Abstract
Members of the ALMT gene family contribute to the Al3+ resistance of several plant species by facilitating malate efflux from root cells. The first member of this family to be cloned and characterized, TaALMT1, is responsible for most of the natural variation of Al3+ resistance in wheat. The current study describes the isolation and characterization of HvALMT1, the barley gene with the greatest sequence similarity to TaALMT1. HvALMT1 is located on chromosome 2H which has not been associated with Al3+ resistance in barley. The relatively low levels of HvALMT1 expression detected in root and shoot tissues were independent of external aluminium or phosphorus supply. Transgenic barley plants transformed with the HvALMT1 promoter fused to the green fluorescent protein (GFP) indicated that expression of HvALMT1 was relatively high in stomatal guard cells and in root tissues containing expanding cells. GFP fused to the C-terminus of the full HvALMT1 protein localized to the plasma membrane and motile vesicles within the cytoplasm. HvALMT1 conferred both inward and outward currents when expressed in Xenopus laevis oocytes that were bathed in a range of anions including malate. Both malate uptake and efflux were confirmed in oocyte assays using [14C]malate as a radiotracer. It is suggested that HvALMT1 functions as an anion channel to facilitate organic anion transport in stomatal function and expanding cells.
Keywords: Channel, HvALMT1, malate, oocytes, organic anion, stomata
Introduction
The ALMT family of membrane proteins possess 5–7 predicted transmembrane domains and the UPF0005 domain of unknown function (Delhaize et al., 2007b). Of the seven ALMT genes that have been investigated to date, most are implicated in Al3+ resistance mechanisms, with TaALMT1 from wheat (Triticum aestivum) being the most thoroughly studied. TaALMT1 is expressed constitutively within root apices and maps to chromosome 4DL in the region of a major quantitative trait locus (QTL) for Al3+ resistance (Sasaki et al., 2004; Raman et al., 2005). Variation in Al3+ resistance among many wheat genotypes is positively correlated with TaALMT1 expression (Raman et al., 2005; Sasaki et al., 2006). The membrane topology of TaALMT1 determined experimentally identifies six transmembrane domains in the N-terminal half of the protein, with both termini located on the extracellular side of the membrane (Motoda et al., 2007). Experimental evidence supports the hypothesis that TaALMT1 is an anion channel located on the plasma membrane that, upon exposure to Al3+, is activated to release malate from the cytoplasm of root cells (Sasaki et al., 2004; Yamaguchi et al., 2005; Zhang et al., 2008). This malate binds Al3+ in the apoplasm to form a non-toxic complex (Delhaize et al., 1993; Ryan et al., 1995). When expressed in Xenopus laevis oocytes TaALMT1 is permeable to malate but not citrate, and shows a degree of specificity for activation by Al3+ (Sasaki et al., 2004). In a more recent study, Pineros et al. (2008a) showed that TaALMT1 expressed in Xenopus oocytes is capable of facilitating anion influx and is permeable to inorganic anions such as nitrate and chloride when external anion concentrations are high. Although TaALMT1 activity was enhanced by Al3+, the protein was still functionally active, albeit at a lower level, in the absence of Al3+ and this was also observed when the gene was expressed in tobacco suspension cells (Zhang et al., 2008). Recently, Ligaba et al. (2009) provided evidence that TaALMT1 activity is regulated by protein kinase C-mediated phosphorylation. They also established that TaALMT1 activity was disrupted when the serine residue at position 384 was replaced with an alanine, and concluded that the serine residue needed to be phosphorylated before Al3+ can activate TaALMT1.
