Abstract
Six MaMADS-box genes have been cloned from the banana fruit cultivar Grand Nain. The similarity of these genes to tomato LeRIN is low and neither MaMADS2 nor MaMADS1 complement the tomato rin mutation. Nevertheless, the expression patterns, specifically in fruit and the induction during ripening and in response to ethylene and 1-MCP, suggest that some of these genes may participate in ripening. MaMADS1, 2, and 3, are highly expressed in fruit only, while the others are expressed in fruit as well as in other organs. Moreover, the suites of MaMADS-box genes and their temporal expression differ in peel and pulp during ripening. In the pulp, the increase in MaMADS2, 3, 4, and 5 expression preceded an increase in ethylene production, but coincides with the CO2 peak. However, MaMADS1 expression in pulp coincided with ethylene production, but a massive increase in its expression occurred late during ripening, together with a second wave in the expression of MaMADS2, 3, and 4. In the peel, on the other hand, an increase in expression of MaMADS1, 3, and to a lesser degree also of MaMADS4 and 2 coincided with an increase in ethylene production. Except MaMADS3, which was induced by ethylene in pulp and peel, only MaMADS4, and 5 in pulp and MaMADS1 in peel were induced by ethylene. 1-MCP applied at the onset of the increase in ethylene production, increased the levels of MaMADS4 and MaMADS1 in pulp, while it decreased MaMADS1, 3, 4, and 5 in peel, suggesting that MaMADS4 and MaMADS1 are negatively controlled by ethylene at the onset of ethylene production only in pulp. Only MaMADS2 is neither induced by ethylene nor by 1-MCP, and it is expressed mainly in pulp. Our results suggest that two independent ripening programs are employed in pulp and peel which involve the activation of mainly MaMADS2, 4, and 5 and later on also MaMADS1 in pulp, and mainly MaMADS1, and 3 in peel. Hence, our results are consistent with MaMADS2, a SEP3 homologue, acting in the pulp upstream of the increase in ethylene production similarly to LeMADS-RIN.
Keywords: Developmental control, ethylene, 1-MCP, peel, pulp, ripening
Introduction
Fruit ripening is a genetically controlled program requiring the co-ordination of fruit softening, colour change, aroma development, sugar accumulation, and a reduction in acid levels. This program engages developmental control components and in climacteric fruit, also components of the ethylene synthesis and response pathways (Theologis, 1992; Lelievre et al., 1997; Alexander and Grierson, 2002; Adams-Phillips et al., 2004; Barry and Giovannoni, 2007; Cara and Giovannoni, 2008; Pech et al., 2008). Climacteric fruit exhibit a peak of respiration coinciding with an increase in ethylene production at the onset of the ripening process.
Recently, facilitated by the cloning of the genes mutated in several ripening-inhibited spontaneous tomato mutants, a cascade of transcription regulators acting upstream of the ethylene pathway has been revealed (Giovannoni, 2004, 2007). MADS-box genes play a major role in the molecular circuit of developmental regulation (Giovannoni, 2001, 2004; Vrebalov et al., 2002; Vrebalov et al., 2009; Itkin et al., 2009). Type II MADS-box proteins, which constitute a group within the family of MADS-box genes, are characterized by the existence of M, I, K, and C domains. The DNA-binding domain (M), located at the N-terminus, and the K domain which is separated by 30 amino acids (aa) of the I domain, are highly homologous among various genes of the family (Theissen et al., 2000; De Bodt et al., 2003). The C region, on the other hand, is highly variable and it was implicated in transcriptional activation in higher order complex formation (Egea Cortines et al., 1999; Garcia-Maroto et al., 2003). Phylogenetic analysis revealed that type II MADS-box orthologues can be subdivided into distinct clades, and members of the same clade tend to have similar expression patterns (De Bodt et al., 2003).
Several MADS-box paralogues have been identified in tomato fruit (Busi et al., 2003; Giovannoni, 2007) and it was suggested that they may be involved in fruit development. The genes TAG1, TAGL2, TAGL11, TAGL12, TAGL1, TDR6, and TDR4 were expressed during the first steps of tomato fleshy fruit development. Overexpression of TAG1 resulted in fleshy expansion and ripening-like cell wall metabolism in sepals, indicating that it is responsible for fruit development (Pnueli et al., 1994). The TAGL2 protein, a SEP homologue, was found to interact with four MADS-box proteins, suggesting that MADS-box genes in the tomato fruit create heterodimers, possibly using the transactivation domain of TAGL2 to activate transcription (Busi et al., 2003). In addition, down-regulation of TAGL2 (TM29), caused parthenocarpic fruit development, indicating that this gene may function as a negative regulator of fruit development (Ampomah-Dwamena et al., 2002). TDR4 belongs to the SQUAMOSA (AP1) clade, and its transcription is controlled, at least in part, by the SQUAMOSA Binding Protein LeSPL-CNR (SQUAMOSA promoter binding protein-like-Colourless Non-Ripening). The LeSPL-CNR gene is silenced by hypermethylation at its promoter in the Cnr mutant, causing a delay in fruit ripening (Manning et al., 2006; Seymour et al., 2008). It is still not yet clear if these additional MADS-box genes are involved in fruit ripening, and the functional analysis of these genes is underway in several laboratories. By contrast, the MADS-box gene, LeRIN was thoroughly investigated. Transgenic tomato plants under-expressing this gene had delayed ripening and a deletion of a segment from the C-terminus, which is the basis of the rin mutant, completely prevented ripening in the homozygous state (Vrebalov et al., 2002). The gene is highly expressed during ripening (Vrebalov et al., 2002) and most likely it controls ethylene production (Kitagawa et al., 2005). Although fruit ripening was blocked in the rin mutant, it still responded to exogenous ethylene, indicating that the machinery responsible for ethylene response is still functional in these plants (Lincoln and Fischer, 1988; Giovannoni, 2001). A potential orthologue of LeRIN, FvMADS9, was isolated from strawberry, and was found to be expressed specifically in fruit (Vrebalov et al., 2002).
Additional MADS-box genes were also discovered in other fruits; however, most of them were suggested to be involved in early fruit development. In apple, six MdMADS genes were classified to the AP1 clade and one to the AG (Yao et al., 1999), and they were found to be expressed during early fruit development. Mutation in apple MdPI, a MADS-box gene that is highly homologous to PI, known to be involved in identity determination of petals and stamen in Arabidopsis, was found to result in parthenocarpic fruit development (Yao et al., 2001). In peach, two MADS-box genes similar to TAG and TAGL1 have been cloned and their expression in tomato show that they might be responsible for fruit development (Tadiello et al., 2009). In grapes, several isolated MADS-box genes were related to fruit development; VvMADS1 and VvMADS5 are homologous to AG and SHP (Boss et al., 2001), VvMADS2 and VvMADS4 are related to SEP, and another one to AGL13 gene (Boss et al., 2001). Similar genes (CanMADS) were also isolated from hot pepper (Sung et al., 2001) and hazelnut (Rigola et al., 1998). MADS-box genes have also been isolated from Chinese pear and there was no difference in their expression in climacteric and non-climacteric cultivars (Yamane et al., 2007). Whether or not these genes may be specifically involved in fruit ripening, remains to be determined.
