branching enzymes was based on the method of Nishi et al. (2001) with minor modification. After electrophoresis the gel was washed twice in 50 mM HEPES, pH 7.0 containing 10% glycerol and incubated at room temperature in a reaction mixture consisting of 50 mM HEPES, pH 7.4, 50 mM glucose-1-phosphate, 2.5 mM AMP, 10% glycerol, 50 U phosphorylase a, 1 mM DTT, and 0.08% maltotriose for 16 h. The bands were visualized with a solution of 0.2% (W/V) I2 and 2% KI.
mRNA expression
RNA from developing endosperm 15 d post anthesis was extracted from barley as described in Li et al. (1999). cDNA for real-time PCR was synthesized from RNA using SuperScript™ Choice System cDNA synthesis kit from Invitrogen according to the manufacturer's instructions. Quantitative PCR was conducted according to Warton et al. (2004) using a Rotor-Gene 6000 (Corbett Research, Sydney, Australia) thermal cycler with real-time detection of fluorescence. PCR was performed in a volume of 20 μl using 1.5 U HotStar Taq™ (Qiagen), 1× PCR reaction buffer containing 15 mM MgCl2 (Qiagen), 0.225 μM of each primer, 0.1 mM dNTP, an additional 1 mM MgCl2, and SYBR green I (Molecular Probes, Australia) at a final concentration of 1:20 000. The PCR cycle consisted of an initial denaturation of 95 °C for 10 min, followed by 45 cycles of 95 °C for 20 s, 63 °C for 20 s, 72 °C for 30 s, and a final extension of 72 °C for 1 min. The fluorescence intensity of SYBR green I was read and acquired at 72 °C after the extension step of each cycle. Actin gene (Accession no. AY14545) was used as a housekeeping gene for normalization of SBE IIa and SBE IIb transcript expression. The primers ZLBactinF (5′-GCCGTGCTTTCCCTCTATG-3′) and ZLBactinR (5′-GCTTCTCCTTGATGTCCCTTA-3′) were used to amplify the actin transcript. Primers for SBE IIa and SBE IIb transcript amplification were designed in such a way that the sequences cross exon regions so as to prevent genomic DNA amplification. The primers ARHv2bF1 (5′-CAGAATGGACAAAGAATCATCCACG-3′) and ARHv2bR1 (5′-GAAAATACATCCATGCCTCCATCG-3′) and ARHv2aF1 (5′-CAATCACTGATGGTGTAACCAAAGG-3′) and ARHv2aR3 (5′-CCTTCATGTTGGTCAATAGCAGC-3′) were used specifically to amplify SBE IIb and SBE IIa transcripts, respectively. Quantification of individual transcripts was performed using ‘Comparative Quantification’ software supplied by Corbett Research for the Rotor-gene 6000.
Starch characterization of mature barley grains
All characterization studies were carried out on purified starch prepared from barley grain following Schulman and Kammiovirta (1991).
Microscopic analyses of starch granules
Purified starch suspension in water was visualized under both normal and polarized light using a Leica-DMR compound microscope to determine the starch granule morphology. Scanning electron microscopy was carried out on a Joel JSM 35C instrument. Purified starches were sputter-coated with gold and scanned at 15 kV at room temperature.
Estimation of amylose content
Amylose content of barley starches was estimated by two different methods: (i) the iodometric method; and (ii) Sepharose CL-2B gel filtration; both methods essentially as described in Regina et al. (2006). For the iodometric method, 2 mg of starch was defatted by incubation with 85% methanol at 65 °C. Dried starch following defatting was dissolved in urea-dimethyl sulphoxide (UDMSO) solution (nine parts DMSO and one part of 6 M urea) at a ratio of 1 ml of UDMSO per 2 mg of starch. A 50 μl aliquot of the starch–UDMSO solution was treated with 20 μl of I2–KI reagent (2 mg iodine, 20 mg potassium iodide ml−1 of water) and made up to a 1 ml volume with water. The optical density was read at 620 nm on 3×200 μl subsamples per replicate analysis and averaged. Standard samples containing amylose ranging from 0% to 100% were used to generate a standard curve. The absorbance of the test samples was converted to percentage amylose using a regression equation derived from the standard samples. For the gel filtration method using Sepharose CL-2B, 10 mg starch dissolved in 1 N NaOH was run through the column using 10 mM NaOH buffer at a pump speed of 1 ml min−1. The separated fractions of starch were assayed using the Starch Assay Kit (Sigma).
Differential alcohol precipitation of starch
Starch was fractionated into amylose, intermediate, and amylopectin fractions using the method of Klucinec and Thompson (1998) with modifications to fractionate a small amount of starch. Granular starch (100 mg) was dispersed in 3 ml of 90% DMSO by heating the mixture in a boiling water bath for 1 h with intermediate vortexing. Following dispersion, 5 vols of ethanol was added and the mixture was incubated at 4 °C for 4 h. The mixture was centrifuged at 6500 g for 15 min at 4 °C. The supernatant was discarded and the pellet was washed by suspending the pellets in 15 ml of ethanol and centrifuging at 6500 g. The washing step was repeated once with ethanol and once with acetone. The acetone precipitated starch was dried at 37 °C overnight.
The dried starch was redispersed in 3 ml of 90% DMSO as above. To this starch 7 vols (21 ml) of an aqueous mixture of 6% 1-butanol and 6% isoamyl alcohol was added while stirring. The mixture was placed in a boiling water bath for another 20 min, followed by leaving in the water bath to cool down to 28 °C. Following incubation at 4 °C overnight, the mixture was gently agitated and then centrifuged at 10 000 g for 15 min at 4 °C. The supernatant was saved and the precipitates were resuspended in 4 ml water by incubating in a boiling water bath for 30 min with intermittent vortexing. Isoamyl acohol and butanol were added to bring both concentrations to 5.2% v/v, the same concentration as the first precipitation. This mixture was then heated, cooled, and centrifuged as described earlier. The resultant supernatant was combined with the first supernatant. The precipitation in butanol/isoamylalcohol was once more repeated and the supernatant from this step was combined with the earlier two supernatants. This supernatant mixture contained the amylopectin fraction.
The precipitate from the butanol/isoamylalcohol precipitation step was dispersed in 3 ml 90% DMSO and then mixed with 21 ml of 6% 1-butanol, heated, cooled, and spun down as described earlier. The supernatant from this step contained the intermediate fraction. The final precipitate containing the amylose fraction was dispersed in 3 ml of 90% DMSO followed by precipitation using 4 vols of ethanol and then washed and dried as described earlier. The amylopectin and intermediate fraction containing supernatants were also precipitated as described for the amylose fraction.
Chain length distribution
Chain length distribution of barley starches was analysed following the method of O'Shea et al. (1998) using a P/ACE 5510 capillary electrophoresis system (Beckman) with argon-LIF detection.