In contrast to TaALMT1, transcript levels of other ALMT1 genes implicated in the Al3+ resistance of other species are increased when roots are exposed to Al3+ (Sasaki et al., 2004; Hoekenga et al., 2006; Ligaba et al., 2006; Fontecha et al., 2007; Collins et al., 2008). Of these, AtALMT1 from Arabidopsis thaliana (Hoekenga et al., 2006), and BnALMT1 and BnALMT2 from rape (Brassica napus; Ligaba et al., 2006) confer Al3+-activated malate efflux when expressed in Xenopus oocytes. In addition to a role in Al3+ resistance, AtALMT1 has recently been implicated in the recruitment of beneficial bacteria to the rhizosphere of Arabidopsis after malate efflux from roots was triggered by foliar infection with Pseudomonas syringae (Rudrappa et al., 2008).
The remaining two ALMT genes characterized to date from maize (Zea mays) and Arabidopsis have different functions and do not appear to contribute to Al3+ resistance. The ZmALMT1 protein of maize is located on the plasma membrane of cells at the root apices of maize. Although ZmALMT1 expression is induced by Al3+ and the protein is weakly activated by Al3+, Pineros et al. (2008b) proposed a role in mineral nutrition because the protein is more permeable to inorganic anions such as sulphate and nitrate than to malate or citrate. Another ALMT gene from Arabidopsis (AtALMT9) is expressed in hypocotyls and leaves. The protein is located at the tonoplast and transports malate out of the cytoplasm and into the vacuole (Kovermann et al., 2007).
This study describes the isolation and characteriztion of an ALMT gene from barley (Hordeum vulgare) named HvALMT1. Although HvALMT1 shares some features with ALMT1 genes implicated in Al3+ resistance, its subcellular location, expression pattern, and chromosomal location indicate roles distinct from Al3+ resistance.
Materials and methods
Plant cultivars and growth conditions
Seeds of barley cultivars Dayton, Golden Promise, Morex, and Zhepi 2 were surface-sterilized in ∼1% (w/v) sodium hypochlorite for 20 min, rinsed thoroughly in water, and pre-germinated in the dark on moist filter paper at room temperature. After germination, seedlings were grown either in pots containing compost mix or in hydroponics at pH 4.3 (Delhaize et al., 2004). When Al3+ was added to the hydroponic solution it was included as 2.5 μM AlCl3·6H2O. For growth in the presence or absence of P, either 100 μM KH2PO4 or no added KH2PO4 were used. To avoid reduced Fe availability which may result from the higher P concentration, the 2 μM FeCl3 was replaced with 20 μM Fe:EDTA in experiments that used 100 μM KH2PO4.
Isolation and cloning of HvALMT1
HvALMT1 was isolated by probing a bacterial artificial chromosome (BAC) library (HV_MBa; Clemson University Genomics Institute; Yu et al., 2000) with the full-length TaALMT1-1 coding region (GenBank accession number AB081803). Hybridization was undertaken for 2 d at 65 °C in a solution of 6× SSC (900 mM NaCl, 90 mM sodium citrate), 50 mM TRIS (pH 8.0), 10 mM EDTA, 5× Denhardt's reagent, 0.2% SDS, and 10% dextran sulphate. The membrane was washed twice with 2× SSC (300 mM NaCl, 30 mM sodium citrate) and 0.1% SDS at 65 °C. The positive BAC clone with the greatest signal intensity (0428J08) was digested with HindIII, and the fragments were subcloned into pBluescript® (Stratagene) and maintained in Escherichia coli (strain XL1 Blue; Stratagene). The nucleotide sequences of TaALMT1-1 and rice OsALMT5 (GenBank accession number AL606598) were aligned and the primers Hv1-1 and Hv1-2 (Supplementary Table S1 available at JXB online) designed from regions of identity. These primers and the plasmid sequencing primers M13F and M13R (Supplementary Table S1; New England Biolabs) were used to obtain the HvALMT1 genomic and coding sequence from subclones.