Banana, like tomato, is a climacteric fruit, which is characterized by an increase in respiration and a burst in ethylene production occurring at the onset of ripening. However, in light of the fact that banana is a monocot, it is still not clear if this fruit uses similar components to those of the tomato eudicot for controlling ripening. In addition, banana ripening differs from that of tomato also because it has pulp and peel, which, based on their altered pattern of ethylene synthesis, suggests that their ripening programmes differ (Clendennen et al., 1997). Ethylene production in peel starts after its production in the pulp and its initiation is dependent on pulp ethylene (Dominguez and Vendrell, 1993). So far, components related to the ethylene control of ripening have been cloned in banana. The expression of several ACC oxidase and synthase genes were correlated with increased ethylene levels (Clendennen et al., 1997; Liu et al., 1999; Pathak et al., 2003). An ethylene receptor (Wu et al., 1999), a CTR1 orthologue (Clendennen et al., 1997) and four EIN3-like genes were isolated (Mbeguié-A-Mbeguié et al., 2008). Coinciding with the biochemical changes observed during banana fruit ripening, differentially-expressed genes were isolated from the pulp (Clendennen and May, 1997; Medina-Suarez et al., 1997) and peel (Drury et al., 1999; Liu et al., 2002) of banana fruit after the initiation of the ripening process. MADS-box genes have been isolated from a Brazilian cultivar and from the cultivars Pisang bregnan and Nanicao, and have been deposited in GenBank (Liu et al., 2009). The expression of a MADS-box gene (GenBank accession 941800 from Pisang) increased after propylene application and this increase was reduced following 1-MCP treatment (Inaba et al., 2007). Also the expression of MADS-box gene cloned from the Brazilian cultivar (GenBank accession DQ060444) was correlated with an increase in ethylene production (Liu et al., 2009). However, so far there has been no comprehensive study of the MaMADS-box genes expressed in banana fruit. In addition, the interaction between ethylene and the expression of fruit MaMADS-box genes has not been fully studied.
In this study, six full-length MADS-box genes from banana (Grand Nain) have been cloned and characterized, and the ability of two of these genes to complement the rin mutation in tomato has been examined. In addition to their expression in various banana organs and during ripening in peel and puld, the interactions between ethylene and the expression induction of these genes have been determined.
Materials and methods
Plant materials and treatments
Banana (Musa acuminata AAA Cavendish subgroup, Grand Nain) grown along the Mediterranean shore in Israel during the winter months, were used in this study. Several stages during banana ripening were identified based on the details described in INIBAP (International Network for Improvement of Banana and Plantains: www.inibap.org): from the first stage of green banana fruit through the third stage of yellow banana with green edges until the seventh stage when brown spots are apparent. Fingers from the upper hand proximal to the trunk were taken from at least five bunches, separated, sprayed with 0.1% thiobendazole, and air-dried. They were packed in aerated polyethylene bags and stored at 20 °C. Samples were taken from pulp and peel, separately. In addition, other banana plant organs were used; bulb core (BC), root (R), pseudo stem (PS), young leaf (YL), male flower (MF), female flower (FF), bract leaf (BL), male flower ovary (MFO), and female flower ovary (FFO).
Ethylene treatment of 10 μl l−1, based on preliminary experiments, was applied on the third day after harvest (DAH) for 18 h. 1-MCP treatments of 0.3 μl l−1 was applied at the onset of an increase in ethylene production (usually on the 8th DAH) for 18 h and samples for the determination of ripening parameters were taken on consecutive days up to 17 DAH.
Characterization of fruit-ripening parameters
Fruit ripening was determined using the following parameters: peel colour, fruit firmness, and carbon dioxide (CO2) and ethylene emission (C2H4). Peel colour from the surface area of the individual upper banana fingers was determined using Minolta CR-300 (Minolta Corporation, New Jersey, USA) and the results are expressed as hue angle (°). Firmness was measured using a Chatillon Force tester (Ametek Inc., Florida, USA), and results are expressed in Newton (N).
Carbon dioxide and ethylene production were determined by sealing a banana finger in a 2 L sealed glass jar at 20 °C for 1 h. Samples were withdrawn from the sealed jars using gas-tight syringes. Carbon dioxide concentrations were determined by a Packard 7500 gas chromatograph (Packard, IL, USA) with a thermal conductivity detector and a CTR-I packed column using helium as a carrier gas. Ethylene concentration was determined with Varian 3300 gas chromatograph equipped with a flame ionization detector and a C-5000 packed column using nitrogen as the carrier gas.
Cloning of MADS box genes
Total RNA was extracted from separated banana peel and pulp and cut into 1 g tissue and frozen in –80 °C. The frozen tissue was pulverized under liquid nitrogen and 100 mg was used with the Spectrum Plant RNA Kit (Sigma, UK). Extraction was done as described (http://www.sigmaaldrich.com/sigma/bulletin/STRN250bul.pdf). The extracted RNA was digested and cleaned with the TURBO DNase kit according to the protocol described at the site (http://www.ambion.com/techlib/prot/bp_1907.pdf). RNA concentrations ranging between 50–350 μg μl−1 and a ratio 1.8–2.0 (of absorbance at 260/280) was obtained. cDNA was prepared using the Verso™ cDNA kit (Thermo Fisher Scientific Inc., USA). Cloning was performed on a mixture of cDNA from peel and pulp of different developmental stages.
The MaMADS1 fragment was cloned by screening a ripe banana pulp cDNA library at low stringency with a full-length probe of LeMADS-RIN. In addition, several fragments of MADS-box genes from banana fruit of various cultivars have been deposited in GenBank http://www.ncbi.nlm.nih.gov/BLAST: a fragment of 378 nucleotides of the cultivar Nanicao (AY463009: corresponding to MaMADS2) and a sequence of 888 nucleotides from the Brazilian cultivar (DQ060444: corresponding to MaMADS5) and three sequences from the Pisang cultivar, one of 931 nucleotides (AY941799: corresponding to MaMADS3), a sequence of 944 nucleotides (AY941800: corresponding to MaMADS4), and a sequence of 615 nucleotides (AY941798: corresponding to MaMADS6). Aided by these sequences, a full-length of MaMADS genes from the cultivar Grand Nain fruit have been cloned by successive reactions using the primers described in Supplementary Table 1 at JXB online. For each of the genes, the primers described in reaction A were used to clone an initial fragment; in reaction B, they were used for completing the 3′ end, and in reaction C, they were used for completing the 5′ end. Both reactions B and C used a RACE technology (5′/3′ RACE Kit 2nd generation, Roche Applied Science). The PCR amplifications were carried out in a Mastercycler gradient apparatus (Eppendorf, Germany) using 40 cycles of denaturing at 95 °C for 30 s, annealing at 52 °C for 60 s and elongation at 72 °C for 60 s. PCR product was further purified from the gel using QIAEX II gel extraction kit (Qiagen Ltd, UK) according to the manufacturer's instruction (http://www1.qiagen.com), and ligated to the pGEM-T® easy vector (Promega Corporation, Madison, USA). The ligation reaction was transformed into JM 100 competent E. coli cells (www.rbcbioscience.com) (RBC Bioscience).