Reducing end assay
The frequency of branching in amylose and amylopectin molecules was determined based on the reducing end assay using dinitrosalicylic acid (DNS) of Bernfeld (1955). Accurately weighed (5 mg) starch was debranched with isoamylase as follows. Starch was suspended in 187.5 μl of water followed by dissolution in 12.5 μl of 2 M NaOH by vigorous vortexing and boiling for 5 min. After cooling of the starch solution, 8 μl of glacial acetic acid was added followed by 25 μl of 1 M sodium acetate and 250 μl of water. For debranching, 2.5 μl of isoamylase (Megazyme) was added to the starch solution and incubated at 37 °C for 2 h. After incubation, the enzyme was inactivated by boiling for 10 min. The debranched starch solution (50 μl) was mixed with 150 μl of DNS solution and incubated at 100 °C for 10 min. DNS solution was prepared by dissolving 5 g of DNS in 100 ml of 2 M NaOH followed by adding di-sodium tartrate solution (150 g dissolved in 250 ml of water) and making up the final volume to 500 ml. After incubation of the starch–DNS solution for 10 min at 100 °C, 180 μl of water was added and the absorbance measured at 540 nm. A calibration curve was generated using malto-triose solution for a concentration ranging from 0–200 nmol. The reducing end of the starch sample was estimated as equivalent malto-triose.
Total starch content
Starch content was determined by the AOAC Method 996.1 using the total starch assay kit supplied by Megazyme (Bray, Co Wicklow, Republic of Ireland).
Starch swelling power
Starch swelling power of gelatinized starch was determined following the small scale test of Konik-Rose et al. (2001) which measures the uptake of water during gelatinization of starch.
Pasting and gelatinization properties
Starch paste viscosity and gelatinization properties were estimated using rapid visco analyser (RVA) and differential scanning calorimetry (DSC), respectively, essentially as described in Regina et al. (2004). The temperature profile for the RVA comprised the following stages: hold at 60 °C for 2 min, heat to 95 °C over 6 min, hold at 95 °C for 4 min, cool to 50 °C over 4 min, and hold at 50 °C for 4 min.
Statistical analysis
Analysis of variance was conducted using the 8th edition of Genstat for Windows (VSN International Ltd, Herts, UK).
Results
Generation of barley transgenic lines using hairpin RNAi constructs of SBE IIa and SBE IIb
Experiments designed to define the degree to which the SBE IIa and SBE IIb genes control the synthesis of α(1→6) linkages in barley endosperm starch were carried out by utilizing a hairpin-RNA-mediated silencing approach. RNAi constructs targeting SBE IIa (hp-SBE IIa) and SBE IIb (hp-SBE IIb) were generated by cloning regions of wheat SBE IIa (GenBank accession no. Y11282) and wheat SBE IIb (GenBank accession no. AY740401) cDNA and have been described in Regina et al. (2006). Briefly, DNA fragments corresponding to exons 1 to 3 of the targeted genes were cloned as inverted repeats, separated by intron 3 of the respective gene. Exons 1 to 3 of wheat SBE IIa are ∼93% identical to that of barley (GenBank accession no. AF064560) and the same exons of wheat SBE IIb are ∼ 92% identical to that of barley (GeneBank accession no. AF064561). These exons of SBE IIa and SBE IIb have an identity of 70% between each other. The inverted repeat region was driven at the 5′ end by a high molecular weight glutenin promoter (D×5 subunit gene, Accession no. X12928) directing expression in the endosperm from an early developmental stage, and terminated at the 3′ end by a nopaline synthase terminator. The constructs were transferred to a barley transformation vector pWBVec8 (Wang et al., 1998) and used for Agrobacterium-mediated transformation of immature embryos.
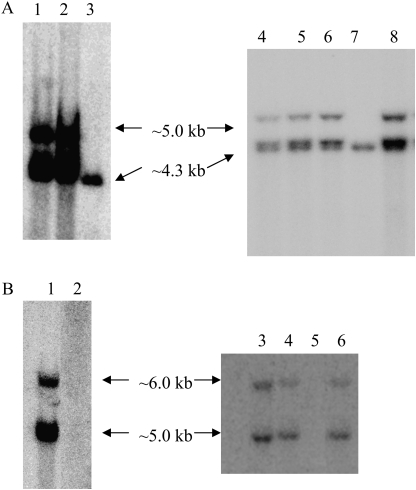
Positive barley transgenic lines from 12 independent transformation events of SBE IIa (hp-SBE IIa lines) and seven events of SBE IIb (hp-SBE IIb lines) were identified by PCR (data not shown). Genomic DNA from individual plants of PCR positive barley hp-SBE IIa and hp-SBE IIb T0 transgenic lines and selected T2 and T3 transgenic lines (based on a reduction in the expression of SBE II isoforms as described later) was hybridized to probes from the intron 3 region of wheat SBE IIa and SBE IIb genes, respectively, that were used to generate the respective constructs. Wheat SBE IIa intron 3 region has an identity of ∼92% with that of the barley SBE IIa gene and wheat SBE IIb intron 3 region has an identity of ∼67% to that of the barley SBE IIb gene. A non-transformed control (WT) showed one band of ∼4.3 kb using a wheat intron 3 SBE IIa probe (Fig. 1A, lanes 3 and 7) and no bands using a wheat intron 3 SBE IIb probe (Fig. 1B, lanes 2 and 6). The results from T0 lines (data not shown) indicated 1–5 copies of transgene in hp-SBE IIa and 1–4 copies in hp-SBE IIb transgenic barley lines.
Fig. 1.
(A, B) Southern hybridization of genomic DNA from barley transgenic lines. ApaI digested genomic DNA extracted from individual plants of transgenic barley lines was hybridized to a probe from the intron 3 region of SBE IIa (A) and SBE IIb (B) genes. (A) Lanes 1 and 2 are two different T2 lines of the same transformation event, IIa 4 and IIa 4.1, lanes 3 and 7 are non transformed control (WT), lanes 4–6 are F2 barley lines of the cross between IIa 4×IIb 4, BC 1, BC 2, and BC 3, and lane 8 is a T3 line of IIa 4. (B) Lane 1 is a T2 line, IIb 4, lanes 2 and 6 are WT, lane 3–5 are same F2 lines as the lanes 4–6 of (A), and lane 7 is a T3 line of IIb 4.
In order to study the effects of the presence of both hp-SBE IIa and hp-SBE IIb transgenes together in barley, a T3 line of hp-SBE IIa (IIa 4) and a T3 line of hp-SBE IIb (IIb 4) were intercrossed and the progeny designated as BC. These two parental lines were shown to have the same Southern banding pattern in both T2 and T3 generations (Fig. 1A, lanes 1 and 8; B, lanes 1 and 7, respectively). The hp-SBE IIa and hp-SBE IIb transgenes segregating in the F2 lines of this cross was shown in the Southern blot (Fig. 1A, B). While the F2 lines BC 2 and BC 3 contained both hp-SBE IIa and hp-SBE IIb transgenes, BC 1 (Fig. 1A, lane 4; B, lane 3) contained only the hp-SBE IIa and the not hp-SBE IIb transgene.
