Abstract
SPATULA is a bHLH transcription factor that promotes growth of tissues arising from the carpel margins, including the septum and transmitting tract. It is also involved in repressing germination of newly harvested seeds, and in inhibiting cotyledon, leaf, and petal expansion. Using a reporter gene construct, its expression profile was fully defined. Consistent with its known functions, SPT was expressed in developing carpel margin tissues, and in the hypocotyls and cotyledons of germinating seedlings, and in developing leaves and petals. It was also strongly expressed in tissues where no functions have been identified to date, including the dehiscence zone of fruits, developing anthers, embryos, and in the epidermal initials and new stele of root tips. The promoter region of SPT was dissected by truncation and deletion, and two main regions occupied by tissue-specific enhancers were identified. These were correlated with eight regions conserved between promoter regions of Arabidopsis, Brassica oleracea, and Brassica rapa. When transformed into Arabidopsis, the B. oleracea promoter drove expression in reproductive tissues mostly comparable to the equivalent Arabidopsis promoter. There is genetic evidence that SPT function in the gynoecium is associated with the perception of auxin. However, site-directed mutagenesis of three putative auxin-response elements had no detectable effect on SPT expression patterns. Even so, disruption of a putative E-box variant adjacent to one of these resulted in a loss of valve dehiscence zone expression. This expression was also specifically lost in mutants of another bHLH gene INDEHISCENT, indicating that IND may directly regulate SPT expression through this variant E-box.
Keywords: Arabidopsis thaliana, auxin, bHLH, carpel development, dehiscence zone, germination, INDEHISCENT, leaf development, SPATULA, transmitting tract
Introduction
Master genes that control developmental decisions in plant morphogenesis are now being revealed. Many of these encode transcription factors that fall into a limited number of families. One large family is made up of basic Helix-Loop-Helix (bHLH) proteins, with 162 members identified in Arabidopsis (Bailey et al., 2003). These are involved in the regulation of diverse processes, including anthocyanin production, trichome development, and light signalling through phytochromes.
Several bHLH proteins are associated with organ morphogenesis. One of these is SPATULA (SPT) which was identified through its requirement for the normal development of carpels (Alvarez and Smyth, 1999, 2002). Loss of SPT function resulted in severe disruption of the septum and internal transmitting tract of the ovary and style, reduction in stigmatic tissues, and lack of fusion of the two carpels in apical regions. These defects are also manifest in the developing silique (Groszmann et al., 2008). Expression of the SPT gene occurs in all of these regions from early in their development (Heisler et al., 2001). SPT seems to activate its target genes, and apparently requires co-activators that are confined to these locations in that mis-expression of SPT elsewhere in the plant mostly had no effect (Groszmann et al., 2008).
SPT function may be associated with variation in auxin levels within the gynoecium. There is evidence that apical–basal patterning of the Arabidopsis gynoecium into the stigma and style, ovary, and the short stem (gynophore) depends on a declining gradient of auxin concentration from apex to base (Nemhauser et al., 2000). Such a gradient is indicated by the strong expression of the auxin responsive element DR5 in apical regions of early developing gynoecia (Benkova et al., 2003). Further, addition of an inhibitor of polar auxin transport to the growing apex of the gynoecium disrupts this patterning, reducing the relative amounts of style and ovary (presumably requiring higher auxin levels) and increasing the gynophore (low levels) (Nemhauser et al., 2000). When the same inhibitor is added to the apex of spatula mutant gynoecia, the phenotype is restored close to the wild type at the apex, suggesting that the chemical block to movement of auxin out of this region now promotes its growth. However, patterning of the remainder of the spt mutant gynoecium is not disrupted by the inhibitor to the extent seen in the wild type, so it may be that SPT function is also involved in the apical to basal transduction of the auxin signal (Nemhauser et al., 2000), or in negatively regulating polar auxin transport down the gynoecium (Ståldal et al., 2008).
Further evidence for an auxin-related role for SPT comes from its possible negative regulation by ETTIN (ETT), an Auxin Response Factor (ARF). It has been proposed that ETT normally perceives auxin concentrations in developing gynoecia and defines the boundaries between style and ovary, and between ovary and gynophore. This conclusion was based on the effect on gynoecium development of partial and full loss of ETT function (Sessions et al., 1997), and its response to polar transport inhibitors (Nemhauser et al., 2000). When both SPT and ETT function are simultaneously disrupted in double mutant plants, the ett disruptions are much weaker as though they depended upon SPT function (Alvarez and Smyth, 1998). This is consistent with the observed ectopic expression of SPT in ett mutant gynoecia (Heisler et al., 2001), and implies that ETT normally negatively regulates SPT expression.
SPT is widely expressed outside the developing flower (Schmid et al., 2005). Its action, if any, in these tissues was not initially associated with any mutant phenotype, suggesting that SPT may often have redundantly acting partners. However, a subsequent study has reported that SPT also plays a role in inhibiting the germination of freshly harvested Arabidopsis seeds (Penfield et al., 2005). This role can be relieved by cold-treating the seeds in the light during imbibition, or by ageing the seeds. Further, it was observed that spt mutant seedlings have larger cotyledons than the wild type, and, later, larger petals. Recently a role for SPT in suppressing leaf growth has also been reported (Ichihashi et al., 2010). Thus evidence is accumulating that SPT plays a broader role than in solely promoting carpel morphogenesis.
In this study, the expression of SPATULA throughout the developing and mature plant has been defined. Using a reporter gene construct, the promoter region was dissected to localize elements controlling tissue-specific expression. Also, promoter regions conserved between Arabidopsis and B. oleracea and B. rapa were identified, and tested for their ability to match AtSPT expression patterns. Three putative Auxin Response Elements (AuxREs) in conserved regions were mutated, but no consequences to SPT expression were detected. However, an E-box element involved in directing SPT expression specifically in the valve margins and dehiscence zones of the silique was uncovered. This expression was dependent on the action of INDEHISCENT, a bHLH transcription factor that may bind to this element to activate SPT expression.
Materials and methods
Plant material
Unless otherwise mentioned, Arabidopsis thaliana Landsberg erecta was used. Seeds of the mutant line indehiscent-12 (a strong allele previously known as houdini-2) were provided by Steve Swain, CSIRO.
Generation of reporter gene constructs
A genomic cosmid clone carrying all of the SPT coding and upstream sequences (Heisler et al., 2001)) was digested with KpnI (at –6253 bp in the 5’ promoter region) and XhoI (at +313 bp in the first exon). This was inserted into a pBluescript vector and then into pBI101.2 (using a SalI site in the polylinker that is compatible with the XhoI site) to generate pSPT-6253:GUS, a translational fusion of SPT with GUS at codon 92 (Heisler et al., 2001) [or codon 76 if translation starts at the second methionine (Groszmann et al., 2008)]. 5′ truncations of the pSPT-6253 insert were generated using convenient restriction sites at –2217 bp (HindIII), –1592 bp (XbaI), –1262 bp (PstI), –357 bp (ClaI), and –180 bp (SpeI) and placed in pBI101.2 as before. Other truncations at –1203 bp, –313 bp, –260 bp, and –221 bp were generated from the pSPT-1262 insert using 5′ primers that generated a HindIII site in a 5′ extension, and a 3′ primer that overlapped the XhoI site used to clone pSPT-1262 (see Supplementary Table S1 at JXB online). PCR products were then cloned and inserted into pBI101.2 as before. Promoter sequences between –633 bp and –180 bp were deleted from pSPT-2217, pSPT-1262, and pSPT-1203 using the SpeI restriction sites at these locations. To generate a deletion from –100 bp to –1 bp in pSPT-1262, a 5′ forward primer that overlapped the PstI site at –1262 bp and a 3′ reverse primer that ended at –100 bp and incorporated an overhanging KpnI site were used to amplify the appropriate fragment which was then inserted into the PstI and KpnI polylinker sites of the minimal GUS promoter plasmid pTATA-GUS (provided by Yuval Eshed and John Bowman), and the GUS cassette transferred to pMLBART using flanking NotI sites to generate pSPT-1262Δ(100-1):GUS. The validity of all clones was checked by sequencing the inserts.