Root tips were harvested from barley cultivar Dayton, homogenized in liquid nitrogen, and RNA extracted using the RNeasy® Plant Mini Kit (Qiagen) including the optional on-column DNase digestion. The cDNA was prepared as detailed by Delhaize et al. (2004), made up to a final volume of 100 μl with deionized water, and then used to obtain the full-length coding sequence by using primers Hv1-3 and Hv1-4 (Supplementary Table S1 at JXB online) in a PCR. The PCR product was subsequently cloned into pGEM® T-easy (Promega) to produce the plasmid pHvALMT1-GEM T-easy and sequenced to confirm the absence of mutations caused by PCR.
Clones corresponding to expressed sequence tags (ESTs) of barley (cv Morex) (GenBank accession numbers BI946959 and BE194233) were obtained from the Clemson University Genomics Institute (HV_SMEl 0002P22 and HV_SMeh 0084L23). The clone corresponding to EST BI946959 was sequenced using the primers M13F, M13R, and Hv1-1. The clone corresponding to EST BE194233 was amplified and sequenced with primers M13F, M13R, Hv1-2, Hv2-1, Hv2-2, Hv2-3, Hv2-4, Hv2-5, and Hv2-6 (Supplementary Table S1 at JXB online) to obtain regions outside of the EST sequence reported on the database.
Allelic variation and chromosomal location of HvALMT1
Genomic DNA was extracted using the method detailed by Sasaki et al. (2004). The following primers were used to obtain the genomic DNA sequence of HvALMT1 from each of the cultivars: Hv1-2, Hv1-3, Hv1-4, Hv1-5, Hv1-6, Hv1-7, Hv1-8, and Hv1-9 (Supplementary Table S1 at JXB online). The genomic DNA from leaves of wheat lines that contain a single pair of barley chromosomes (Islam et al., 1981) was extracted using the sodium perchlorate method detailed by Lagudah and Appels (1991) and used in a PCR to localize the gene to a barley chromosome.
Analysis of HvALMT1 expression level
Barley plants were grown in hydroponics in a growth cabinet set at 22 °C with a 16 h photoperiod at 400 μmol m−2 s−1 photon irradiance and a 16 °C night temperature. For the Al3+ treatments, roots and shoots were sampled for HvALMT1 expression after 4 d of growth. For the minus P treatment, 13-day-old seedlings were sampled to determine HvALMT1 expression 6 d after the withdrawal of P from the nutrient solution. For expression in barley spikes the floral structures including awns were harvested at various stages of development.
Tissues frozen in liquid nitrogen were homogenized, RNA purified, and cDNA prepared as detailed earlier. Expression of HvALMT1 was determined by real-time quantitative reverse transcription-PCR (qRTPCR) using the RotorGene 2000 thermal cycler (Corbett Research) and the method of Delhaize et al. (2004). The primers qRTPCR-1 and qRTPCR-2 (Supplementary Table S1 at JXB online) were used for the detection of HvALMT1. The barley gene for glyceraldehyde-3-phosphate dehydrogenase (HvGAPDH) was used as an internal reference and was amplified using the primers qRTPCR-3 and qRTPCR-4 (Supplementary Table S1; Burton et al., 2004). The products from the qRTPCR were confirmed as the correct gene by analysis of the melt curve, gel electrophoresis, and sequencing. The RotorGene 6 comparative quantitation software (Corbett Research) was used to determine HvALMT1 gene expression values relative to HvGAPDH.