Suppression of the expression levels of SBE IIa and SBE IIb genes in developing barley endosperm
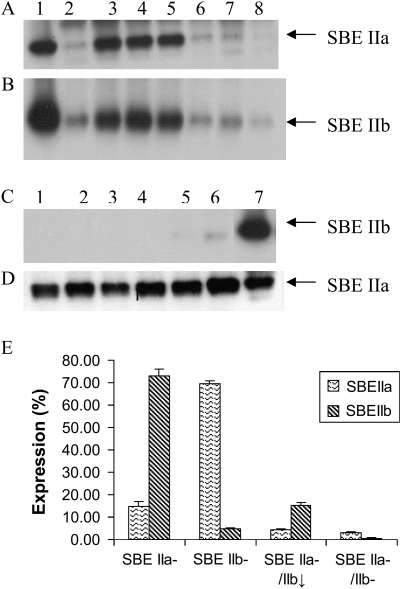
Individual endosperm samples from 7–10 developing T1 grains from 12 hp-SBE IIa and seven hp-SBE IIb T0 barley plants were analysed for protein expression at 15 d post anthesis (dpa) by immunoblotting using antibodies specific for SBE IIa or SBE IIb. Previously it was shown that these antibodies were isoform specific and do not cross react (Regina et al., 2005). The level of SBE IIa or SBE IIb expression in individual T1 endosperms of respective transgenic lines varied from near wild-type levels, through to weak expression (1–20%) and to no apparent expression (<1%). Figure 2A shows the segregation of SBE IIa expression in T1 endosperms from the hp-SBE IIa line, IIa 4. Figure 2C shows the segregation of SBE IIb expression in T1 endosperms from the hp-SBE IIb line, IIb 4. In some of the hp-SBE IIa lines, including the line IIa 4, SBE IIb expression was also reduced in developing endosperms with a reduced SBE IIa level (Fig. 2B). SBE IIa expression was unaltered in all the seeds analysed in which SBE IIb expression was essentially completely silenced by hp-SBE IIb (Fig. 2D).
Fig. 2.
Immunodetection of SBE IIa and SBE IIb in developing endosperms of barley transgenic lines. (A, B) Segregation of SBE IIa (A) and SBE IIb (B) expression in seven T1 developing endosperms (lanes 2–8) from the T0 barley hp-SBE IIa transgenic line IIa 4 as shown by immunoblotting using anti-SBE IIa and anti-SBE IIb antibodies, respectively. Lane 1 is WT. (C, D) Segregation of SBE IIb (C) and SBE IIa (D) expression in six T1 developing endosperms (lanes 1–6) from the T0 barley hp-SBE IIb transgenic line IIb 4 as shown by immunoblotting using anti-SBE IIb and anti-SBE IIa antibodies, respectively. Lane 7 is WT. (E) The relative levels of expression of SBE IIa and SBE IIb in selected barley transgenic lines, SBE IIa–, SBE IIb– , SBE IIa–/SBE IIb↓, and SBE IIa–/SBE IIb–. Immunoblots of non-denaturing PAGE scanned and the band intensities of the images measured using the TotalLab software package (Nonlinear Dynamics Ltd, Newcastle, UK). The expression of SBE IIa and SBE IIb in each transgenic line is expressed as a percentage of the level of expression of the respective isoform in WT.
Ten F3 developing endosperms each from the lines BC 2 (containing the hp-SBE IIa and hp-SBE IIb transgenes) and BC 1 (containing the hp-SBE IIa transgene only) were also analysed for SBE II expression levels (data not shown). SBE IIa and SBE IIb expression were both significantly reduced in all the ten endosperms analysed in BC 2 with the levels decreased to ∼ 4.5% and ∼0.8%, respectively, compared to WT expression, whereas in BC 1 which contained only hp-SBE IIa transgene, the level of SBE IIa in all the ten endosperms studied was reduced to ∼15% of WT while the SBE IIb expression remained close to normal.
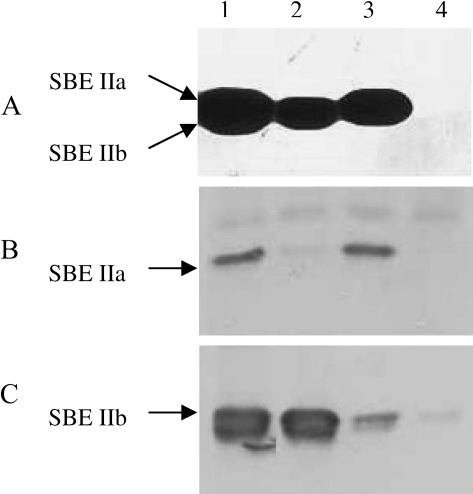
Four individual homozygous transgenic lines were selected for further characterization to represent lines where SBE IIb alone is suppressed (designated as SBE IIb–), SBE IIa alone is suppressed (designated as SBE IIa–), SBE IIa is suppressed with a concomitant reduction in SBE IIb (designated as SBE IIa–/SBE IIb↓), and both SBE IIa and SBE IIb isoforms are reduced (designated as SBE IIa–/SBE IIb–). From among all the lines analysed by Western blotting, these selected individuals exhibited the lowest expression levels of the respective isoform/s. The homozygous condition of these lines was decided based on no segregation of protein expression analysed from nine individual endosperms for at least two subsequent generations. The level of expression of SBE II isoforms in these lines in comparison with WT is shown in Fig. 2E. SBE II enzyme activity was assayed by zymogram in three of these lines (Fig. 3). The SBE IIa and SBE IIb isoform specific activities were confirmed by immunoblotting using anti-SBE IIa and anti-SBE IIb antibodies. The results revealed no SBE IIa or SBE IIb activity in the SBE IIa–/SBE IIb– line (Fig. 3, lane 4). In the SBE IIa– line, only SBE IIb activity and no SBE IIa activity (Fig. 3, lane 2) was detected and in the SBE IIb– line, only SBE IIa and no SBE IIb activity (Fig. 3, lane 3) was detected. However, weak bands of SBE IIa and SBE IIb were detected in the immunoblots in SBE IIa– and SBE IIb–, lines respectively. An activity assay was not conducted in the SBE IIa–/SBE IIb↓ line.
Fig. 3.
Comparison of branching enzyme in barley transgenic lines. Electrophoresis of soluble proteins (15 μg well−1) from 15 dpa endosperms was performed as described in the Materials and methods. (A) Activity staining of branching enzymes. After electrophoresis the gel was washed twice and incubated with maltotriose and visualized with iodine. (B, C) Immunoblots. The membranes were probed with SBE IIa antibody (B) and SBE IIb antibody (C). The samples were: lane 1, WT; lane 2, SBE IIa–; lane 3, SBE IIb–; lane 4, SBE IIa–/SBE IIb–.