Plant transformation and GUS staining
All transformations of Arabidopsis thaliana were carried out in Landsberg erecta, except for pSPT-6253:GUS which were first inserted into Columbia plants and then backcrossed three times to Landsberg erecta. Plants were transformed by the floral dip method, and transformants selected for kanamycin resistance (for pBI101.2-based plasmids), or Basta resistance (for pMLBART based plasmids). Plants were stained for expression of the uidA [β-glucuronidase (GUS)] reporter gene using 2 mM X-Gluc and 3 mM K3Fe(CN)6 and 3 mM K4Fe(CN)6. The latter two components were included to reduce intercellular movement of the blue stain precipitate. If staining was very strong, the ferricyanide and ferrocyanide concentrations were increased to 6 mM. If weak, they were reduced to 0 mM or 0.5 mM. In most cases, between 10 and 30 independent transformants were screened, and staining patterns recorded if they were present in the majority of those independent transformants that showed some staining.
Stained material was observed as whole mounts, or in thin sections. The latter were obtained by embedding fixed material in Paraplast plus or by using a JB-4 plastic embedding kit (ProSciTech), sectioning at 7–8 μm, and viewing under light or dark field optics. GUS product appears pink under the latter conditions (unless very abundant in which case it appears blue).
Identification and cloning of SPT orthologues in Brassica oleracea and B. rapa
Using –6253 to +85 bp of the AtSPT sequence as the query, BLASTN searches were performed against the TIGR B. oleracea shot gun sequence database and B. rapa BAC sequences (accessed through: http://brassica.bbsrc.ac.uk/). SPT orthologues were identified through matching the available downstream amino acid sequence with that of AtSPT and SPT of other species (Groszmann et al., 2008). Promoter sequence for B. oleracea SPT (formally named BolC.SPT.a (Østergaard and King, 2008), but will be called BoSPT from now on) up to –2660 bp was obtained through sequencing of two partially overlapping shot gun sequencing clones BOMRY82 (GenBank BZ512670) and BOMKN39 (GenBank BH708336) obtained from Horticulture Research International, Wellesbourne, UK. Regions of the BoSPT promoter sequence upstream of –2660 bp were obtained directly from the sequence of shot gun clones oej25e04.b1 (GenBank BH988149) and oee36f02.b1 (GenBank BZ002153) respectively. Brassica rapa SPT sequences, BraA.SPT.a (BrSPTa) and BraA.SPT.b (BrSPTb), were derived from BAC clones AC232512 and CU695342, respectively. Regions conserved between AtSPT, BoSPT, BrSPTa, and BrSPTb (>70% nucleotide sequence identity) were identified initially through the BLASTN search, and then aligned using Clustal W and refined manually.
Promoter sequence from B. oleracea (BoSPT) was cloned from the two partially overlapping BAC genomic clones BOMRY82 and BOMKN39. By using primers with extensions to create new restriction sites (see Supplementary Table S1 at JXB online), the 1857 bp SPT promoter region from BOMRY39 along with the 5′ UTR and 31 codons of the first exon was amplified and translationally fused to GUS in the plasmid pRITA using 5′ KpnI and 3′ HindIII sites, creating pBoSPT-1857. The 803 bp of promoter sequence 5′ to this was amplified from the other BAC, BOMRY82, creating 5′ XhoI and 3′ KpnI sites. This was then inserted into the XhoI and KpnI sites of pBoSPT-1857 to generate pBoSPT-2660. The promoter:GUS cassettes were then removed from pRITA using NotI and cloned into the plant transformation vector pMLBART.
Site-directed mutagenesis of promoter sequences
To generate mutations of putative Auxin Response Elements (AuxREs) and adjacent E-boxes in the SPT promoter region, the insert pSPT-1262:GUS in pBluescript was used as the starting point. Mutations were incorporated into primers that overlapped a specific targetted element, both forward and reverse primers in each case (see Supplementary Table S1 at JXB online). The sequence flanking each site in the 5′ direction was then amplified using the reverse primer and an upstream forward primer overlapping the 5′ HindIII site of pSPT-1262:GUS. Sequences flanking the site in the 3′ direction were amplified using the forward primer and a downstream reverse primer overlapping the XhoI site at the 3′ end of pSPT-1262:GUS. The two products (one 5′ to the target site and one 3′ to it, each now carrying the mutation) were then mixed, denatured, and annealed at the overlapping targeted site, and a full-length version of pSPT-1262:GUS created by Taq polymerase. This product, which is now mutant for the target site, was then amplified by PCR using the outer flanking primers, and the product inserted into pBI101.2 using the HindIII and XhoI sites at its ends as before. Shorter versions of these pSPT-1262mut:GUS constructs (pSPT-1262Δ(633-180)mut, pSPT-357mut, and pSPT-180mut) were generated by the same strategies that were used to generate equivalent wild-type constructs.
Results
SPT is widely expressed in developing tissues
To map the expression of SPATULA throughout the developing plant, a sequence of 6253 bp upstream of the start of transcription to 313 bp downstream was translationally fused to the GUS reporter gene and transformed into wild-type plants. Careful comparison of reporter gene expression with that recorded by in situ hybridization of SPT mRNA in developing flowers (Heisler et al., 2001) indicated that 6253 bp of upstream region is sufficient to match the endogenous expression pattern.
Reporter gene expression occurred in the inflorescence meristem and floral primordia from the earliest stages (Fig. 1A), becoming localized to newly arising carpels, stamens, and petals at stages 6–7 (floral and fruit stages are from Smyth et al., 1990). In the carpels, expression was limited to medial regions at stage 7, then to the inner (adaxial) side where the septum arises at stages 8–9 (Fig. 1B). Expression continued in the developing septum (Fig. 1C), becoming confined to the transmitting tract of the septum and extending within the style, and including the developing stigmatic papillae (Fig. 1D), but decreasing in these regions as the gynoecium matured at stage 13. In the valves, expression in the valve margins was seen as early as stage 9 (Fig. 1B), in the vasculature from stage 11 (Fig. 1C), and new weak expression occurred transiently throughout the walls commencing at stage 12 and fading by the end of stage 13 (not shown). SPT expression was also detected in ovule primordia as they arose at stage 9, continuing as they developed (Fig. 1C, D).