BioEdit Sequence Alignment Editor software (v. 7.0.9) was used for the DNA alignment. The phylogenetic analysis was performed using the tools in the site (http://www.phylogeny.fr/version2_cgi/index.cgi), which enables this analysis by PhyML, a software implementing a new method for building phylogenies from DNA and protein sequences using maximum likelihood, and the tree was drawn by TreeDyn (Dereeper et al., 2008). Primer Express software (v. 2.0) was used for design of primers for quantitative RT PCR (Q-RT-PCR) and for cloning (http://www.appliedbiosystems.com/support/apptech/#).
Expression analysis of MaMADS-box genes
Gene expression was determined by Quantitave RT PCR (Q-RT-PCR). The sequence of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and a ribosomal gene (AY821550 and EU433925, respectively) were used as a reference for equalizing the levels of RNA. Forward and reverse primers for the references genes are: 5′-GCAAGGATGCCCCAATGT-3′ and 5′-AGCAAGACAGTTGGTTGTGCAG-3′ for GAPDH, 5′-GCGACGCATCATTCAAATTTC-3′ and 5′-TCCGGAATCGAACCCTAATTC-3′ for the ribosomal gene. Primers and cDNA concentrations used for the reactions were predetermined as described, to enable a linear and highly efficient response (http://www.abgene.com/downloads/article-SYBRoptimise.pdf). The reaction mixture contained forward and reverse primers (see Supplementary Table 2 at JXB online) and Power SYBR Green PCR Master mix (Applied Biosystems, USA) in a 20 μl total sample volume. Reactions were run in triplicate on a Rotor-Gene 3000 PCR machine (Corbett Life Research, Australia) using 35 cycles of 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 20 s. The results represent one experiment out of at least two independent samplings, for which, usually two preparations of cDNA were examined. Data obtained were analysed with Rotor-Gene 6 software, and by the qBase quantification Software (http://medgen.ugent.be/qbase/). Data are expressed according to the ΔΔCT method. To enable presentation of all the different transcripts in response to either ethylene or 1-MCP, expression is expressed as a percentage increase of the lowest sample.
Complementation of rin mutants
Tomato plants used in complementation studies were homozygous for the rin mutation in cultivar Ailsa Craig and grown in greenhouses under standard conditions with 16/8 h day/night.
Full-length cDNAs of MaMADS1 and MaMADS2 were cloned into the plant transformation vector pBI121 and transformed into rin/rin tomato as described in Vrebalov et al. (2002). Kanamycin-resistant transformants were grown to maturity in the greenhouse and confirmed for transgene integration by DNA gel-blot analysis using the nptII gene (Kan resistance) as probe. Seeds were saved and all subsequent analyses for ripening complementation were performed on T1 progeny which either contained or segregated out the transgene, as confirmed by DNA gel-blot analysis. A total of 11 and 17 independent transgenic lines were generated for MaMADS1 and MaMADS2, respectively.
Results
Isolation of MADS-box genes from banana fruit
Six full-length transcripts were cloned from the banana fruit of the cultivar Grand Nain of the Cavendish subgroup (Fig. 1). Each one of them exhibited a difference in the amino acid (aa) sequence, when compared to the homologuos partial sequences from other other banana cultivars deposited in GenBank, (data not shown). Alignment of the putative aa of the six genes showed high similarity in the two regions: 1 to about 55 aa–M domain, and around 95 to about 170 aa–K domain (Fig. 1A). The genes were highly variable in their C domain. Phylogenetic analysis of these genes with MADS-box genes from various clades revealed that the genes MaMADS1–4 belong the SEP3 clade and indeed they contain the SEP3 motif (Fig. 1B) (Malcomber and Kellogg, 2005). The genes MaMADS2 and 4 show the highest similarity between them and the highest to LeMADS-RIN (Table 1). The genes MaMADS5 and MaMADS6 belong to the AGAMOUS (AG) and PISTILLATA (PI) clades, respectively. These genes indeed contain the respective domains (Fig. 1 C, D) (Skipper et al., 2006). Comparing the C-terminus of all the banana genes to that of LeMADS-RIN from tomato showed lower homology to Le-MADS-RIN than when the whole gene is compared (Table 1). (Similarities are: 30% for MaMADS1, 35% for MaMADS2, 28% for MaMADS3, 32% for MaMADS4, 20% for MaMADS5, and 15% for the gene MaMADS6). The genes isolated have shown the highest homology to the corresponding genes: MaMADS1: 75% similarity to AOM1 (AAQ83834 from Asparagus officinalis) (Caporali et al., 2000), MaMADS2 and MaMADS4: 88% and 84% similarity, respectively, to MADS-box (AAQ03226 from Elaeis guineensis) (Adam et al., 2006), MaMADS3: 78% similarity to AGL9a (ABK35281from Crocus sativus). MaMADS5: 85% similarity to MADS (BAD83772 from Asparagus virgatus), MaMADS6: 88% similarity to PISTILLATA-like protein (ABB92623 Alpinia oblongifolia) (Gao et al., 2006).
Fig. 1.
Isolation of MaMADS-box genes from Cavendish banana (cultivar Grand Nain). (A) Putative proteins alignment of MaMADS1–6 sequences isolated from banana fruit and the similarity among them. The gene sequences were deposited in http://www.ncbi.nlm.nih.gov/BLAST as EU869307, EU869306, EU869308, EU869309, EU869310, and EU869311, respectively. (B) Phylogenetic analysis of MaMADS1–4 with other SEP3 clade genes. The analysis was performed on the full-lengths of the individual genes. The SEP3 motif for each of the genes is presented on the right. The genes included in this alignment are: AoAOM1 (AAQ83834), SISEP3 (BAD10945.1), LItuAGL9 (AAX15920.1), PaAGL9.1 (AAX15923.1), HcSEP1 (BAC80253.1), PsM6 (AAX69068), DcSEP1 (AAZ95252.1), AdMADS (CAA48859.1), SaMADSD (CAA69916.1), PsMADS (CAA11258.1), PhFBP26 (AAF19164.1), NsMADS3 (AAD39034.1), LeTM5 (AAP57413.1), EcAGL9 (AAX15918.1), DeCDM44 (AAO22982.1), BpMADS (CAB95648.1), DEFH200 (CAA64743.1), PtMADS6 (AAO49811.1), AtSEP (AAT46095.1), VvMADS4 (AAM21344.1), AmDEFH72 (CAA64742.1). (C) Phylogenetic analysis of MaMADS5 with other AG genes. The existence of AG motifs for all the genes is shown. The genes included in this alignment are: DtSEEDSTICK (AAY86365.1), DnMADS2 (ABQ08574.1), AvMADS (BAD83772.1), HoMADS1 (AAF08830.2). (D) Phylogenetic analysis of MaMADS6 with other PI genes. The existence of PI motifs for all the genes are shown. The genes included in this alignment are: AoPI (ABB92623.1), P9PI (AAV28175.1), PI10 (AAV28490.1), HPI2 (AAD22494.2), PI1 (ABG90945.1). Lines underneath the genes mark the genes isolated from banana. Stars and dots indicate identity and similarity, respectively.
Table 1.