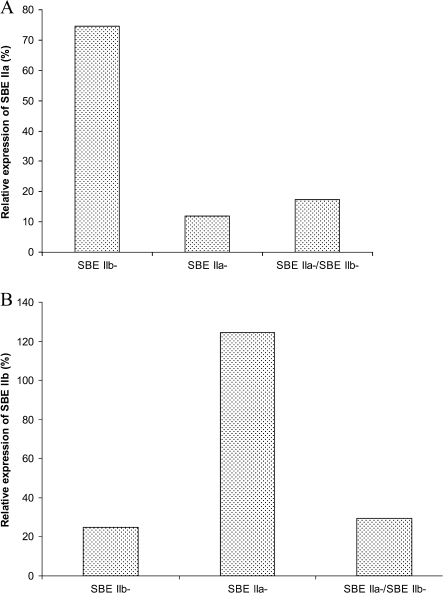
Transcript levels of SBE IIa and SBE IIb in endosperm of barley transgenic lines
The expression of SBE IIa and SBE IIb transcripts in SBE IIb–, SBE IIa–, and SBE IIa–/SBE IIb– were estimated through comparative quantitation using real-time PCR (see the Materials and methods). SBE IIa and SBE IIb transcript levels from each of the transgenic lines were normalized with the level of actin transcript from the respective lines. The level of each SBE II transcript was expressed as a percentage in relation to the expression of the respective enzyme transcript in WT (Fig. 4). The results showed that the level of both SBE IIa and SBE IIb transcripts were reduced to ∼25% of WT in the SBE IIa–/SBE IIb– line. In the SBE IIa– line the transcript level of SBE IIa was reduced to ∼18% of WT, whereas the expression of SBE IIb was ∼ 120% of the WT. In the SBE IIb– line the transcript level of SBE IIb was ∼22% of WT, whereas SBE IIa expression was ∼ 72% of the wild type.
Fig. 4.
Transcript expression in transgenic barley lines. Expression of SBE IIa (A) and SBE IIb (B) transcripts in transgenic barley lines compared with the non-transformed control (WT) determined by comparative quantitation using real-time PCR. The expression of the SBE II isoform is normalized to expression of the house-keeping gene actin from respective lines. The average of two biological replicates is presented in the figure.
Altered starch granule morphology in barley endosperm when both SBE IIa and SBE IIb were suppressed
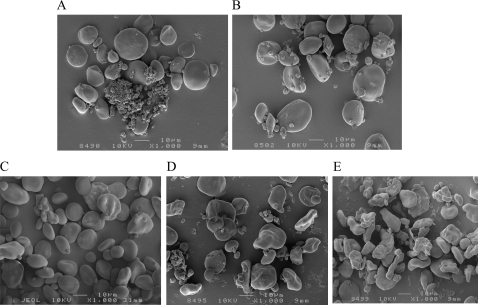
The morphology of endosperm starch granules from mature grains of barley transgenic lines containing the hp-SBE II transgenes was analysed by scanning electron microscopy. Starch granules from the SBE IIb– line (Fig. 5B) appeared to have similar morphology as that of WT (Fig. 5A). Starch granules with altered morphology were occasionally observed in the SBE IIa– line (Fig. 5C). A significant alteration in starch granule morphology was observed in the SBE IIa–/SBE IIb↓ (Fig. 5D) and SBE IIa–/SBE IIb– (Fig. 5E) lines, with the distortion being more extreme in the latter.
Fig. 5.
Scanning electron micrographs of isolated starch granules (1000× magnification). (A) WT, (B) SBE IIb–, (C) SBE IIa–, (D) SBE IIa–/SBE IIb↓, and (E) SBE IIa–/SBE IIb– lines.
Starch granules from the SBE IIa–/SBE IIb↓ and SBE IIa–/SBE IIb– lines with altered granule morphology revealed a significant loss of birefringence (which is indicative of a loss of crystallinity) with fewer granules showing birefringence and an overall reduction in intensity of birefringence when visualized under polarized light (Table 1). On average, ∼40% of the starch granules in SBE IIa–/SBE IIb↓ endosperm did not display birefringence in contrast to <3% in WT seeds. The loss of birefringence in the endosperm starch granules of SBE IIa–/SBE IIb– line was >90%. In the case of the the SBE IIa– or SBE IIb– lines the starch granules did not show any statistically significant alteration with regard to the level of birefringence compared to WT.
Table 1.
Birefringence of starch granules and amylose content of starch from barley transgenic lines
| Line no. | Targeted enzyme | % Starch granules witha |
Mean amylose content (%)c | ||
| No BFb | Partial BF | Full BF | |||
| Wild type | Non-transformed | 2.4 | 4.6 | 93.1 | 28.5 |
| SBE IIb– | SBEIIa | 1.8 | 5.3 | 92.9 | 30.5 |
| SBE IIa– | SBEIIb | 1.5 | 9.1 | 89.4 | 38.1 |
| SBE IIa–/SBE IIb↓ | SBEIIa and SBEIIb | 38.9 | 26.3 | 34.8 | 65.8 |
| SBE IIa–/SBE IIb– | SBEIIa and SBEIIb | 94.1 | 3.3 | 2.7 | 89.3 |
| LSD (5%) | – | 11.6 | 4.5 | 12.9 | 4.6 |
Mean of three microscopic fields.
Birefringence.
Mean of three replicates.
Elevation of amylose content in barley grains when both SBE IIa and SBE IIb are suppressed
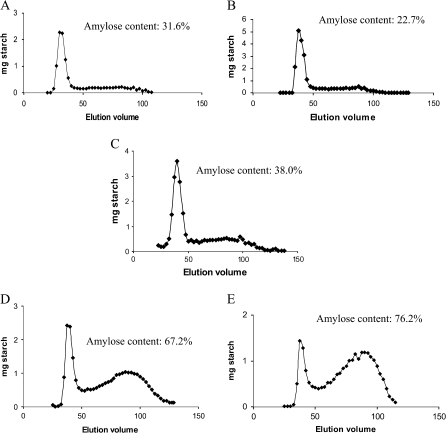
The amylose content of barley transgenic lines with reduced expression of SBE IIa and/or SBE IIb was analysed by an iodometric method (Table 2). No significant difference in amylose content was observed in the SBE IIb– line (30.5%) compared to WT (28.5%). An elevation in amylose content was observed in lines in which SBE IIa expression was reduced. While there was only a slight increase (38.1%) when SBE IIa alone was reduced (SBE IIa– line) compared to WT, the increase was more dramatic when both SBE IIa and SBE IIb were reduced with amylose contents of 65.8% and 89.3% for SBE IIa–/SBE IIb↓ nd SBE IIa–/SBE IIb– lines, respectively. Similar increases were noticed by gel filtration using a Sepharose CL-2B column. The amylose contents estimated by this method for SBE IIa–/SBE IIb↓ was 67.2% (Fig. 6D) and SBE IIa–/SBE IIb– was 76.2% (Fig. 6E) whereas that of SBE IIa– was 38.0% (Fig. 6C), SBE IIb– 22.7%, (Fig. 6B), and WT 31.6% (Fig. 6A). A strong negative correlation (r= –0.989) was observed between amylose content and percentage of starch granules with full birefringence.
Table 2.