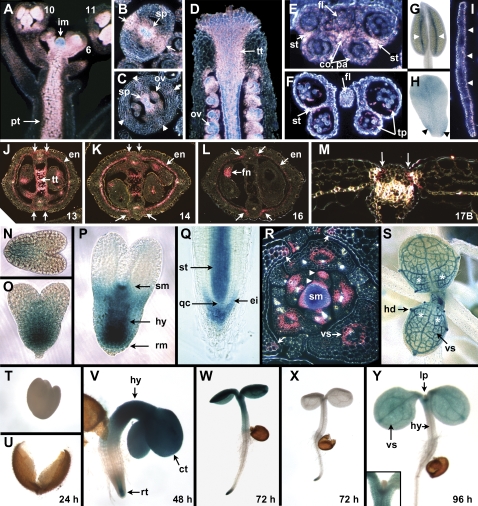
Fig. 1.
Expression patterns of SPT assessed by the pSPT-6253:GUS reporter gene. (A) Inflorescence (longitudinal section), showing heavy staining in the inflorescence meristem (im), developing gynoecia and stamens of young buds (developmental stages indicated), and the pith of the stem (pt). (B) Ovary of stage 9 flower (transverse section), with staining visible in medial regions (arrows), and in the developing lobes of the septum (sp). (C) Ovary of stage 11 flower (transverse section), with staining in the septum (sp), newly arising ovules (ov), and weak staining in the vasculature of the wall (arrowheads). (D) Apex of the gynoecium of a stage 12 flower (longitudinal section), showing heavy staining of the transmitting tract (tt) including the stigma, internal style, and setpum. Developing ovules (ov) are also stained. (E, F) Transverse sections of stamens from flowers at stage 9 (E) and 12 (F), including the anther and filament (fl). Expression occurs in lateral regions where stomia will develop (st). It is also present in the connective (co) and parietal cells (pa) at stage 9 (E), and in the degenerating tapetum (tp) at stage 12 (F). (G) Stamen at stage 13 showing expression in the stomia where dehiscence will soon occur (arrowheads). (H) Petal from a stage 10 flower, showing moderate GUS staining in the upper blade and basal margin regions where the claw is developing (arrowheads). (I) Mature petal (transverse section), with staining visible in the adaxial (upper) epidermis (arrowheads). (J–M) Developing siliques (transverse sections) at the indicated stages, showing staining in the dehiscence zone commencing from late stage 13 to stage 17B (arrows). At maturity (stage 17B), this is localized in the separation layer adjacent to the refringent lignified layer of the valve margins (M). Expression also occurs in layer b of the endodermis (en), falling away as it becomes lignified by stage 17B (M). Transmitting tract (tt) expression is seen at stages 13 (J) and 14 (K) only, while expression occurs in the funiculus (fn) and chalaza of maturing seeds (L). (N–P) Developing embryos (whole mounts), showing staining in basal regions from the early (N) and later (O) heart stage. At the torpedo stage (P), staining is heavier in the root meristem (rm), central hypocotyl (hy), and the newly arising SAM (sm). (Q) Root tip (whole mount), with staining in the epidermal (and columellar) initials (ei), and in the newly arising stele (st), but not in the quiescent centre (qc). (R) Transverse section of the shoot apical meristem and leaf primordia of a 12–14-d-old plant. Strong expression can be seen in the meristem (sm), and in adaxial and abaxial medial regions of leaf primordia (arrowheads). Later, expression is associated with the vasculature (vs), and the basal margins of older leaves (arrows). (S) Third and fourth leaves of a 12–14 d-old seedling (whole mount), with heavy expression in the vasculature (vs) and hydathodes (hd), and weaker, more uniform staining throughout the basal regions (asterisks). Spots of expression in stomata can be seen on the petioles of older leaves. (T–Y) Seedlings sampled daily after imbibition. At 24 h, no staining is visible in the embryo (T), here dissected from the already split testa (U). At 48 h, heavy staining occurs in the cotyledons (ct), hypocotyl (hy), and root tip (rt) (V). At 72 h, staining continues in these regions (W). Cotyledon and hypocotyl staining is not seen in plants carrying the shorter pSPT-1262:GUS promoter region (X). At 96 h, cotyledon staining continues especially in the vasculature (vs), but it is weak in the hypocotyl (hy) (Y). New leaf primordia (lp) do not show strong expression of SPT (see inset in Y).
Of the other floral organs, reporter expression was recorded in developing stamens from their inception, becoming localized to two lateral regions of the anther (not shown). It continued in these domains where stomia will later develop (Fig. 1E), and in the mature stomia themselves (Fig. 1G). It was also expressed in the connective from stage 9 (Fig. 1E), and in the tapetum until it degenerated at stages 12–13 (Fig. 1F). The filament carried a stained vascular strand throughout most of its development. Developing petals were also weakly stained from late stage 7/early stage 8 (not shown), with expression continuing throughout the expanding blade and basal margins of the claw (Fig. 1H). It was confined to the upper (adaxial) epidermis at stage 12 (Fig. 1I). SPT reporter gene expression was also present in the floral receptacle and nectaries but not the sepals.
After anthesis (stage 13) and fertilization, GUS staining decreased in the septum, valves, and later in the ovules where it became confined to the distal funiculus and adjacent chalazal region (Fig. 1J–L). One striking increase from late stage 13 was the localization of GUS staining in the developing endocarp within the valves, specifically the inner (b) layer (Fig. 1J–L). This faded as these cells became lignified at stage 17B (Fig. 1M). Strong expression was also visible in the valve–replum boundary region from stage 13 (Fig. 1J–L), more broadly at first but eventually becoming localized to the separation layer of the dehiscence zone during early stage 17B. GUS staining was still visible in the separation layer in mature green siliques (stage 17B), adjacent to the newly lignified cells of the valve margins (Fig. 1M).
Vegetative expression occurred in many but not all tissues. It was observed in the developing embryo from the transition stage through the heart stage, confined to the basal half (Fig. 1N, O). By the torpedo stage it had become concentrated in the developing root meristem and the procambium of the future hypocotyl, and in the newly arisen shoot apical meristem (Fig. 1P). In germinating seeds (Fig. 1T–Y), expression was first detected 2 d after imbibition of aged seeds (Fig. 1V), and was strong throughout the hypocotyl and cotyledons, and later in the cotyledon's vasculature (Fig. 1Y). It faded from the hypocotyl after around 4 d (Fig. 1Y). Expression also appeared from the second day in two regions of the developing root tip (Fig. 1V). These were the epidermal initials and the maturing epidermis of the proliferation zone (still covered by lateral root cap cells), and the stele, from immediately above the quiescent centre through the elongation zone but fading in the differentiation zone. This pattern was maintained throughout all growing primary and secondary root tips (Fig. 1Q).
As seedlings developed, strong expression was observed in the shoot meristem (Fig. 1R), and later in the inflorescence meristem as well (Fig. 1A; see also Heisler et al., 2001). It extended down through the rib zone into the pith (Fig. 1A). Expression in newly arising leaf primordia did not extend throughout (Fig. 1Y), but was localized to two medial regions, adjacent to and distant from the meristem (Fig. 1R; see Supplementary Fig. S1 at JXB online for serial sections flanking Fig. 1R). As leaf primordia developed, expression was associated with the developing vasculature, and in basal margins (Fig. 1R; see Supplementary Fig. S1 at JXB online). In older developing leaves, continuous expression was seen in basal regions (Fig. 1R, S), and it also occurred in the phloem, hydathodes, and stomata (see Supplementary Fig. 1S at JXB online), falling away as the leaves matured.