Comparison between the different MaMADS-box genes and LeRIN
| MaMADS1 | MaMADS2 | MaMADS3 | MaMADS4 | MaMADS5 | MaMADS6 | LeRIN | |
| MaMADS1 | 71 | 52 | 69 | 48 | 42 | 56 | |
| MaMADS2 | 71 | 54 | 88 | 41 | 37 | 62 | |
| MaMADS3 | 52 | 54 | 55 | 44 | 43 | 55 | |
| MaMADS4 | 69 | 88 | 55 | 41 | 40 | 60 | |
| MaMADS5 | 48 | 41 | 44 | 41 | 43 | 49 | |
| MaMADS6 | 42 | 37 | 43 | 40 | 43 | 43 | |
| LeRIN | 56 | 62 | 55 | 60 | 49 | 43 |
Numbers indicate percentage homology at the amino acid level.
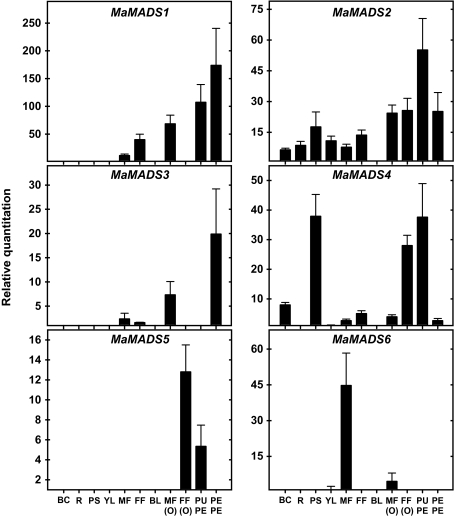
The expression of these genes in vegetative and reproductive tissues has been examined (Fig. 2). MaMADS1–5 genes are highly expressed in either peel or pulp, however, MaMADS4 and 5 are also highly expressed in other reproductive tissues. MaMADS4 and MaMADS5 are expressed in the female flower ovary (FF-O), and only MaMADS4 is also expressed in the vegetative tissue pseudo stem (PS). The genes MaMADS1 and 2 are highly expressed in peel and pulp, however, MaMADS2 is also expressed in other tissues at lower levels. By contrast, MaMADS6 is highly expressed in the male flower (MF), more than in fruit. This analysis shows that, in peel, MaMADS1 and 3 are predominant during the climacteric stage, while, in pulp, MaMADS2, 4, and 5 are most highly expressed.
Fig. 2.
Expression patterns of MaMADS genes in various plant organs. BC, bulb core; R, root; PS, pseudostem; YL, young leaf; MF, male flower; FF, female flower; BL, bract leaf; MF(O), male flower ovary; FF(O), female flower ovary; PUPE, pulp at climacteric peak; PEPE, peel at climacteric peak. Expression was determined by Q-RT- PCR as relative quantification. The specificity of the primers was determined for each of the genes (see Table 1, Materials and methods), and the expression of both ribosomal RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as reference genes. Results are of a representative experiment, and are an average of three repetitions ±SD. The values of relative quantification obtained were 105 higher.
MaMADS-box genes expression in peel and pulp during normal ripening
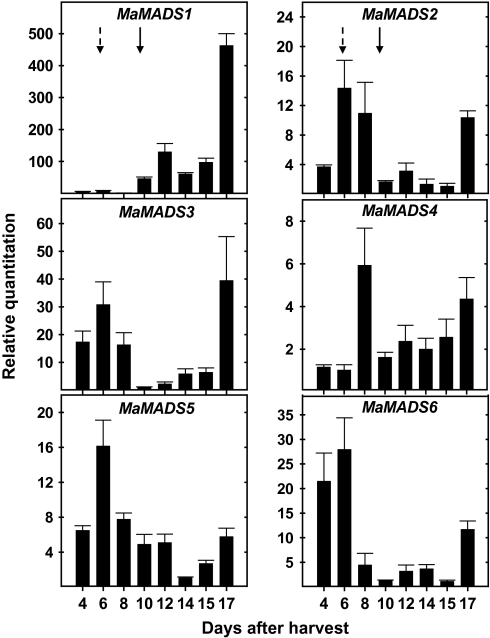
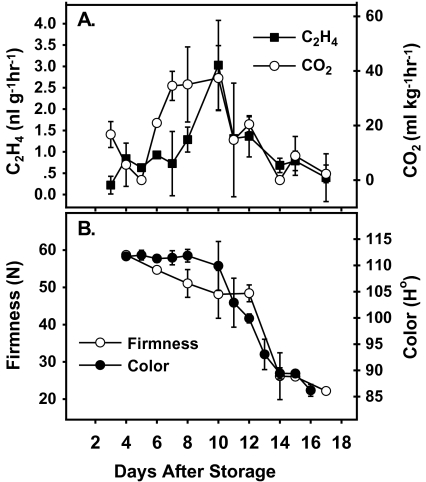
The LeMADS-RIN which was proven to be involved in fruit ripening, exhibited elevated expression at the onset of ripening (Vrebalov et al., 2002). Therefore, the expression patterns of all the banana fruit MaMADS genes have been determined during ripening to examine which of the genes shows similar expression to that of LeMADS-RIN (Figs 4, 5). Production of ethylene and carbon dioxide in whole fruit and ripening parameters of colour and firmness have been determined following harvest (Fig. 3). Carbon dioxide climacteric production preceded the peak of ethylene, and it coincided with a reduction in firmness (Fig. 3A, B). Changes in colour (from green to yellow) appeared 4 d later and started following the ethylene peak (Figs. 3A, B).
Fig. 4.
Dynamic changes in MaMADS-box genes expression in pulp tissue during ripening. The expression was determined in fruit from the upper hand according to details described in Fig. 2. The increase in CO2 production appeared 6 d after harvest (DAH) (broken arrow) and the ethylene peak in these fruits was detected by 10 DAH (full arrow). Results are of a representative experiment, and represent an average of three repetitions ±SD.
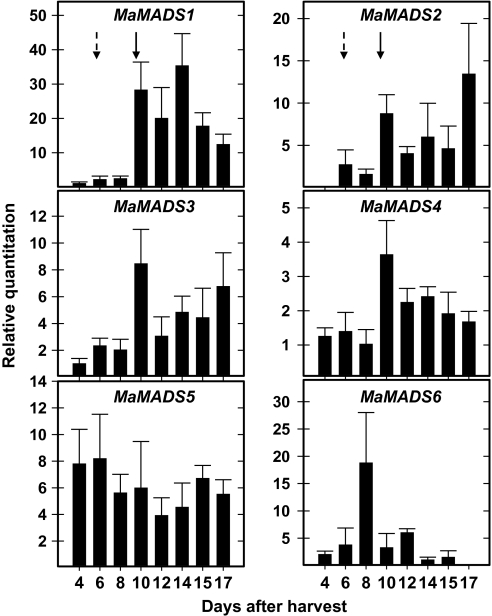
Fig. 5.
Dynamic changes in MaMADS-box genes expression in peel tissue during ripening. The expression was determined in fruit from the upper hand according to details described in Fig. 2. An increase in CO2 production appeared 6 d after harvest (DAH) (broken arrow) and the ethylene peak in these fruits was detected at 10 DAH (full arrow). Results are of a representative experiment, and represent an average of three repetitions ±SD.
Fig. 3.
Description of ethylene and carbon dioxide production (A) and ripening parameters (B) in whole fruit. The parameters were determined in banana from the upper hand immediately following harvest and at consecutive days during storage at 20 °C and 75% RH. Ripening parameters determined were peel colour (Ho angle) and firmness (N).