Branching frequency of amylose and amylopectin from barley transgenic lines
| Line no. | Enzyme targeted | Branching frequencya |
|
| Fractions separated by sepharose CL 2B gel filtration | |||
| Amylopectin (nmol equivalent reducing ends mg−1 starch) | Amylose (nmol equivalent reducing end mg−1 starch) | ||
| Wild type | Non transformed | 136.2 | 4.5 |
| SBE IIb– | SBE IIb | 130.8 | 13.3 |
| SBE IIa– | SBE IIa | 131.0 | 7.3 |
| SBE IIa–/SBEIIb– | SBE IIa and SBE IIb | 45.6 | 7.3 |
| LSD (0.05) | 15.56 | 3.8 | |
Mean of three replicates.
Fig. 6.
Sepharose CL 2B gel chromatogram of starch from barley transgenic lines. (A) WT, (B) SBE IIb–, (C) SBE IIa–, (D) SBE IIa–/SBE IIb↓, and (E) SBE IIa–/SBE IIb–. Starch molecules separated by gel filtration are assayed using starch assay kit (Sigma). Amylose content estimated by this method as a percentage of total starch is shown on respective graphs.
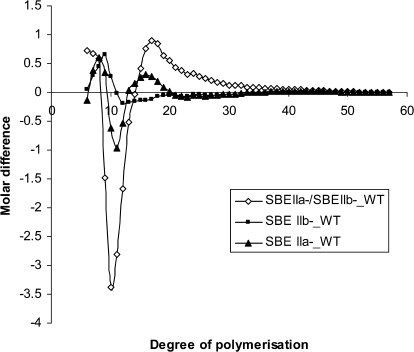
Chain length distribution of amylopectin is altered in barley endosperm when SBE IIa and /or SBE IIb are suppressed
Starch from the barley transgenic lines and the control was fractionated into amylose, amylopectin, and intermediate fractions by the differential alcohol precipitation method of Klucinec and Thompson (1998) (see the Materials and methods). In their study, the three fractions separated from corn starch using this method have been shown to bear the properties expected of amylose, amylopectin, and intermediate molecules. The nature of the amylose and amylopectin fractions that were separated from barley starches was confirmed by their iodine binding property (data not shown). The chain length distribution of isoamylase de-branched amylopectin fraction thus generated was determined by fluorophore assisted carbohydrate electrophoresis (FACE). The molar difference plots in which the normalized chain length distributions of WT subtracted from that of the transgenic lines are given in Fig. 7. The chain length distribution varied considerably depending on the type of SBE II isoform/s reduced. In the SBE IIb– line in which only SBE IIb was reduced, there was a reduction in the proportion of chains of DP 12–18 and an increase in the proportion of shorter chains of DP 7–10. In the SBE IIa– line where only SBE IIa was reduced, the proportion of chains of DP 10–12 decreased while that of DP 7–9 and 14–18 increased. In the SBE IIa–/SBEIIb– line, where both isoforms were reduced, there was a significant reduction in the chains of degree of polymerization (DP) 9–13 whereas there was a considerable increase in the proportion of chains of DP 6–8 and 15–30. The line SBE IIa–/SBE IIb↓ was not included in this particular experiment, however, a previous comparison of FACE profiles of debranched starch of SBE IIa–/SBE IIb↓ produced a chain length distribution profile similar to that of the SBE IIa–/SBE IIb– line, although the changes were more extreme in the SBE IIa–/SBE IIb– line compared to the SBE IIa–/SBE IIb↓ (data not shown).
Fig. 7.
Chain length profile comparison of starches from barley transgenic lines with respect to that of WT. The percentage of total mass of the individual oligosaccharides from starches from respective non-transformed controls is subtracted from the corresponding values from starches from transgenic lines. Samples are SBE IIa– (filled triangles), SBE IIa–/SBE IIb– (open diamonds), SBE IIb– (filled squares).
For the starch characterization studies described in the rest of the paper, the line SBE IIa–/SBE IIb↓ was not included due to the scarcity of starch sample from this line.
Frequency of α(1→6) branching altered in barley endosperm starch
The frequency of α(1→6) branch points in amylose and amylopectin fractions separated by Sepharose CL-2B column gel chromatography from barley transgenic lines was estimated by isoamylase debranching of starch followed by measuring the reducing ends thus created by a reducing end assay (see the Materials and methods), and expressed as nmols of equivalent reducing ends per mg of starch derived from maltotriose standards. There was a significant reduction in the frequency of branching in amylopectin molecules in SBE IIa–/SBE IIb– starch with a reducing end equivalent value of 45.6 nmol mg−1 starch compared to the WT which gave a value of 136.2 nmol mg−1 starch (Table 2). The branching frequency of amylose molecules from the transgenic starches revealed that both lines with reduced expression of SBE IIa (SBE IIa– and SBE IIa–/SBE IIb–) did not vary statistically in comparison to WT, however, amylose from the SBE IIb– line exhibited a significant increase in the branching frequency with a value of 13.3 nmol mg−1 starch compared to the WT (4.5 nmol) and both the SBE IIa reduced lines (7.3 nmol for both SBE IIa– and SBE IIa–/SBE IIb– lines). These results were also supported by the branching frequency determined on the amylose and amylopectin fractions separated by the differential alcohol precipitation method of Klucinec and Thomson (1998) (data not shown).
Changes in starch functional properties with reduction in SBE IIa and SBE IIb in barley grains
Starches from SBE IIa suppressed lines showed a significantly reduced starch swelling volume determined by a small-scale starch swelling power test compared to the control, with a more severe reduction of swelling in SBE IIa–/SBE IIb– starch than in SBE IIa– starch. The value was reduced from 14.4 in WT to 12.8 in SBE IIa– starch, and further reduced to 5.0 in SBE IIa–/SBE IIb– starch (Table 3). However, the trend was found to be in the opposite direction when SBE IIb alone was reduced and, in this line, the starch swelling power increased to a value of 15.6 (Table 3).
Table 3.
Endosperm starch swelling power, 100 kernel weight, total starch content, and DSC starch gelatinization parameters of transgenic barley lines
| Line no. | Starch swelling powera | 100 kernel weight (g)a | Starch content (% of whole meal flour)a | DSC parameters (Peak 1)a |
|||
| Onset °C | Peak °C | End °C | ΔHj/g | ||||
| Wild type | 14.4 | 5.00 | 60.0 | 59.4±0.05 | 63.0±0.34 | 69.8±0.38 | 5.4±0.51 |
| SBE IIb– | 15.6 | 3.99 | 54.7 | 61.2±0.07 | 64.8±0.30 | 70.8±0.53 | 5.8±0.44 |
| SBE IIa– | 12.8 | 4.21 | 56.9 | 60.9±0.14 | 64.8±0.32 | 70.9±1.51 | 4.8±0.44 |
| SBE IIa–/SBEIIb– | 5.0 | 3.77 | 45.4 | 59.27±0.65 | 71.31±0.89 | 80.43±0.32 | 2.51±0.38 |
| LSD (0.05) | 1.0 | 0.06 | 2.4 | – | – | – | – |
Mean of three replicates.