SPT expression is controlled by two main subregions of the upstream promoter sequence
To define the tissue-specific regulatory elements controlling these patterns within the 6253 bp upstream region, it was subdivided by successive deletions of the 5′ end (Fig. 2). The first deletion removed 4036 bp, leaving 2217 bp upstream. This resulted in three major changes in the staining profile (Table 1). First, staining in some tissues was lost, including the valve and endocarp of the gynoecium, the petal blade, the nectaries, the floral receptacle, and the pith. Vegetative staining in the hydathodes and stomata was also now undetectable. Elements essential for expression in these regions are apparently localized to this 4 kb upstream region (Fig. 2). Next, the general level of staining of the other tissues was reduced (Table 1), suggesting general enhancers of expression also occur here. Finally, stronger expression was now seen in the basal margins of leaves, in the claw of petals, and newly in the basal margins of sepals (Table 1). It seems likely that this expression is normally repressed to varying degrees (partially in leaves and petals; totally in sepals) by silencers in the region now deleted (Fig. 2).
Fig. 2.
Localization of enhancer and silencer elements in the promoter region of SPT. The 6253 bp upstream region together with 313 bp of the 5′ UTR and first exon were translationally fused with GUS, and the consequences of successive 5′ truncations on expression patterns compared with those of the full-length construct (Table 1). The sites of the 5′ ends are indicated above the line. The consequences of internal deletions of the region between –2217 bp and –633 bp, and between –100 bp and +313 bp, were also assessed. The deduced locations of tissue-specific enhancers, general enhancers, and tissue-specific silencers of expression are indicated. [Note that (i) the hypocotyl and cotyledon enhancers in the upstream region could occur between –2217 bp and –1262 bp; (ii) all tissue-specific enhancers in the promoter proximal region may lie 3’ to –180 bp rather than –221 bp; (iii) those tissue-specific enhancers between –221 and –1 bp listed in the lower group confer expression which is repressed by tissue-specific silencers further upstream; and (iv) the location of the enhancer element for leaf primordium expression was not mapped.] (This figure is available in colour at JXB online.)
Table 1.
Expression patterns of 6253 bp of the SPT promoter region fused to the GUS reporter gene (first data column)
| bp upstream of full-length cDNA | −6253 | −2217 | −1592 | −1262 | −1203 | −357 | −313 | −260 | −221 | −180 |
| No. transformants | 21 | 29 | 11 | 13 | 9 | 20 | 8 | 11 | 17 | 21 |
| No. with staining | 16 | 25 | 7 | 10 | 6 | 13 | 6 | 9 | 14 | 12 |
| Enhancer regiona | ||||||||||
| −2217 to –6253 | ||||||||||
| Valve* | +++ | − | − | − | − | − | − | − | − | − |
| Endocarp | +++ | − | − | − | − | − | − | − | − | − |
| Petal blade* | +++ | − | − | − | − | − | − | − | − | − |
| Nectary | +++ | − | − | + | − | − | − | − | − | + |
| Receptacle | +++ | − | − | − | − | − | − | − | − | − |
| Hypocotyl/cotyledon | ++++ | ND | ND | − | ND | ND | ND | ND | ND | − |
| Pith* | +++++ | − | − | − | − | − | − | − | − | − |
| Hydathode | ++++ | − | − | − | − | − | − | − | − | − |
| Stomate | +++ | − | − | − | − | − | − | − | − | − |
| Enhancer regiona | ||||||||||
| −1 to –221 | ||||||||||
| Dehiscence zone* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + |
| Funiculus/ovule* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + |
| Tapetum* | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++ | +++ | ++ | + |
| Connective* | ++++ | +++ | ND | +++ | +++ | ++ | + | + | + | + |
| Root tip epidermis* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + | + |
| SAM* and IM* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + |
| Developing gynoecium* | ++++ | +++ | ND | +++ | +++ | ++ | + | + | + | − |
| Septum/transtract* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | − |
| Stigma* | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − |
| Stomium* | +++++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | − |
| Root tip stele | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + | − |
| Leaf vasculature* | ++++ | ND | ND | ND | ND | ND | ++ | − | − | − |
| Leaf primordium | ++++ | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Embryo | ++++ | ND | ND | +++ | ND | ND | ND | ND | ND | ND |
| Silencing regions (various) | ||||||||||
| Petal claw | + | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | +++ | ++ | − |
| Basal sepal margin | − | +++ | ND | +++ | +++ | +++ | ++ | ++ | ++ | − |
| Basal leaf margin | + | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | − |
| Apex of filament | − | − | +++ | +++ | ND | +++ | ND | ND | ++ | +++ |
| Leaf epidermis | − | − | − | − | − | ND | ++ | ++ | ++ | − |
| Root epidermis | − | − | − | − | − | − | − | − | +++ | − |
Also shown are staining patterns in a series of 5’ truncations of this construct, with the location of the 5’ end of the truncation shown at the head of each column. Tissues are organized into categories depending on the deduced location of their specific enhancer elements. Relative staining levels are indicated by the number of + symbols; no detectable staining is indicated by a – symbol. ND: not determined.
Transcripts detected by in situ hybridization (Heisler et al., 2001) shown by an asterisk.
Next, four further truncations were made, successively removing 5′ sequences and leaving 1592, 1262, 1203, and 357 bp from the start of transcription (Fig. 2). Most staining patterns and intensities were unchanged compared with the 2217 bp promoter region (Table 1). There was one striking difference–expression in the hypocotyl and cotyledons of germinating seedlings was no longer detected in the 1262 bp construct (Fig. 1X). This region was not assessed in the 2217 bp construct or in the other deletions, so it can only be concluded that the controlling region lies somewhere upstream of 1262 bp. The one new expression site seen was the apex of the stamen filament (Table 1), apparently silenced by sequences in the –1592 to –2217 bp region.
Finally, four further 5′ truncations were created, leaving 313, 260, 221, and 180 bp of upstream sequence (Figs 2, 3). No tissue expression sites were lost down to the 221 bp construct, although staining intensity was reduced in some, suggesting loss of general enhancers (Table 1). However, the 180 bp driver generated markedly weaker staining throughout, and expression could not be seen in the early gynoecium, or later in the septum, transmitting tract, and stigma. It was also no longer seen in the stomium of maturing anthers, or in the developing root stele, although it was still present in the newly arising epidermis of the root tip. New expression was observed in the mature epidermis of leaves and roots (Table 1), suggesting that silencers of these patterns occur upstream of 313 bp for leaves and 221 bp for roots.
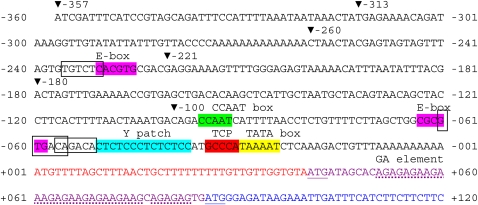
Fig. 3.