The patterns of expression of the various genes in the peel and pulp were unique to the tissue examined. An increase in expression of most of the genes correlated with CO2 production in the pulp and with ethylene production in the peel (Figs 4, 5), except the expression of MaMADS6 in pulp which exhibited high expression during the green stage immediately after harvest and prior to any increase in ethylene or CO2 production (Fig. 4). Expression of MaMADS6 in peel was only slightly induced, but prior to the onset of the ethylene peak (Fig. 5). Also the expression of MaMADS5 in peel was not increased in parallel to ethylene or CO2 overproduction and was constant during the fruit ripening period (Fig. 5). The increased expression of MaMADS2, 3, and 5 in pulp paralleled the increase in CO2 production and that of MaMADS4 followed. The increase in MaMADS1 expression in pulp, on the other hand, appeared at the onset of ethylene production and a major increase occurred late, after harvest, when the banana was completely ripe (stage 7). This induction was correlated with a second wave of increased expression in the genes MaMADS2, 3, and 4 (Fig. 4), indicating that there are two stages of MaMADS-box participation in ripening.
In the peel, the increase in expression of the genes MaMADS1, 2, 3, and 4 was correlated with the increase in ethylene production and, while MaMADS3 and 2 remained high, that of MaMADS1 and 4 decreased gradually during ripening (Figs 3, 5). Most notably, is that the expression levels of MaMADS2 in peel were lower than those in pulp (Figs 3, 4, 5).
Interactions between ethylene and MaMADS-box gene expression
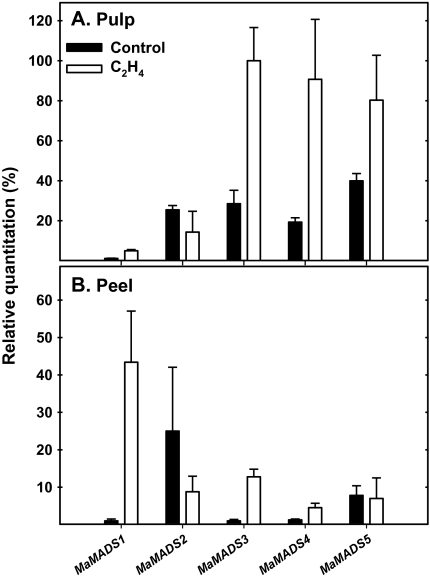
Since most of the gene transcripts were induced in parallel with the onset of ripening, the interactions between ethylene and MaMADS-box genes expression has been studied following the application of ethylene or 1-MCP (Figs 6, 7). Ethylene application at the green stage increased endogenous ethylene production (data not shown). This treatment also increased MaMADS3, 4, and 5 in the pulp and MaMADS1 and 3 in the peel. However, this treatment did not increase the expression of MaMADS2 either in the pulp or in the peel (Fig. 6).
Fig. 6.
Response to exogenous ethylene of MaMADS-box genes in peel and pulp. Banana fruits were treated with 10 μl l−1 ethylene 4 d after harvest for 18 h. The expression was determined for control and ethylene-treated samples relative to other samples during fruit ripening and the results of day 5 after harvest are presented.
Fig. 7.
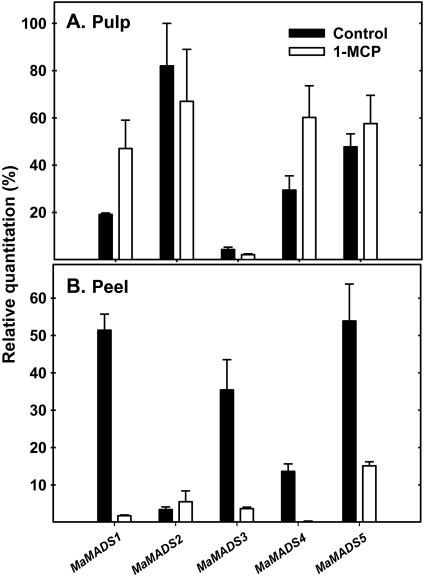
Response to 1-MCP of MaMADS-box genes in peel and pulp. Banana fruits were treated with 0.3 μl l−1 1-MCP at the onset of ethylene peak (8 d after harvest for 18 h). The expression was determined for control and 1-MCP-treated samples relative to other samples during fruit ripening and results of day 10 after harvest are presented.
1-MCP was applied prior to the ethylene peak (8 d following harvest in Fig. 7). This aimed to show a direct effect of ethylene on MaMADS-box gene expression at the onset of ripening. 1-MCP treatment on the 8th day reduced ethylene and CO2 production (data not shown). This treatment decreased the expression of MaMADS1, 3, 4, and 5 in the peel, and, for MaMADS1 and 3, this represents a reciprocal image of the ethylene effect (Fig. 6). However, in the pulp, this treatment increased the expression levels of MaMADS1 and 4, suggesting that at this stage of ripening, these genes are under the negative control of ethylene. Also, this treatment, similar to that observed for ethylene, did not affect the expression levels of MaMADS2 (Fig. 7).
Analysis of tomato plants expressing MaMADS1 and MaMADS2 genes
As a first step towards functional analysis, and since banana transformation is difficult and time-intensive, we transformed into the homozygous rin mutation of tomato cultivar Ailsa Craig, 35S-driven constructs of both MaMADS1 and MaMADS2. A similar strategy was used to validate the function of the tomato LeMADS-RIN gene (Vrebalov et al., 2002). MaMADS2 was specifically selected for complementation of rin due to its closest sequence similarity to LeMADS-RIN and to the fact that it is not affected by ethylene and MaMADS1 due to its high expression in peel and pulp.
Transgenic T1 generation plants and non-transgenic siblings were confirmed for transgene overexpression in mature fruit (data not shown) and monitored for ripening parameters. No changes in ethylene, softening, and carotenoid accumulation in ripening were detected in transgenic rin/rin fruits (Fig. 8), nor were any abnormalities noted in floral or fruit development. Application of exogenous ethylene to these transgenic fruit also had no impact on their development.
Fig. 8.
Expression of MaMADS genes in rin/rin tomato fruit did not complement ripening. Ailsa Craig wild type, nearly isogenic rin/rin, and transgenic rin/rin tomato fruit were tagged at anthesis and designated as breaker stage at the same age as wild-type fruit which showed the first signs of colour change. Fruit were harvested 10 d post-breaker stage and photographed. Fruit are shown from (A) Ailsa Craig wt, (B) Ailsa Craig nearly isogenic for rin/rin, (C) rin/rin T1 lines over-expressing MaMADS1 (line 15 on the left and line 18 on the right), (D) rin/rin T1 lines over-expressing MaMADS2 (line 3 on the left and line 8 on the right). (This figure is available in colour at JXB online.)