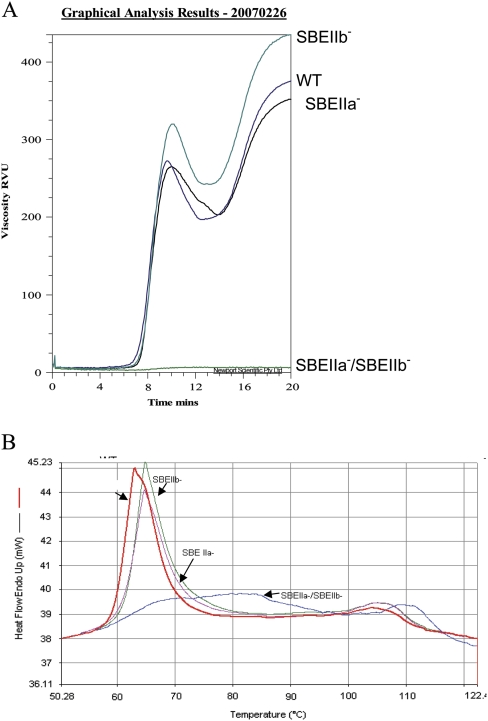
Analysis of viscosity parameters determined by a rapid visco analyser (RVA) revealed that starch from SBE IIa–/SBE IIb– failed to paste at the temperature profile that was used for RVA analysis (Fig. 8A). SBE IIa– had an RVA profile very similar to that of WT, whereas starch from the SBE IIb– line exhibited a higher peak (320 RVU) and final viscosity (435 RVU) compared to WT, which showed a peak viscosity of 272 RVU and a final viscosity of 375 RVU.
Fig. 8.
RVA and DSC studies on starches from barley transgenic lines. (A) RVA profile of starches from WT, SBE IIa–, SBE IIb–, and SBE IIa–/SBE IIb–. (B) DSC profile of starches from WT, SBE IIa–, SBE IIb–, and SBE IIa–/SBE IIb–.
Thermal properties of starch from transgenic lines were studied by differential scanning calorimetry (DSC). Comparative profiles of barley transgenic lines and WT are shown in Fig. 8B. The profile showed two peaks. Peak 1 represents the transition induced by melting of the amylopectin crystallites and peak 2 represents the transition due to dissociation of the amylose–lipid complex (Soulaka and Morrison, 1985). The SBE IIa–/SBE IIb– starch showed a much flatter thermal transition profile compared to other starches and had a higher peak (71.3 °C) and end of gelatinization (80.4 °C) temperatures represented in peak 1 compared to other starches (Fig. 8B; Table 3). Starches of both SBE IIa– (64.8 °C) and SBE IIb– (64.8 °C) also showed a slight increase in the gelatinization peak temperature compared to WT (63.0 °C). Gelatinization enthalpy was significantly lower in SBE IIa–/SBE IIb– starch compared with starches from other lines. With regard to the amylose–lipid dissociation peak (peak 2) also, the SBE IIa–/SBE IIb– starch showed higher peak and end temperature values (data not shown).
Alterations in kernel weight and total starch content of barley transgenic lines
A statistically significant reduction in 100 kernel weight and total starch content in mature grains was noticed in all transgenic lines compared to the WT, with a more extreme reduction in the SBE IIa–/SBE IIb– line in both cases (Table 3). The value of 100 kernel weight was reduced from 5.00 g in WT to 4.21 g in SBE IIa–, 3.99 g in SBE IIb–, and 3.77 g in the SBE IIa–/SBE IIb– line. The total starch content was reduced from 60% in WT to 56.9% in SBE IIa–, 54.7% in SBE IIb–, and 45.4% in SBE IIa–/SBE IIb– line.
Discussion
The objective of this study was to define the contribution of individual isoforms of starch branching enzymes to α(1→6) linkage formation in starch in barley endosperm. The synthesis of α(1→6) linkages is important to the fine structure of amylopectin and amylose, and the amount of amylopectin synthesized, relative to amylose. The roles of SBE IIa and SBE IIb in determining amylose content, chain length distribution, and branching frequency of starch in barley was examined. The study revealed that, in barley, a reduction in the expression of both SBE IIa and SBE IIb was required to elevate the amylose content to >65% from ∼28% in WT, whereas when either of the isoforms was reduced individually there was either no change (in the case of the SBE IIb reduced line) or a slight increase (in the case of the SBE IIa reduced line) in amylose content. The study also revealed that both SBE IIa and SBE IIb play specific roles in shaping the fine structure of amylose and amylopectin either directly or indirectly.
The fine structure of amylopectin in high amylose barley was significantly altered with respect to amylopectin chain length distribution. When SBE IIa was reduced either by itself or in combination with a reduction in SBE IIb there was an increase in the proportion of the two sets of chains, one that of very short chains (<DP 9) and the other that of long chains (>DP 15). The proportion of medium DP chains showed a significant reduction in both these lines compared to WT. A similar change was observed in wheat (Regina et al., 2006). This observation differed from that of high amylose maize and potato where an increased proportion of only long chains was reported (Shi et al., 1998; Schwall et al., 2000). When the SBE IIb isoform was reduced in barley, the chain length profile again differed from both that of WT as well as that of SBE IIa– and SBE IIa–/SBE IIb– starches. The results clearly revealed that both SBE IIa and SBE IIb isoforms played distinct roles in determining the chain length distribution: suggesting the requirement of SBE IIa for the synthesis of chains of DP 10–12, while SBE IIb was required to synthesize chains of DP 12–18. There might also be some functional overlap between SBE IIa and SBE IIb, for example, in the case of synthesis of DP 12. The results also indicated some type of functional interaction between SBE IIa and SBE IIb because when both were missing there was greater reduction of DP 9–13, and the effect on DP 15–18 changed to an increase as against a decrease in this DP range when SBE IIb alone was reduced.
One of the primary effects of altered SBE activity will be on the distribution or frequency of branches (Thompson, 2000; Yao et al., 2004). Since both amylopectin and amylose are branched molecules, the frequency of branching in both these molecules was studied after separating the fractions through either a Sepharose CL-2B column or by differential alcohol precipitation. The results indicated that there was a significant reduction in the number of branches in the amylopectin molecules when both the SBE II isoforms were reduced simultaneously. When either of the isoforms was reduced there was no significant alteration in the number of branches in the amylopectin molecules. The more intriguing observation was the increased frequency of branching in the amylose fraction when SBE IIb expression was reduced (Table 2). Prediction of the effects of down-regulation of individual enzymes involved in starch biosynthesis is difficult because of the pleiotrophic effects on protein complex formation (Tetlow et al., 2004). It may be that SBE IIb has an inhibitory role on SBE I in vivo in barley and in the absence of SBE IIb, SBE I is active in branching long-chained starch molecules. Morell et al. (1997) showed an increased affinity of SBE I in vitro for amylose over amylopectin. Interestingly increased branching of the amylose fraction is not observed when both SBE IIa and SBE IIb are missing. When the activity of both SBE II isoforms are absent, as in the case of the SBE IIa–/SBE IIb– line, SBE I may be required to act on both amylopectin and amylose and hence the extent of branching in this line is not to the level that was observed when SBE IIb alone was reduced. However, no difference was observed in SBE I activity between the transgenic lines in our in vitro assay (data not shown). This is under further investigation using a range of substrates including purified starch from developing endosperm from each of the transgenic lines. Eliminating or reducing the expression of SBE I did not yield any noticeable phenotype in wheat or maize, although in those studies branching of amylose was not investigated. Increased branching of amylopectin in maize was reported by Yao et al. (2004) due to the absence of SBE I in an SBE IIb mutant background, the authors indicating the possibility of SBE I inhibiting SBE IIa in vivo. It is understood that much more research needs to be carried out in this area before a clear picture emerges.