Transient expression of a HvALMT1::GFP fusion
The plasmid pHvALMT1-GEM T-easy was digested with EcoRI and the fragment that contained HvALMT1 ligated into pYES2 (Invitrogen). Homologous recombination in yeast was used to produce an in-frame fusion of green fluorescent protein (GFP) with the 3′ end of the full HvALMT1 cDNA as follows: primers Hv1GFP-1 and Hv1GFP-2 (Supplementary Table S1 at JXB online) were used to amplify the GFP-coding region (sGFPS65T) from pGFP-ART7 (derived from pMNG1004; Upadhyaya et al., 1998) and to introduce a SmaI site into the construct. The 5′ end of the PCR product contained a sequence matching the 3′ end of HvALMT1 without the stop codon and in-frame with the GFP sequence. The 3′ end of the PCR product matched a region downstream of HvALMT1 in pHvALMT1-YES2. These sequences allowed the product to undergo homologous recombination when introduced in yeast, thereby inserting GFP at the 3′ end of HvALMT1. Yeast (Sacchromyces cerevesiae) was transformed according to Gietz et al. (1995), the resulting plasmid was extracted from transformed yeast (Hoffman and Winston, 1987), digested with BamHI and SmaI, and the fragment containing HvALMT1::GFP was cloned into the expression vector pUbi.tml (Wang and Waterhouse, 2000). This was digested with NotI and the fragment that contained HvALMT1::GFP was cloned into the pWBVec8 binary vector (Wang et al., 1998). The HvALMT1::GFP fragment was sequenced after cloning into pUbi.tml to confirm that the fusion was in-frame. A construct containing a constitutively expressed GFP from the ubiquitin promoter (pPSUbiGFP) was used as a control (Schunmann et al., 2004).
Transient expression in leek (Allium ampeloprasum var. porrum) was undertaken using a method modified from Delhaize et al. (2007a). Between 3 μg and 6 μg of DNA was precipitated onto the gold particles before the leek was bombarded in a vacuum of ∼85 kPa using a rupture disk pressure of 6200 kPa in the PDS-1000/He Biolistic® Particle Delivery System (BioRad). Images were captured after 6–28 h using a Leica TCS SP2 confocal laser scanning microscope and associated software.
Production of a HvALMT1 promoter::GFP fusion
The positive BAC clone used to isolate HvALMT1 (0428J08) also contained genomic sequence flanking the coding region. Initial sequencing of HvALMT1-positive subclones (derived from clone 0428J08) using the primers M13F and M13R revealed sequence upstream of the HvALMT1 coding region, enabling the design of primers Hv1GFP-3 and Hv1GFP-4 (Supplementary Table S1 at JXB online). These primers were used to amplify 2906 nucleotides upstream of the HvALMT1 ATG start site and to introduce a 5′ PacI and 3′ AscI site. The PCR was conducted using Proofstart™ DNA Polymerase (Qiagen) with the manufacturer's protocol and the product was digested with PacI and AscI. The digest was purified using the QIAquick® PCR Purification Kit (Qiagen) and cloned into the pHvPht1::GFP-Vec8 vector (Schunmann et al., 2004) from which the HvPht1 promoter had been removed by digestion with PacI and AscI. The full sequence of the cloned promoter region was obtained by sequencing with the primers Hv1GFP-5, Hv1GFP-6, Hv1GFP-7, and Hv1GFP-8 (Supplementary Table S1). The vector was then introduced into Agrobacterium tumefaciens strain AGL1 by tri-parental mating. Barley cultivar Golden Promise was transformed as detailed by Tingay et al. (1997), except that embryos were not injured by particle bombardment before being co-cultivated with the Agrobacterium. The initial transformants (T0) were planted to soil and the next generation of seedlings (T1) was used to localize GFP fluorescence.
GFP fluorescence in stomatal guard cells of transgenic plants expressing the promoter::GFP construct was quantified with a fluorescence microscope (Leica MZFLIII) and AnalySIS Five software (Olympus-Soft Imaging Systems, Germany). The green wavelengths were first separated from the images and converted into a grey scale that allowed the intensity to be measured. One end of the ‘dumbbell’ structure of a guard cell was selected with an ellipse tool, and the average grey value of this selection was then determined. An equivalent region immediately adjacent to the guard cell was taken as a ‘blank’ value which was subtracted from the raw data. The effect of abscisic acid (ABA) on GFP fluorescence was determined by spraying leaves with a 1 mM solution of ABA containing 0.05% Triton X-100 according to the method of Raskin and Ladyman (1988). Control plants were sprayed with a solution that contained 0.05% Triton X-100. Seedlings were grown in the dark for 6 d before treatments were imposed as this allowed GFP fluorescence to be quantified in the absence of strong autofluorescence from chlorophyll.