Discussion
Gene structures and similarity to LeRIN
The full-length cDNAs of six MADS-box genes have been cloned from the banana fruit Grand Nain cultivar. The genes are highly divergent in their C-terminus. This is common among MADS-box genes even those arising from gene duplications. The differences between the genes may have arisen due to mutations that may lead to new functions (Maere et al., 2005). The genes MaMADS2 and MaMADS4 show the highest similarity, even in their I region (Fig. 1A), which is responsible for partner selection (Garcia-Maroto et al., 2003); however, the role of the I region in dimerization of these two genes has not been clarified yet. Phylogenetic analysis of these genes with MADS-box genes of various clades indicate that MaMADS6 belongs to the PI clade, MaMADS5 to the AG clade, and MaMADS1, 2, 3, and 4 belong to the SEP3 clade and indeed these genes contain the typical motifs for each of the groups (Fig. 1) (Malcomber and Kellogg, 2005; Skipper et al., 2006). Neither of the genes cloned were highly similar to the LeMADS-RIN which encodes a component of the developmental control of ripening and belongs to the SEP4 clade. Nevertheless, the highest similarities of the LeMADS-RIN C-terminus are to that of MaMADS2 and 4 genes, which are only 35% and 32%, respectively. It has been determined before that orthologues of SEP4 are missing from non-core eudicots, monocots, and basal angiosperms (Malcomber and Kellogg, 2005), and might be the reason for not finding SEP4 homologue in banana.
Genes of these clades have been isolated in other fruits; the PI homologue was found to be mutated in an apple cultivar undergoing parthenocarpic development (Yao et al., 2001) and AG genes, which are involved with carpel development, were isolated from apple fruit (Yao et al., 1999), grapes (Boss et al., 2001), peach (Tadiello et al., 2009), and tomato (TAG1, TAG11, and TAGL1) (Busi et al., 2003). SEP genes with high expression in fruit have also been identified in tomato (Giovannoni, 2007). The genes belonging to the SEPALLATA (SEP) clades often retain similar functional capacity (Zahn et al., 2005) and they appear to contribute to the creation of multimeric complexes (Honma and Goto, 2001). In addition, MADS-box genes belonging to other clades have been cloned from tomato; like the gene TDR6 which belongs to the AP3 group, and MC, TAGL2 and TDR4 which belong to the SQ/AP1 clade (Vrebalov et al., 2002; Busi et al., 2003). Studies in Arabidopsis and tomato revealed that AG together with SEP are responsible for carpel development (Seymour et al., 2008; Vrebalov et al., 2009) and it is possible that MaMADS5 fulfil similar function in banana fruit development, possibly aided by MaMADS6, the PI homologue.
MADS-box genes have been cloned from a genome wide expression profile of another monocot, rice, and 75 MADS-box genes have been identified (Arora et al., 2007). Protein alignment between the MaMADS and the rice MADS-box genes deposited in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) revealed that MaMADS1, MaMADS2, and MaMADS4 have about 70% homology to MADS-box transcription factor 8 (Os09g32948.1) and MaMADS3 has about 73% homology to MADS-box transcription factor 6 (Os02g45770.1) all classified as SEP3. MaMADS5 has about 66% homology to MADS-box transcription factor 3 (Os01g10504.3) classified as AG, while MaMADS6 has a similar homology to MADS-box transcription factor 4 (Os05g34940.2) classified as PI. It is still not clear if these genes in rice have any function in ovary development and carpel maturation.
Expression patterns of MaMADS genes and their possible involvement in ripening
In many cases, MADS-box genes are not expressed exclusively in one tissue, but are recruited for different functions (Theissen et al., 2000; Garcia-Maroto et al., 2003; Immink et al., 2003). Among the banana fruit MADS-box genes, only MaMADS1–3 are expressed in fruit tissues at their highest levels, but the other genes are expressed at higher levels in other tissues (Fig. 2). The MaMADS5 gene belonging to the AG clade, besides being expressed in fruit, was also expressed in the female flower ovary (Fig. 2), and MaMADS6, belonging to the PI clade, was expressed mainly in male flowers (Fig. 2), however, it was also expressed to some degree in the pulp and peel (Figs 4, 5), and the levels in pulp were even higher after harvest (Fig. 4). Also, among the SEP3 genes, MaMADS4 was expressed in the pseudo-stem as well as in the female flower ovary (Fig. 2). Indeed, genes belonging to the SEP clade have been found to be expressed mainly in inflorescences and act as redundant genes in flower development, and are even found to be expressed in vegetative tissue (Malcomber and Kellogg, 2005). In tomato too, some of the genes that were expressed in fruit were also expressed in other organs. TAGL12 was found not to be specific to fruit. TDR6, TDR4, and TAG1 were also previously described to be involved in flower development (Busi et al., 2003). In rice, another monocot, although MADS-box genes showed expression in reproductive tissue, they had a general tendency also to be expressed in vegetative tissues (Arora et al., 2007).
The expression patterns during fruit ripening are very similar in the pulp for the genes MaMADS2–5, and in peel for MaMADS1–4 (Figs 4, 5). Similar expression of genes, especially if they are from the same clade, had been suggested to show that they have a redundant function (Purugganan et al., 1995; Theissen et al., 2000; Immink et al., 2003). These gene products may create heterodimers during the ripening process and different heterodimers may be created in the pulp and peel. Changes in the expression patterns of the various genes in the pulp and in peel during ripening (Figs 4, 5) suggest that the transcription complexes created during ripening are constantly changing.
Usually there is a parallel increase in ethylene and CO2 at the onset of ripening in climacteric banana fruit (Domínguez and Vendrell, 1994) and in some studies it was reported that an increase in ethylene preceded the increases in respiration, when fruit were exposed to propylene (McMurchie et al., 1972), or during natural ripening (Burg and Burg, 1965). In the current experiments, the CO2 increase preceded the increase in ethylene by three days. It is interesting to note, that in several experiments that were performed in the winter months, the increase in CO2 preceded that of ethylene by one day and up to three days, however, in the summer months the increase in ethylene and CO2 appeared together (data not shown). The spatial separation between the increase in respiration and ethylene, which existed in these experiments, enabled us to determine that the increase in gene expression in the pulp of MaMADS2–5 correlated with a CO2 increase, and preceded ethylene increase. However, in experiments where ethylene and CO2 increased concurrently, the increase in gene expression did not precede the ethylene peak (data not shown). In the peel, on the other hand, the increase in expression of MaMADS1, 2, 3 and 4 occurred later and in parallel with the increase in ethylene production in whole bananas. These results fit the suggestion that ripening starts in the pulp and then progresses to the peel (Dominguez and Vendrell, 1993). Moreover, our results support the idea that the initiation of climacteric respiration is not dependent on ethylene (Pech et al., 2008). The decrease in firmness that was initiated before the ethylene peak (Fig. 3), supports the idea that the initiation of some ripening processes start before the ethylene peak.
Interactions between ethylene and MADS-box gene expressions
In this study, exogenous ethylene and 1-MCP have been used to determine the involvement of ethylene in the MaMADS gene expression. Application of ethylene advanced the climacteric peak, and increased respiration, while 1-MCP inhibited ethylene production (data not shown), as has been reported previously (Golding et al., 1998; Zhang et al., 2006; Liu et al., 2009). Using ethylene, it was possible to determine that MaMADS3, 4, and 5 in the pulp, and MaMADS1 and 3 in the peel are regulated by ethylene (Fig. 6). Since MaMADS3 is induced by ethylene in the peel and pulp, it is suggested that its expression is controlled by an ethylene-induced transcription factor common to both peel and pulp. However, MaMADS4 and 5 in the pulp and MaMADS1 in the peel are most likely controlled by tissue-specific ethylene-induced transcription factors. These results may explain why, during the climacteric peak, MaMADS4 and 5 are elevated mainly in the pulp and MaMADS1 mainly in the peel in parallel to the climacteric peak. This further emphasizes the difference in ripening programmes which exist in the peel and in pulp.