The changes in starch structure due to altered expression of branching enzymes also influenced the functionality of starch as indicated by changes in starch swelling power, thermal, and pasting properties. In agreement with previous findings, swelling of starch showed an inverse relationship to amylose content (Shi et al., 1998). A broader gelatinization temperature range for high amylose barley starch was revealed, similar to that of the high amylose maize starches (Shi et al., 1998). The melting peak of the amylose–lipid complex was also higher in high amylose barley. The changes in DSC profile observed in SBE IIa–/SBE IIb– starch could be attributed to the structural changes in amylopectin, together with the increased proportion of amylose leading to the suppression of hydration and the swelling of starch. Studies on low amylopectin starch (LAPS) maize suggested the possibility of the formation of amylose double helices that require high temperature and energy to disorder leading to a high gelatinization temperature (Shi et al., 1998). Crystallinity and double helix structure studies are yet to be done on the high amylose barley line described here. Our study suggests that the pasting properties of starch could also be influenced by a combination of the content and the structure of both glucan components. The high amylose barley starch produced by combined suppression of SBE IIa and SBE IIb did not paste at the temperature profile used for the RVA experiment. The SBE IIb reduced starch showed an increased starch swelling power and peak viscosity compared to the control, despite the fact that both these starches have a similar amylose content. The subtle alterations in the amylose and amylopectin structure, including the increased branching frequency in amylose when SBE IIb was reduced, might have contributed to the elevated swelling and peak viscosity in this line.
The importance of the SBE IIa in controlling the amylose content in wheat was established (Regina et al., 2006), however, all the wheat lines with a significant reduction in SBE IIa expression showed a concomitant reduction in SBE IIb. In this study, RNAi constructs targeting specific isoforms of branching enzymes have successfully suppressed the respective enzyme expression in barley. In some of the SBE IIa reduced lines, there was a concomitant reduction in SBE IIb, whereas in some other lines, there was no reduction in SBE IIb. The respective gene cassettes used in SBE IIa and SBE IIb RNAi constructs for barley transformation were the same as those used for wheat transformation described in Regina et al. (2006) due to the high level of homology between wheat and barley genes (see Results). The sequence of SBE IIb used in the hp-SBE IIb construct has one stretch of 21 nt with 100% identity in barley SBE IIa cDNAs and two regions of 21 nt with ∼70% identity as determined by the software Compare of GCG Package (Genetics Computer Group, Inc, Wisconsin). Similarly, the sequence of SBE IIa used in the hp-SBE IIa construct has one stretch of 21 nt with 95% identity in barley SBEIIb cDNAs and four regions with 70–80% identity. Evidence from the wheat work has shown that the cross suppression was not due to the RNAi mechanism directly as shown by the normal level of accumulation of SBE IIb transcripts in SBE IIa targeted lines. In this study, the transcript analysis through real-time PCR has shown that the reduction in the level of transcript is conspicuous only in the isoform targeted. Possible explanations given for cross suppression in wheat were either due to post transcriptional mechanisms or decreased SBE IIb stability when SBE IIa was reduced due to changes in protein complexes described by Tetlow et al. (2004). The current results from the barley work have indicated that SBE IIb can remain stable in the absence of SBE IIa (as observed in some of the lines). Hence a third possible explanation for cross suppression is epigenetic modifications consequential to the inactivation of SBE IIb. RNA silencing resulting in the formation of silent chromatin, characterized by histone modifications and dense DNA methylation has recently been revealed in plants (Ingelbrecht et al., 2006).
Abolishing SBE I activity resulted in no major impact on starch properties (Safford et al., 1998; Blauth et al., 2002; Regina et al. 2004), although, in rice, minor changes in the fine structure of amylopectin has been reported (Satoh et al., 2003). In maize and rice, suppression of SBE IIb was required in order to obtain a high amylose phenotype (Garwood et al., 1976; Mizuno et al., 1993). However, in a previous study in wheat (Regina et al., 2006) and in this study in barley, suppression of SBE IIb alone resulted in only minor changes in amylose content, suppression of SBE IIa was required for significant changes in amylose content. In maize mutations abolishing the expression of SBE IIa led to no significant change in the endosperm amylose content or starch structure (Blauth et al., 2001, 2002). These differences can be related to differences in relative gene expression in these species. In maize, SBE IIb is the predominant isoform in the endosperm which is at least 50 times the abundance of SBE IIa (Gao et al., 1997). In rice, SBE IIb is the major isoform in the endosperm (Yamanouchi and Nakamura, 1992). In barley endosperm there is approximately equal activity of SBE IIa and SBE IIb in the endosperm (Sun et al., 1998). These results suggest that there is a high degree of functional overlap between the SBE IIa and SBE IIb isoforms such that the control of α(1→6) linkage formation is driven by the relative expression levels of the two genes in the cereal endosperm. However some non-overlapping functionality is also indicated by the specific alterations in the fine structure of the starch molecules when each of the isoforms is reduced. The reason for the generally observed duplication of starch biosynthetic enzyme isoforms in monocots (eg SBE IIa, SBE IIb, SS IIa, SS IIb, SS IIc, SS IIIa, SS IIIb etc) is not only to allow differential expression in tissue and developmental stages, but perhaps also to satisfy the need for altered enzymatic functionality.
The production of high amylose barley provides an opportunity to explore a range of additional end-uses and benefits of this crop. Increased amylose content is one method of producing ‘resistant starch’: starch that resists breakdown in the small intestine of humans, surviving to pass into the large bowel where it is fermented, releasing short chain fatty acids that have a range of benefits for bowel health and may protect against the risk of colo-rectal cancer (Topping and Clifton, 2001; Topping et al., 2003). Other compositional changes are yet to be analysed in the high amylose barley described here.
Acknowledgments
The authors acknowledge Jean-Phillippe Ral for scientific advice on starch characterization, Oscar Larroque for starch chain length analysis, Peggy Kooij for conducting the starch swelling power test, and Rosemary White for microscopy.