Expression in Xenopus laevis oocytes
To prepare for expression in X. laevis oocytes, pHvALMT1-YES2 was digested with BamHI and NotI, and the fragment containing HvALMT1 was cloned into the GATEWAY® entry vector pENTR11 (Invitrogen). This fragment then underwent recombination with pGEMHE:DEST using the Gateway® LR clonase II reaction (Invitrogen) as per the manufacturer's protocol.
Electrophysiology experimental conditions
The plasmid expressing HvALMT1 was linearized using NheI and used as a template for capped copy RNA (cRNA) transcription using a Message Machine® T7 kit (Ambion®). Oocytes from X. laevis were isolated and maintained using a modification of the method described by Virkki et al. (2006). Oocytes were injected with 46 nl of cRNA (0.7 μg μl−1) encoding HvALMT1 or RNase-free water using a micro-injector (Nanoject II automatic nanolitre injector, Drummond Scientific). After injection, the oocytes were incubated at 18 °C in Ca-Frog Ringer's solution which was replaced daily. Experiments were undertaken 2 d after injection of the oocyte.
Recordings were performed on both control oocytes (water injected) and oocytes expressing HvALMT1 that had been incubated in 10 mM malate–BTP (bis-TRIS propane) overnight. It was subsequently found that the currents were similar whether oocytes were pre-incubated in malate or not (data not shown).
The composition of the basic recording solution was as used by Kovermann et al. (2007) with modifications to reveal malate-dependent currents and also the selectivity or permeability of the transporter for other organic anions. These solutions consisted of 10 mM malic acid, citric acid, fumaric acid, or MES added to the basic solution containing 0.3 mM CaCl2, adjusted to a constant osmolarity of 220 mosmol kg−1 with mannitol and buffered with BTP to either pH 7.5, 5.5, or 4.5. The methods described by Sasaki et al. (2004) were used to assess whether HvALMT1 was activated by Al3+. Briefly, oocytes were injected with either HvALMT1 cRNA or H2O (control) and incubated for 48 h in ND88 buffer at pH 7.6. The oocytes were subsequently injected with 46 nl of 0.1 M Na-malate and transferred to ND88 pH 4.5 1–4 h prior to patch clamping. Bathing solutions for the experiment were adjusted to pH 4.5 and cells monitored for 6 min after addition of Al3+. Whole-cell currents were recorded under constant perfusion and temperature (22 °C) with a GeneClamp 500 amplifier (Axon Instruments) using conventional two-electrode voltage clamp techniques. From a holding potential of –40 mV the voltage was stepped between 60 mV and –140 mV in 20 mV increments. The duration of each voltage pulse was 0.5 s, with a 1.0 s resting phase between successive voltage steps. The output was digitized and analysed using a Digidata 1322A-pCLAMP 8 data acquisition system (Axon Instruments). Recording electrodes filled with 3.0 M KCl had resistances of between 0.5 MΩ and 1.2 MΩ. Data analysis was performed using the Clampfit software (version 8.2; Molecular Devices Corporation) and GraphPad software (GraphPad Software Inc.) to construct current–voltage curves.
Experimental conditions for the influx and efflux of radioactive malate
For influx experiments, the oocytes expressing HvALMT1 and control oocytes were incubated in Ca-Frog Ringer's solution containing radioactive malate (L-1,4 (2,3) [14C]malic acid; Amersham) at a final malate concentration of 2 mM. The oocytes were placed in a 96-well plate with each well containing 100 μl of solution. After 30 min, the oocytes were placed in scintillation vials and digested with 0.1 N HNO3 before being counted in a scintillation counter (Tri-Carb® 2100 TR Liquid Scintillation Analyser; Packard).