The analysis of gene expression following 1-MCP applied at the onset of ethylene burst (Fig. 7), demonstrated that the expression of both MaMADS4 and MaMADS1 is negatively regulated by ethylene at the climacteric stage in pulp tissue. It is possible that early after harvest MaMADS4 is induced by ethylene, but, during the climacteric peak, its expression is inhibited by ethylene. For MaMADS1, the expression increased by several-fold following the climacteric peak, suggesting that the inhibition observed during the climacteric peak is removed when ethylene is reduced. On the other hand, in the peel, MaMADS1, 3, and 5 are reduced by 1-MCP, further strengthening the conclusion that at least MaMADS1 and 3 are ethylene-induced. Negative regulation of genes by ethylene at the climacteric peak has been demonstrated before in tomato (Hoeberichts et al., 2002). Moreover, 1-MCP increased ethylene production in tomato when applied at the breaker stage and also in banana when applied at the yellow transition stage (Pelayo et al., 2003). In another study, application of 1-MCP to propylene-treated fruit revealed that 1-MCP increased ethylene production concomitantly with MaACS1 in the pulp and not in the peel (Inaba et al., 2007). It is possible that, in the pulp, in comparison to peel, there is an additional control component of ethylene production which is responsible for a reduction in ethylene production and MaMADS4 is involved in this control.
The unique expression of MaMADS2 suggests that this gene plays an important role in banana fruit ripening
Sequence similarity, in combination with similar temporal and spatial expression patterns, is necessary to establish orthologous relationships between MADS-box genes (Arora et al., 2007). In the case of MaMADS2 and LeMADS-RIN, it is clear that the two genes have very low similarity especially in their C-terminus, however, the expression pattern during ripening is very similar; both genes are induced at the onset of the climacteric (Fig. 4) (Vrebalov et al., 2002).
Several studies on the involvement of MADS-box genes in flowering suggest that the functional similarity is not necessarily related to sequence similarity and the function of specific MADS-box genes can be taken by another type of gene. For instance, in rice, another monocot, Arabidopsis-related genetic switch systems control floral transition, but they are based on different MADS-box transcription factors (Andersen et al., 2004). Similarly, a SEP3 from lily, a monocot species, did not possess the predicted E class function during floral organ development, when expressed in Arabidopsis (Tzeng et al., 2003). While neither MaMADS2 nor MaMADS1 was able to complement the tomato rin mutation, this result does not exclude a similar role for these genes in banana fruit ripening. Indeed, the ethylene-independent expression pattern of MaMADS2 during banana fruit ripening suggests that this gene may regulate ripening and act upstream of the ethylene response pathway. Transgenic banana with reduced levels of MaMADS2 expression will be required to test this hypothesis.
In summary, our results suggest that several MaMADS-box genes may function during the climacteric and some may be induced by ethylene, but MaMADS2 is most likely serves as an upstream regulator, since it is not affected by ethylene. Our results also clearly support previous suggestions that ripening initiates in the pulp, possibly via an increase in CO2 and ethylene produced by the pulp initiates ripening of the peel. It is also suggested that different components of the MaMADS-box genes participate in the ripening programs, all along the paper of these two tissues.
Supplementary Material
Acknowledgments
Contribution from the Agricultural Research Organization, The Volcani Center, Bet Dagan, Israel. No. 550/09. The research was supported by BARD (The United States-Israel Binational Agricultural Research and Development Fund) research Grant Award No. IS-3803-05.
References
- Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear J. MADS box genes in oil palm (Elaeis guineensis): patterns in the evolution of the SQUAMOSA, DEFICIENS, GLOBOSA, AGAMOUS, and SEPALLATA subfamilies. Journal of Molecular Evolution. 2006;62:15–31. doi: 10.1007/s00239-005-0333-7. [DOI] [PubMed] [Google Scholar]
- Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends in Plant Science. 2004;9:331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D. Ethylene biosynthesis and action in tomato:a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiology. 2002;130:605–617. doi: 10.1104/pp.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CH, Jensen CS, Petersen K. Similar genetic switch systems might integrate the floral inductive pathways in dicots and monocots. Trends in Plant Science. 2004;9:105–107. doi: 10.1016/j.tplants.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor K. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242–263. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C, Giovannoni JJ. Ethylene and fruit ripening. Journal of Plant Growth Regulation. 2007;26:143–159. [Google Scholar]
- Boss PK, Vivier M, Matsumoto S, Dry IB, Thomas MR. A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Molecular Biology. 2001;45:541–553. doi: 10.1023/a:1010634132156. [DOI] [PubMed] [Google Scholar]
- Burg SP, Burg EA. Relationship between ethylene production and ripening in banana. Botanical Gazette. 1965;126:200–204. [Google Scholar]
- Busi MV, Bustamante C, D'Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. MADS-box genes expressed during tomato seed and fruit development. Plant Molecular Biology. 2003;52:801–815. doi: 10.1023/a:1025001402838. [DOI] [PubMed] [Google Scholar]
- Caporali E, Spada A, Losa A, Marziani G. The MADS box gene AOM1 is expressed in reproductive meristems and flowers of the dioecious species Asparagus officinalis. Sexual Plant Reproduction. 2000;13:151–156. [Google Scholar]
- Cara B, Giovannoni J. Molecular biology of ethylene during tomato fruit development and maturation. Plant Science. 2008;175:106–113. [Google Scholar]
- Clendennen SK, Kipp PB, May GD. The role of ethylene in banana fruit ripening. In: Kanelis AK, editor. Biology and biotechnology of the plant hormone ethylene. Kluwer Academic Publishers; 1997. pp. 141–148. [Google Scholar]
- Clendennen SK, May GD. Differential gene expression in ripening banana fruit. Plant Physiology. 1997;115:463–469. doi: 10.1104/pp.115.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Van de Peer Y, Theissen G. And then there were many: MADS goes genomic. Trends in Plant Science. 2003;8:475–483. doi: 10.1016/j.tplants.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, Vendrell M. Ethylene biosynthesis in banana fruit: evolution of EFE activity and ACC levels in peel and pulp during ripening. Journal of Horticultural Science. 1993;68:63–70. [Google Scholar]
- Dominguez M, Vendrell M. Effect of ethylene treatment on ethylene production, EFE activity and ACC levels in peel and pulp of banana fruit. Postharvest Biology and Technology. 1994;4:167–177. [Google Scholar]
- Drury R, Hortensteiner S, Donnison I, Bird CR, Seymour GB. Chlorophyll catabolism and gene expression in the peel of ripening banana fruits. Physiologia Plantarum. 1999;107:32–38. [Google Scholar]
- Egea Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO Journal. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X-M, Xia Y-M, Li Q-J. Isolation of two putative homologues of PISTILLATA and AGAMOUS from Alpinia oblongifolia (Zingiberaceae) and characterization of their expression. Plant Science. 2006;170:674–684. [Google Scholar]