Sequence of the SPT promoter region from –357 bp to +120 bp. The site of the start of transcription (+1 bp) is from GenBank entry AU237757 (Seki et al., 1998), and is 18 bp longer than that proposed by Heisler et al. (2001). The commencement of translation may occur at the first methionine codon at +41 (Heisler et al., 2001), or the second at +86 bp (Groszmann et al., 2008) (both underlined). A putative TATA box, a GCCCA motif bound by TCP transcription factors, a Y patch, putative auxin response elements (boxed) and nearby E-boxes, a CCAAT box, and a possible GA element in the translated region (dotted underline) are indicated. The ends of 5′ truncations are indicated with triangles. (For comparison with equivalent B. oleracea and B. rapa sequences, see Supplementary Fig. S1 at JXB online.) (This figure is available in colour at JXB online.)
To test if the 3′ part of this region, from –100 bp to –1 bp and containing a putative TATA box, Y patch, and CCAAT box (Fig. 3), was essential for expression, it was deleted in the pSPT-1262:GUS construct, and fused to a minimal 35S:GUS construct. 15 transformed lines were obtained, and none showed detectable expression (except in pollen grains where the minimal 35S:GUS construct alone drives expression).
Thus a second regulatory subregion was identified lying between –1 bp and –221 bp (Fig. 2). Some tissue-specific elements were confirmed in the region downstream of 180 bp, and others possibly from –180 bp to –221 bp. Enhancers of these, and several other tissue-specific silencers, also occurred in the interval between –180 bp and –357 bp. The region between –1 bp and –100 bp was required for any expression.
The region between –633 and –1203 bp carries redundant enhancers
To test the role of elements upstream of –180 bp, the region from this site up to –633 bp (both being SpeI sites) was deleted within the pSPT-2217:GUS construct to yield pSPT-2217(Δ633-180):GUS. It was predicted that the expression would resemble that of the –180 bp construct in that all the tissue-specific and general enhancers upstream of –180 bp would be lost. However, the pattern of expression in 11 different inserts was indistinguishable from that recorded for the full-length pSPT-2217:GUS insert lines. Thus it seems likely that the region between –633 and –2217 bp carries regulatory elements that act redundantly with those lying between –180 and –633 bp (with the 5′ limit in the latter likely to be –357 bp as no enhancers were identified in the –357 to –633 bp region) (Fig. 2). These redundant elements were further constrained to downstream of –1203 bp because successive truncation of the 5′ end of the deletion construct pSPT-2217(Δ633-180):GUS to –1262 bp, and to –1203 bp, had no effect on the expression pattern.
It is possible that certain tissue-specific enhancer elements occur in the short –180 to –221 bp region (lost in the –180 bp to –633 bp deletion constructs), and these redundantly occur in the –633 bp to –1203 bp region. However, another possibility, perhaps more likely, is that these tissue-specific enhancers are in fact located in the –1 bp to –180 bp region, and that expression levels were below the level of detection in the –180 bp GUS reporter plants. Redundant general enhancers such as those present in the –633 bp to –1203 bp region may be necessary to boost their expression to detectable levels. Further tests are required to distinguish these possibilities.
Eight regions in the SPT upstream sequence of Arabidopsis are conserved in Brassica oleracea and B. rapa
Conservation of promoter regions can indicate the location of conserved regulatory elements. Eight conserved regions were identified in the 6253 bp upstream region of AtSPT when aligned with equivalent regions of SPT orthologues from B. oleracea and B. rapa (Fig. 4; see Supplementary Fig. S2 at JXB online). Three of these occurred in the region between –6253 bp and –2217 bp where general enhancers and tissue-specific elements were identified. The other five were localized to the region from just upstream of –1262 bp to –1 bp where all other regulatory elements were mapped (Fig. 2).
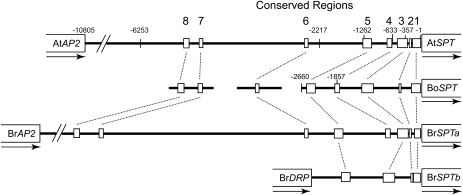
Fig. 4.
Regions in the promoter of SPT in Arabidopsis (AtSPT) that are conserved in the promoters of orthologous genes in B. oleracea (BoSPT), and B. rapa (BrSPTa and BrSPTb). Orthology was confirmed by comparisons of the translated regions (to the extent available). The 5’ ends of two reporter gene constructs of BoSPT at –2660 bp and –1857 bp are indicated. The gene immediately upstream of AtSPT and BrSPTa is APETALA2. In BrSPTb it is a Disease Resistance Protein, suggesting that a rearrangement has occurred near this gene since duplication. The full sequence of the B. oleracea promoter region was not available. Sequences of the conserved regions are provided in Supplementary Fig. S1 at JXB online.
To test if the B. oleracea promoter region that included these five conserved sequences could drive floral expression in the same locations in transgenic Arabidopsis, a reporter gene construct was generated carrying 2660 bp of the B. oleracea promoter translationally fused to GUS, and eight independent transgenic Arabidopsis lines were scored. In general, expression levels were higher, although the patterns were similar to those from the equivalent –1262 bp Arabidopsis reporter. For example, developing carpel and stamen primordia were strongly labelled (Fig. 5A), and gynoecium expression continued in the early medial regions (Fig. 5B) and later in the developing transmitting tract (Fig. 5C, D). Two main differences in staining pattern were noted. First, B. oleracea sequences drove expression in four abaxial zones of the medial and lateral regions of the valve wall outside of vascular strands from stages 10–13 (e.g. Fig. 5C, D). Second, dehiscence zone expression was not seen at any stage in the pBoSPT-2660:GUS lines (Fig. 5E, F). Stamen expression patterns were very similar to pSPT-1262:GUS (Fig 5G), although expression in pollen grains was seen in the B. oleracea constructs. A subsequent deletion of 803 bp creating a shorter B. oleracea promoter construct, pBoSPT-1857:GUS, revealed the presence of general enhancers as expression was less intense although spatially similar (not shown). Region 5 and half of region 4 reside within this deleted region (Fig. 4), the majority of which overlaps a section of the AtSPT promoter identified as containing general redundant enhancers (Fig. 2).
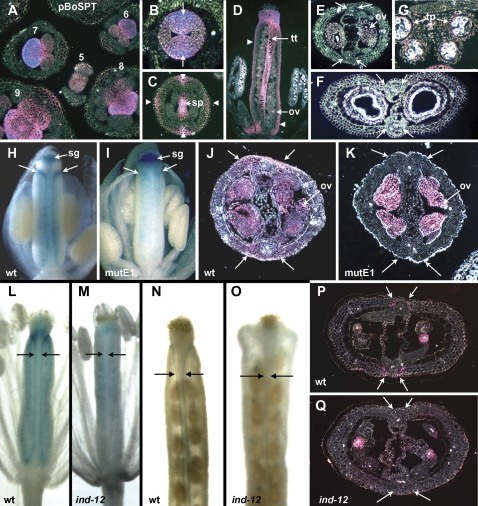
Fig. 5.