References
- Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annual Review of Plant Biology. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Bernfeld P. Amylases, α and β. Methods in Enzymology. 1955;1:149–158. [Google Scholar]
- Bhattacharyya MK, Martin C, Smith A. The importance of starch biosynthesis in the wrinkled seed shape character of peas studied by Mendel. Plant Molecular Biology. 1993;22:525–531. doi: 10.1007/BF00015981. [DOI] [PubMed] [Google Scholar]
- Blauth SL, Kim KN, Klucinec J, Shannon JC, Thompson DB, Guiltinan M. Identification of Mutator insertional mutants of starch-branching enzyme 1 (SBE1) in Zea mays L. Plant Molecular Biology. 2002;48:287–297. doi: 10.1023/a:1013335217744. [DOI] [PubMed] [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan M. Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiology. 2001;125:1396–1405. doi: 10.1104/pp.125.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvalle D, Planchot V, Ball SG, D'Hulst C. Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. The Plant Cell. 2006;18:2694–2709. doi: 10.1105/tpc.105.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergason V. High amylose and waxy corns. In: Hallauer AR, editor. Specialty corns. Boca Raton: CRC Press; 1994. pp. 97–110. [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ. Independent genetic control of maize starch-branching enzymes IIa and IIb. Plant Physiology. 1997;114:69–78. doi: 10.1104/pp.114.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood DL, Shannon JC, Creech RG. Starches of endosperms possessing different alleles at the amylose-extender locus in Zea mays. Cereal Chemistry. 1976;53:355–364. [Google Scholar]
- Ingelbrecht IL, Mirkov TE, Dixon AG, Menkir A. Epigenetic lessons from transgenic plants. In: Teixeira da Silva JA, editor. Floriculture, ornamental and plant biotechnology. Global Science Books Ltd; 2006. pp. 88–97. [Google Scholar]
- Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, Jeffcoat R, Safford R. A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterization of multiple forms of SBE A. The Plant Journal. 1999;18:163–171. doi: 10.1046/j.1365-313x.1999.00441.x. [DOI] [PubMed] [Google Scholar]
- Klucinec JD, Thompson DB. Fractionation of high-amylose maize starches by differential alcohol precipitation and chromatography of the fractions. Cereal Chemistry. 1998;75:887–896. [Google Scholar]
- Konik-Rose CM, Moss R, Rahman S, Appels R, Stoddard F, McMaster G. Evaluation of the 40 mg Swelling Test for measuring starch functionality. Starch-Stärke. 2001;53:14–20. [Google Scholar]
- Kosar-Hashemi B, Irwin JA, Higgins J, Rahman S, Morell MK. Isolation, identification and characterization of starch-interacting proteins by 2-D affinity electrophoresis. Electrophoresis. 2006;27:1832–1839. doi: 10.1002/elps.200500400. [DOI] [PubMed] [Google Scholar]
- Li Z, Rahman S, Kosar-Hashemi B, Mouille G, Appels R, Morell MK. Cloning and characterization of a gene encoding wheat starch synthase I. Theoretical and Applied Genetics. 1999;98:1208–1216. [Google Scholar]
- Mizuno K, Kawasaki T, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T. Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. Journal of Biological Chemistry. 1993;268:19084–19091. [PubMed] [Google Scholar]
- Morell MK, Blennow A, Kosar-Hashemi B, Samuel MS. Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiology. 1997;113:201–208. doi: 10.1104/pp.113.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding the biosynthesis of the amylopectin crystal. Plant Physiology. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides-efficiency of labelling and high resolution separation. Carbohydrate Research. 1998;307:1–12. [Google Scholar]
- Rahman S, Li Z, Abrahams S, Abbott DC, Appels MK, Morell MK. Characterization of a gene for the major isoform of starch branching enzyme I in the wheat endosperm. Theoretical and Applied Genetics. 1999;98:156–163. [Google Scholar]
- Rahman S, Regina A, Li Z, Mukai Y, Yamamoto M, Kosar-Hashemi B, Abrahams S, Morell MK. Comparison of starch-branching enzyme genes reveals evolutionary relationships among isoforms. Characterization of a gene for starch-branching enzyme IIa from wheat D genome donor Aegilops tauschii. Plant Physiology. 2001;125:1314–1324. doi: 10.1104/pp.125.3.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M. High amylose wheat generated by RNA-Interference improves indices of large bowel health in rats. Proceedings of the National Academy of Sciences, USA. 2006;103:3546–3551. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Li Z, Pedler A, Mukai Y, Yamamoto M, Gale K, Sharp P, Morell MK, Rahman S. Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta. 2005;222:899–909. doi: 10.1007/s00425-005-0032-z. [DOI] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Li Z, Rampling L, Cmiel M, Gianibell C, Konik-Rose C, Larroque O, Rahman S, Morell MK. Multiple isoforms of starch branching enzyme I in wheat: lack of the major SBE-I isoform does not alter starch phenotype. Functional Plant Biology. 2004;31:591–601. doi: 10.1071/FP03193. [DOI] [PubMed] [Google Scholar]
- Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, Strongitharm BH, Russel A, Gidley MJ. Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydrate Polymer. 1998;35:155–168. [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Naoko F, Nakamura Y. Starch-branching enzyme 1-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiology. 2003;133:1–11. doi: 10.1104/pp.103.021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA. Production of very-high-amylose potato starch by inhibition of SBE A and B. Nature Biotechnology. 2000;18:551–554. doi: 10.1038/75427. [DOI] [PubMed] [Google Scholar]
- Schulman AH, Kammiovirta K. Purification of barley starch by protein extraction. Starch. 1991;43:387–389. [Google Scholar]
- Shi Y-C, Capitani T, Trzasko P, Jeffcoat R. Molecular structure of a low-amylopectin starch and other high-amylose maize starches. Journal of Cereal Science. 1998;27:289–299. [Google Scholar]
- Soulaka AB, Morrison WR. The amylose and lipid contents, dimensions, and gelatinsation characteristics of some wheat starches and their A-and B-granule fractions. Journal of the Science of Food and Agriculture. 1985;36:709–718. [Google Scholar]
- Sun C, Sathish P, Ahlandsberg S, Deiber A, Jansson C. Identification of four starch-branding enzymes in barley endosperm: partial purification of forms I, IIa and IIb. New Phytologist. 1997;137:215–222. doi: 10.1046/j.1469-8137.1997.00796.x. [DOI] [PubMed] [Google Scholar]
- Sun C, Sathish P, Ahlandsberg S, Jansson C. The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiology. 1998;118:37–49. doi: 10.1104/pp.118.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. The Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DB. On the non-random nature of amylopectin branching. Carbohydrate Polymer. 2000;43:223–239. [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fleg S, Wang M, Thornton S, Brettel R. Agrobacterium tumefaciens-mediated barley transformation. The Plant Journal. 1997;11:1369–1376. [Google Scholar]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiological Reviews. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Topping DL, Morell MK, King RA, Li Z, Bird AR, Noakes M. Resistant starch and health-Himalaya 292 a novel barley cultivar to deliver benefits to consumers. Starch. 2003;55:539–545. [Google Scholar]
- Wang M-B, Li Z, Matthews PR, Upadhyaya NM, Waterhouse PM. Drew RA, editor. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Proceedings of the International Symposium of Biotechnology of Tropical and Subtropical Species. Acta Horticulturae. 1998;461:401–407. [Google Scholar]
- Warton K, Foster NC, Gold WA, Stanley KK. A novel gene family induced by acute inflammation in endothelial cells. Gene. 2004;342:85–95. doi: 10.1016/j.gene.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Yao Y, Thompson DB, Guiltinan MJ. Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiology. 2004;136:3515–3523. doi: 10.1104/pp.104.043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi H, Nakamura Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant and Cell Physiology. 1992;33:985–991. [Google Scholar]