- Garcia-Maroto F, Carmona M-J, Garrido J-A, Vilches-Ferron M, Rodriguez-Ruiz J, Lopez Alonso D. New roles for MADS-box genes in higher plants. Biologia Plantarum. 2003;46:321–330. [Google Scholar]
- Giovannoni JJ. Molecular biology of fruit maturation and ripening. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:170–180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Current Opinion in Plant Biology. 2007;10:283–289. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Golding JB, Shearer D, Wyllie SG, McGlasson WB. Application of 1-MCP and propylene to identify ethylene-dependent ripening processes in mature banana fruit. Postharvest Biology and Technology. 1998;14:87–98. [Google Scholar]
- Hoeberichts FA, Van Der Plas LHW, Woltering EJ. Ethylene perception is required for the expression of tomato ripening-related genes and associated physiological changes even at advanced stages of ripening. Postharvest Biology and Technology. 2002;26:125–133. [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Immink RG, Ferrario S, Busscher Lange J, Kooiker M, Busscher M, Angenent GC. Analysis of the petunia MADS-box transcription factor family. Molecular Genetics and Genomics. 2003;268:598–606. doi: 10.1007/s00438-002-0781-3. [DOI] [PubMed] [Google Scholar]
- Inaba A, Liu X, Yokotani N, Yamane M, Lu W-J, Nakano R, Kubo Y. Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. Journal of Experimental Botany. 2007;58:1047–1057. doi: 10.1093/jxb/erl265. [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAGAMOUS-LIKE1 is a component of the fruit ripening regulatory network. The Plant Journal. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ito H, Shiina T, Nakamura N, Inakuma T, Kasumi T, Ishiguro Y, Yabe K, Ito Y. Characterization of tomato fruit ripening and analysis of gene expression in F1 hybrids of the ripening inhibitor (rin) mutant. Physiologia Plantarum. 2005;123:331–338. [Google Scholar]
- Lelievre JM, Latche A, Jones B, Bouzayen M, Pech JC. Ethylene and fruit ripening. Physiologia Plantarum. 1997;101:727–739. [Google Scholar]
- Lincoln JE, Fischer RL. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiology. 1988;88:370–374. doi: 10.1104/pp.88.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu B, Hu L, Li M, Su W, Wu J, Yang J, Jin Z. Involvement of a banana MADS-box transcription factor gene in ethylene-induced fruit ripening. Plant Cell Reports. 2009;28:103–111. doi: 10.1007/s00299-008-0613-y. [DOI] [PubMed] [Google Scholar]
- Liu P, Goh C, Loh C, Pua E, Liu P, Goh CJ, Loh CS, Pua EC. Differential expression and characterization of three metallothionein-like genes in Cavendish banana (Musa acuminata) Physiologia Plantarum. 2002;114:241–250. doi: 10.1034/j.1399-3054.2002.1140210.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Shiomi S, Nakatsuka A, Kubo Y, Nakamura R, Inaba A, Liu XJ. Characterization of ethylene biosynthesis associated with ripening in banana fruit. Plant Physiology. 1999;121:1257–1265. doi: 10.1104/pp.121.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proceedings of the National Academy of Sciences, USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends in Plant Science. 2005;10:427–435. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Mbeguié-A-Mbeguié D, Hubertb O, Fils-Lycaonc B, Chilletd M, Baurense F-C. EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande Nain) Physiologia Plantarum. 2008;133:435–448. doi: 10.1111/j.1399-3054.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL. Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature. 1972;237:235–238. doi: 10.1038/237235a0. [DOI] [PubMed] [Google Scholar]
- Medina-Suarez R, Manning K, Fletcher J, Aked J, Bird CR, Seymour GB. Gene expression in the pulp of ripening banana. Plant Physiology. 1997;115:453–461. doi: 10.1104/pp.115.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Asif MH, Dhawan P, Srivastava MK, Pravendra N. Expression and activities of ethylene biosynthesis enzymes during ripening of banana fruits and effect of 1-MCP treatment. Plant Growth Regulation. 2003;40:11–19. [Google Scholar]
- Pech JC, Bouzayen M, Latché A. Climacteric fruit ripening: ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Science. 2008;175:114–120. [Google Scholar]
- Pelayo C, Vilas-Boas EVdB, Benichou M, Kader AA. Variability in responses of partially ripe bananas to 1-methylecyclopropene. Postharvest Biology and Technology. 2003;28:75–85. [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. The Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigola D, Pe ME, Fabrizio C, Me G, Sari Gorla M. CaMADS1, a MADS box gene expressed in the carpel of hazelnut. Plant Molecular Biology. 1998;38:1147–1160. doi: 10.1023/a:1006022524708. [DOI] [PubMed] [Google Scholar]
- Seymour G, Poole M, Manning K, King JG. Genetics and epigenetics of fruit development and ripening. Current Opinion in Plant Biology. 2008;11:58–63. doi: 10.1016/j.pbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Skipper M, Johansen LB, Pedersen KB, Frederiksen S, Johansen BB. Cloning and transcription analysis of an AGAMOUS- and SEEDSTICK ortholog in the orchid Dendrobium thyrsiflorum (Reichb. f.) Gene. 2006;366:266. doi: 10.1016/j.gene.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Sung SK, Moon YH, Chung JE, Lee SY, Park HG, An G. Characterization of MADS box genes from hot pepper. Molecules and Cells. 2001;11:352–359. [PubMed] [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, Caporali E, Colombo L, Rotino GL, Trainotti L, Casadoro G. A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. Journal of Experimental Botany. 2009;60:651–661. doi: 10.1093/jxb/ern313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter KU, Saedler H. A short history of MADS-box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Theologis A. One rotten apple spoils the whole bushel: the role of ethylene in fruit ripening. Cell. 1992;70:181–184. doi: 10.1016/0092-8674(92)90093-r. [DOI] [PubMed] [Google Scholar]
- Tzeng T-Y, Hsiao C-C, Chi P-J, Yang C-H. Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiology. 2003;133:1091–1101. doi: 10.1104/pp.103.026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Matas Arroyo AJ, McQuinn R, Chung M, Poolen M, Rose J, Seymour G, Grandillo S, Giovannoni J, Irish VF. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. The Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Do Y, Huang P, Wu HT, Do YY, Huang PL. Plant gene register PGR 99-016. Nucleotide sequence of a cDNA encoding ethylene receptor from banana fruits (Accession no. AF113748) Plant Physiology. 1999;119:805–806. [Google Scholar]
- Yamane M, Abe D, Yasui S, Yokotani N, Kimata W, Ushijima K, Nakano R, Kubo Y, Inaba A. Differential expression of ethylene biosynthetic genes in climacteric and non-climacteric Chinese pear fruit. Postharvest Biology and Technology. 2007;44:220–227. [Google Scholar]
- Yao J, Dong Y, Morris BA. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proceedings of the National Academy of Sciences, USA. 2001;98:1306–1311. doi: 10.1073/pnas.031502498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Kvarnheden A, Morris B. Seven MADS-box genes in apple are expressed in different parts of the fruit. Journal of the American Society of Horicultural Science. 1999;124:8–13. [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE. dePamphilis CW, Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M-J, Jiang Y-M, Jiang W-B, Liu X-J. Regulation of ethylene synthesis by 1-methylcyclopropene. Food Technology and Biotechnology. 2006;44:111–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.