Expression patterns of SPT reporter gene constructs in Arabidopsis. (A–G) Expression driven by the B. oleracea promoter pBoSPT-2660:GUS. (A) Inflorescence (transverse section), showing strong expression in regions of young flower primordia that will develop as stamens and carpels (floral stage indicated). (B) Gynoecium at stage 8 (transverse section) with strong medial expression (arrows). (C) Gynoecium at stage 11 (transverse section), with septum (sp) staining, and new expression in outer medial and lateral regions (arrowheads). (D) Gynoecium at stage 12 (longitudinal section), with staining in the transmitting tract (tt) and ovules (ov). Ovary wall staining is indicated by arrowheads. (E, F) Developing fruits (transverse sections) at stages 14–15 (E) and 17A (F), with staining in ovules (ov) at the earlier stage (E), but no staining visible in valve margins (arrows) where the dehiscence zone will arise. (G) Anther at stage 10–11 (transverse section), with staining in the tapetum (tp). (H–K) Expression driven by pSPT-1262:GUS, and pSPT-1262mutE1:GUS. (H, I) Buds at stage 12 (whole mounts, some sepals, petals, and stamens removed), showing that expression occurs in the carpel margins (arrows) in the normal –1262 construct (H) but not in the mutated version (I). Expression is visible in the stigma (sg) of both constructs. (J, K) Siliques at around stage 13 (transverse sections) show ovule (ov) expression in both (J, K), but in the valve margins only in the unmutated version (arrows) (J). (L–Q) Expression of the full-length promoter (pSPT-6253:GUS) in wild-type and indehiscent-12 mutant plants. Expression occurs in the valve margins of wild-type plants (arrows) but not in ind-12 plants. Expression in developing ovules and seeds is present in both. (L, M) Young siliques of stage 14 flowers (whole mounts, some sepals, petals, and stamens removed); (N, O) maturing siliques at stage 17A (whole mounts); (P, Q) maturing siliques at stage 17A (transverse sections).
Thus the carpel and stamen expression patterns of SPT in Arabidopsis can mostly be generated by the four regions of the –1262 bp promoter that are conserved with two Brassica species.
Site-directed mutagenesis reveals that SPT expression in the dehiscence zone is associated with a variant E-box
There is indirect evidence that SPT expression is associated with auxin levels. These are likely to be transmitted through Auxin Response Factors (ARFs). To test this, putative Auxin Response Elements (AuxREs) (TGTCTC, or close variants), normally bound by ARFs, were identified in three locations in the –1262 bp upstream sequence, and subjected to site-directed mutagenesis (Fig. 6). These were selected because they occurred with adjacent or overlapping E-box (CANNTG) elements, or variants of them (Fig. 6B). As such, they were considered to be potentially associated with auxin responsiveness, given that similar nearby ‘constitutive’ elements (especially CACGNN) were found to be required for auxin inducibility acting through two AuxREs in the GH3 promoter region of soybean (Ulmasov et al., 1995). Two of the AuxREs, in locations 1 and 3, were fully conserved in Brassica promoter sequences (see Supplementary Fig. S2 at JXB online).
Fig. 6.
Details of site directed mutagenesis of elements at three locations in the SPT promoter region. (A) Putative Auxin Response Elements (AuxREs), and nearby E-boxes (or variants), were individually mutated (indicated by X). They were generated in a full-length 1262 bp promoter GUS fusion, or in truncated or deleted versions of it. Expression patterns were recorded in the indicated number of independent transformants that showed any staining. (B) AuxREs and E-boxes in the wild-type sequence (boxed, upper sequences), and the mutational changes made (lower sequences). *Anti-sense sequence.
First, the strongly conserved 5′ half of the AuxREs, TGT, was modified in each of the putative elements (Fig. 6B), both in full-length pSPT-1262:GUS constructs and a range of shorter versions (Fig. 6A). These mutagenized constructs were then transformed into plants, and expression patterns in above-ground tissues carefully compared with the control pSPT1262:GUS plants. No differences were seen.
Next, the putative E-boxes adjacent to each putative AuxRE were disrupted by site-directed mutagenesis (without affecting the AuxRE) (Fig. 6B) and transformed into plants. Again, no staining differences were seen in lines carrying the modified elements near the AuxREs at –1249 bp (location 3) and –236 bp (location 2). However, in insertion lines carrying the modified element near the overlapping AuxREs at –68 bp (location 1), all staining of the dehiscence zone was abolished, although other staining patterns were apparently unaffected (Fig. 5I, K compared with Fig. 5H, J). This occurred in five different insertion lines of constructs driven by the 1262 bp promoter region, and in 10 mutated insertion lines carrying the 180 bp promoter region (Fig. 6A).
SPT expression in the dehiscence zone requires INDEHISCENT function
E-boxes are bound by bHLH transcription factors, and one bHLH factor gene that is expressed in the dehiscence zone of siliques is INDEHISCENT (IND) (Liljegren et al., 2004). As such, it is a candidate to bind to the atypical E-box near the –68 bp AuxREs at location 1 and activate SPT expression. To test this possibility, SPT expression was assessed in indehiscent-12 mutant plants.
Strikingly, all expression of SPT in the dehiscence zone was lost (Fig. 5L–Q), although expression in other regions was unaffected. This loss of expression occurred in mature ind-12 mutant siliques that do not dehisce (Fig. 5O, Q), so it is possible that the loss of the separation layer where SPT is normally expressed precludes its expression at this stage. However, it was also absent at the earlier stages (e.g. Fig. 5M) where SPT is expressed at valve margins in the wild type (Fig. 5L), but where the dehiscence zone has not yet differentiated. Thus it seems that IND function is required for SPT expression specifically in this region.
Discussion
Structure of the SPATULA promoter region
The core SPATULA promoter extends 100 bp upstream of the transcription start site (Fig. 3). It contains a putative TATA box 30 bp upstream, a Y patch of unknown function that is commonly found in this vicinity in plant promoters (Yamamoto et al., 2007), and a CCAAT box associated with the transcription of genes widely expressed in proliferating tissues (Mantovani, 1998; Romier et al., 2003). A possible GA element, identified recently in ∼20% of plant promoter regions (Yamamoto et al., 2009), also occurs but in the transcribed region. Several regulatory elements with a more specific action also occur, and an E-box variant that is required for SPT expression in the developing dehiscence zone of the gynoecium and fruit has been identified. Another likely element is the GCCCA sequence adjacent to the putative TATA box. This matches box II that is recognized by several TCP transcription factors (Kosugi and Ohashi, 1997; Trémousaygue et al., 2003). Again, these seem to drive expression in cycling cells. All these elements are conserved in the promoter regions of three SPT orthologues in Brassica species (see Supplementary Fig. S2 at JXB online).
Immediately upstream of this core (to –221 bp), there is expression evidence for additional tissue-specific enhancers, including those driving expression in developing carpels and transmitting tract. All tissue-specific expression is strengthened by generally acting enhancers from –180 bp upstream to –357 bp. These general enhancers can be substituted with redundant enhancers occurring considerably further upstream (from –633 bp to –1203 bp). Identification of the enhancers will require further dissection of this region, possibly focusing on Brassica homologous regions 4 and 5 located within a general enhancer region of BoSPT (see Supplementary Fig. S2 at JXB online).
There is a large almost silent region from –1203 bp to –2217 bp, but between here and –6253 bp lie further important tissue-specific enhancers driving expression in the valves of the gynoecium, the hypocotyl, and cotyledons of germinating seedlings, and developing petals among other tissues. Further general enhancers occur here as well.
The next gene upstream of SPT is the floral organ identity gene APETALA2. This lies a further 4 kbp upstream (its 3′ UTR ends at –10 805 bp). We have not tested whether this region influences SPT expression. Likewise, we have not tested the role of sequences downstream of the site of the SPT-GUS translational fusion (codon 92), including six introns and the 3′ UTR. The few differences seen using a reporter gene from those reported using in situ hybridization mapping of SPT mRNA may be due to controlling elements outside the –6253 to +313 bp region, or interference from the SPT amphipathix helix present in the translational fusion. Differences include patterns in the valve of newly opened flowers (stage 13) where GUS expression throughout initially matches in situ mapping (Heisler et al., 2001), but then during late stage 13 appears as strong expression in layer b of the developing endoderm. Overall, however, our reporter gene profiles mostly match in situ expression results, available from developing inflorescences and flowers (Heisler et al., 2001), and they are consistent with results from micro-array profiles (Schmid et al., 2005).
Scattered through the SPT promoter are a series of tissue-specific silencers of expression, similar in action to those identified in other plant regulatory regions (Schauer et al., 2009). Without these, expression driven by the promoter region from –1 bp to –221 bp occurs strongly in regions that normally lack SPT transcript, or accumulate it to much lower levels. For example, expression in the basal margins of leaves, sepals, and petals occurs strongly unless the –2217 to –6253 bp region is present. These three tissues share developmental properties, and they may carry a factor that interacts with a common silencing element present in this promoter region. Silencers of expression in the epidermis of leaves (between –1203 to –313 bp), roots (–260 to –221 bp), and the apex of the stamen filament (–2217 bp to –1592 bp) were also detected. It would be interesting to determine if these silencers have evolved to ensure appropriate development of these tissues by repressing SPT expression within them.
SPATULA expression occurs in a subset of proliferating and maturing tissues
The conserved CCAAT box and TCP binding site in the core promoter region suggest that SPT may be expressed wherever cell replication is occurring. This is true for the newly developing gynoecium, stamens, petals, leaves, and for the newly arising epidermis and stele of the root tip, for example. However, SPT is not expressed in other proliferating tissues such as the initiating sepals and the apical half of early embryos. Further, there is strong expression in the central zone of the shoot apical meristem (see also Heisler et al., 2001; Yadav et al., 2009) even though the rate of cell proliferation there is relatively low.
On the other hand, some maturing tissues that are not actively proliferating do display SPT expression. These include the endocarp, the maturing petal blade, the nectaries and receptacle of older flowers, the hypocotyl and cotyledons of the germinating seedling, the pith of the stem, and the hydathodes and stomates of leaves. Strikingly, these are mostly controlled by the 2.2–6.2 kbp upstream region, and an interesting question is how many different tissue-specific elements are involved. Answers will require further dissection of this large promoter region, including analysis of the three locations conserved with Brassica species.
SPATULA expression and its function in developing fruits, seedlings, leaves and petals
Loss of SPT function results in defects in carpel development, and these, coupled with its expression pattern, indicate that SPT targets include genes responsible for the production and differentiation of tissues that arise from carpel margins (Alvarez and Smyth, 1999, 2002; Heisler et al., 2001). SPT expression in developing siliques has now been mapped in the dehiscence zone and, ultimately, in the separation layer, as well as the maturing layer b of the endocarp. No loss of function defects have been reported in these tissues to date, and it may be that other redundantly acting genes are involved. For the separation layer, these could include ALCATRAZ, a recently duplicated sister of SPT (Rajani and Sudaresan, 2001). Potential targets of SPT in the separation layer include enzymes associated with dissolution of cellular interconnections, and these could also be involved in the stomium of anthers and the funiculus of maturing seeds, where SPT is also expressed.
Different types of targets seem to be involved in seedlings where SPT apparently acts to inhibit germination of fresh seeds unless cold and light treated (Penfield et al., 2005). SPT expression could not be detected in aged seeds one day after imbibition, at which time the testa was mostly split. However, it was strongly expressed in cotyledons, hypocotyls, and the root tip after 2 d when the radicle was mostly emerging (Liu et al., 2005). This is consistent with the peak of expression observed at 2 d by quantitative RT-PCR (Penfield et al., 2005). Cotyledon size in germinating seedlings is also influenced by SPT in that loss of SPT function resulted in larger cotyledons (at least under red light), whereas they were smaller in plants ectopically expressing SPT (Penfield et al., 2005). This growth suppression may be causally associated with the inhibition of germination. Loss of SPT function also resulted in larger petals with larger cells (Penfield et al., 2005), again consistent with our observed SPT expression throughout petal development and later in the adaxial epidermis. Cotyledon and petal blade expression are both dependent on the distant upstream region of the promoter, so they may be under common regulatory control.
A recent report extends the role of SPT to suppression of the expansion of another lateral organ, the leaf (Ichihashi et al., 2010). Leaves are somewhat larger in spt mutants, with additional cells of the same size as the wild type, and smaller in 35S:SPT plants. Mutant leaves have the same geometrical shape as normal, and differences may arise through an increase in the size of maturing leaf primordia, perhaps associated with the abaxial and adaxial zones of early SPT expression we have observed here.
Association of SPATULA expression with auxin
There is genetic evidence that SPT expression is negatively regulated by the auxin response factor ETTIN (see Introduction). However, targeted mutagenesis of three putative auxin response elements in the SPT promoter region yielded negative results in that no changes in the pattern of SPT expression were detected. It is true that the AuxRE at location 3 (–1243 to –1238 bp) falls in a silent region (Fig. 2), but the other two occur in active regions. It remains possible that SPT expression is controlled by ETTIN and/or other Auxin Response Factor proteins, possibly at other potential AuxREs not tested here (e.g. at positions –155 bp to –160 bp, or –99 bp to –102 bp; Fig. 3), and tests of their direct binding to SPT promoter sequences by yeast one hybrid and chromatin immunoprecipitation methods would be worthwhile.
The recent discovery of genes controlling two pathways of auxin biosynthesis has allowed a comparison of their expression patterns with that of SPT. The YUCCA1 (YUC1) and YUC4 genes, encoding flavin mono-oxygenases, are required for normal gynoecium development, and both are expressed in newly arising primordia (Cheng et al., 2006). The TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) gene and its relative TAR2, which are also required for normal gynoecium development, are also expressed in newly arising gynoecia (Stepanova et al., 2008). Strikingly, TAA1 is strongly expressed later in the developing medial regions, and TAR2 in the lateral valves, coincident with sites of SPT expression. Also, root tip expression of TAR2 in the newly arising stele and in cells distal to the quiescent centre closely parallels that of SPT (Stepanova et al., 2008).
It may be that SPT expression is directly responding to auxin biosynthesis in developing gynoecia. Alternatively, SPT may be involved in the generation of auxin. It may promote this in the gynoecial apex through activation of genes including members of the STYLISH (STY) family. Evidence that STY genes lie downstream of SPT is the ability of 35S:STY1-GR to rescue style defects in spt mutant plants (Ståldal et al., 2008), and the finding that ectopic STY2 expression can be induced by SPT providing it carries the VP16 activation domain (35S:SPT-VP16) (Groszmann et al., 2008). Once activated, the STYLISH genes may promote auxin biosynthesis as there is evidence that STY1 activates expression of the auxin biosynthetic gene YUC4 (Sohlberg et al., 2006). Whichever way SPT expression and auxin production may be related at the apex of gynoecia, SPT's association with auxin in more basal regions is apparently different (see Introduction).
Potential regulation of SPATULA expression by INDEHISCENT
In this study, evidence was obtained that SPT expression is positively regulated by INDEHISCENT, a bHLH transcription factor required for development of the dehiscence zone of the silique (Liljegren et al., 2004). SPT expression was specifically abolished in this zone, both in ind mutant plants, and in transgenic plants in which a potential IND binding site in the SPT promoter was mutated. However, it should be noted that, although this putative binding site is also strongly conserved in two Brassica species (see Supplementary Fig. S1 at JXB online), the B. oleracea promoter did not drive expression of a reporter gene in the dehiscence zone of Arabidopsis plants. It may be that co-activators required in Brassica are absent in Arabidopsis, and further experimental tests in Brassica plants are needed.
bHLH proteins bind to their DNA recognition sites as dimers, and most bHLH proteins recognize the symmetrical E-box (CANNTG) (or one form of this, the G-box CACGTG). However, the variant E-box involved here (5′-CGCGTG-3′ in the sense strand, or 5′-CACGCG-3′ in the antisense strand) differs from the usual by one base (underlined). Even so, precedents exist for such a variant to be recognized by bHLH dimers, including the hairy transcription factor of Drosophila (Ohsako et al., 1994) and the Tclf5 protein of mouse (Siep et al., 2004). Furthermore, the non-symmetrical nature of the binding site indicates that it may be bound by a bHLH heterodimer. Again there are precedents for this, including Tango, a bHLH-PAS protein from Drosophila that heterodimerizes with two other Drosophila bHLH-PAS proteins, Single-minded and Trachealess (Sonnenfeld et al., 1997; Zelzer et al., 1997). Thus one possibility is that IND and a bHLH partner bind to this non-canonical E-box to up-regulate SPT transcription. bHLH proteins that interact with E-boxes carry a conserved glutamate at position 9 in their basic region that makes contact with the CA component of the E-box. The fact that IND has alanine in this position instead of glutamate suggests that it might interact with the non-canonical CG half.
IND has three close bHLH relatives named HECATE that play a role in determining earlier transmitting tract development (Gremski et al., 2007). However, it seems unlikely that they regulate SPT expression, at least solely through this variant E-box, because transmitting tract expression still occurs when it is in mutant form.
IND regulates dehiscence zone development, at least for the late developing separation layer, by promoting the movement of auxin out of the precursor cells (Sorefan et al., 2009). SPT is specifically expressed in separation layer cells, and it may act here in a parallel manner to its proposed earlier role in interpreting auxin levels in gynoecium development (Nemhauser et al., 2000). It will now be of interest to test potential interactions between SPT and IND and their involvement with auxin.
Supplementary data
The following supplementary data are available at JXB online.
Supplementary Table S1. Primers used for PCR in this study.
Supplementary Fig. S1. Serial transverse sections though the shoot apical meristem and developing leaves of a 12–14-d-old plant, showing expression conferred by the pSPT-6253:GUS reporter gene.
Supplementary Fig. S2. Alignment of eight conserved regions of the SPT promoter regions of AtSPT (Arabidopsis thaliana), BoSPT (Brassica oleracea), and BrSPTa and BrSPTb (Brassica rapa).
Supplementary Material
Acknowledgments
We thank Marcus Heisler, John Alvarez, and other colleagues for stimulating advice and interest, and John Arvanitakis for technical assistance. Steve Swain kindly provided ind-12 mutant seeds, and John Bowman and Yuval Eshed donated cloning vectors. Sandy Floyd and John Bowman provided access to microscopy facilities and advice in their use. This work was supported in part by the Australian Research Council, grant A19927094.
References
- Alvarez J, Smyth DR. Genetic pathways controlling carpel development in Arabidopsis thaliana. Journal of Plant Research. 1998;111:295–298. [Google Scholar]
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. International Journal of Plant Sciences. 2002;163:17–41. [Google Scholar]
- Bailey P, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. The Plant Cell. 2003;15:2497–2501. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenase controls the formation of floral organs and vascular tissues in Arabidopsis. Genes and Development. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007;134:3593–3601. doi: 10.1242/dev.011510. [DOI] [PubMed] [Google Scholar]
- Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. The Plant Journal. 2008;55:40–52. doi: 10.1111/j.1365-313X.2008.03469.x. [DOI] [PubMed] [Google Scholar]
- Heisler MGB, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;128:1089–1098. doi: 10.1242/dev.128.7.1089. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant and Cell Physiology. 2010;51:252–261. doi: 10.1093/pcp/pcp184. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. The Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SL, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Liu P-P, Koizuka N, Homrichhausen TM, Hewitt JR, Martin RC, Nonogaki H. Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. The Plant Journal. 2005;41:936–944. doi: 10.1111/j.1365-313X.2005.02347.x. [DOI] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Research. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development. 2000;127:3877–3888. doi: 10.1242/dev.127.18.3877. [DOI] [PubMed] [Google Scholar]
- Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes and Development. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- Østergaard L, King GJ. Standardized gene nomenclature for the Brassica genus. Plant Methods. 2008;4:10. doi: 10.1186/1746-4811-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse E-M, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control of seed germination through the bHLH transcription factor SPATULA. Current Biology. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Current Biology. 2001;11:1941–1922. doi: 10.1016/s0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. Journal of Biological Chemistry. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, Curtis MD, Grossniklaus U. Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. The Plant Journal. 2009;59:987–1000. doi: 10.1111/j.1365-313X.2009.03928.x. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison T, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J. A gene expression map of Arabidopsis development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Seki M, Carnici P, Nishiyama Y, Hyayshizaki Y, Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. The Plant Journal. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McCall A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Siep M, Sleddens-Linkels E, Mulders S, van Eanennaam H, Wassengaar E, Van Cappellen WA, Hoogerbrugge J, Grooteoed JA, Baarends WM. Basic helix-loop-helix transcription factor Tcfl5 interacts with the Calmegin gene promoter in mouse spermatogenesis. Nucleic Acids Research. 2004;32:6425–6436. doi: 10.1093/nar/gkh979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz L, Kowalczyk M, Sandberg G, Sundberg E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. The Plant Journal. 2006;47:112–123. doi: 10.1111/j.1365-313X.2006.02775.x. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature. 2009;459:583–586. doi: 10.1038/nature07875. [DOI] [PubMed] [Google Scholar]
- Ståldal V, Sohlberg JJ, Eklund DM, Ljung K, Sundberg E. Auxin can act independently of CRC, LUG, SEU, SPT, and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytologist. 2008;180:798–808. doi: 10.1111/j.1469-8137.2008.02625.x. [DOI] [PubMed] [Google Scholar]
- Stepanova A, Robertson-Hoyt J, Yun J, Benavente LM, Xie D-Y, Doležel K, Schlereth A, Jürgens G, Alonso JA. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B. Internal telomeric repeats and ‘TCP’ domain protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. The Plant Journal. 2003;33:957–966. doi: 10.1046/j.1365-313x.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. The Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proceedings of the National Academy of Sciences, USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:67. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Yoshitsugu T, Sakurai T, Seki M, Shinozaki K, Obokata J. Heterogeneity of Arabidopsis core promoters revealed by high density TSS analysis. The Plant Journal. 2009;60:350–362. doi: 10.1111/j.1365-313X.2009.03958.x. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Wappner P, Shilo B- Z. The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes and Development. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.