For efflux experiments, both oocytes expressing HvALMT1 and control oocytes were injected with 46 nl of radioactive malate (Amersham) and placed in 500 μl of buffer (Ca-Frog Ringer's) at pH 4.5 or 7.5. After either 2 min or 5 min the buffer was sampled and replaced with fresh buffer. At the end of the experiment, the sampled buffers and the digested oocytes were placed in scintillation vials and counted in a scintillation counter. For both experiments data were subjected to analysis of variance (ANOVA) and treatment means compared by least significant differences (LSD; P=0.05) using GenStat (11th edition; www.vsni.co.uk). Where required, non-normally distributed data were logn transformed prior to analysis.
GenBank accession numbers
Genbank accession number(s) for (i) HvALMT1 genomic sequences are FJ195909, FJ195908, FJ195911, and FJ195910; (ii) HvALMT2 partial genomic sequences are FJ200746, FJ200747, and FJ200748; (iii) the HvALMT1 promoter is FJ195911; and (iv) the HvALMT1 coding sequence is EF424084.
Results
Cloning of HvALMT1
The TaALMT1 coding region was used to probe a BAC library derived from the barley cultivar Morex. The most strongly hybridizing clone was sequenced and a putative gene 2271 bp long was identified. This sequence consisted of six exons and five introns with a coding region of 1368 bp (Supplementary Fig. S1 at JXB online). The predicted protein, named HvALMT1, consists of 455 amino acid residues with 66% identity to TaALMT1. HvALMT1 possesses features common to the ALMT1 family including a UPF0005 domain and seven transmembrane regions (predicted by SOSUI v1.11, http://bp.nuap.nagoya-u.ac.jp/sosui/; Supplementary Fig. S1).
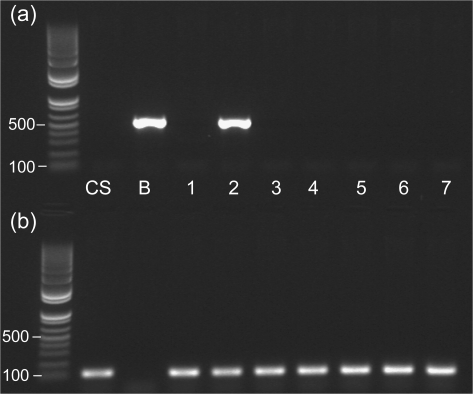
The genomic sequence of HvALMT1 in cultivars Golden Promise, Dayton, Morex, and Zhepi2 (GenBank accession numbers FJ195909, FJ195908, FJ195911, and FJ195910, respectively) revealed three alleles differing at four single nucleotide polymorphisms (SNPs; Supplementary Fig. S2 at JXB online). Only one of these SNPs alters the predicted protein sequence from an alanine at residue 431 in cultivars Dayton (Al3+-resistant), Golden Promise (Al3+-sensitive), and Zhepi2 (Al3+-sensitive) to a threonine in cultivar Morex (Al3+-sensitive). Analysis of wheat–barley addition lines established that the HvALMT1 gene was located on chromosome 2H (Fig. 1).
Fig. 1.
Chromosomal localization of HvALMT1 in wheat–barley addition lines by PCR. PCR was performed with (a) HvALMT1-specific (qRTPCR-1 and qRTPCR-2; 514 bp product) and (b) TaALMT1-specific (TaALMT1R and TaALMT1F; 107 bp product) primers on genomic DNA of wheat cultivar Chinese Spring (CS), barley cultivar Betzes (B) (both parents of the wheat–barley addition lines), and wheat–barley addition lines containing barley chromosomes 1–7. The 100 bp and 500 bp bands are indicated for the DNA size markers (leftmost lanes).
A BLAST search of the NCBI database with the HvALMT1 sequence identified two ESTs of ALMT-like genes from barley. One was derived from floral spikes of the cultivar Morex infected with Fusarium graminearum (GenBank accession number BI946959). When the clone containing this EST was sequenced it was found to be 98.8% identical to HvALMT1 over 1022 nucleotides. The small discrepancy may be due to errors in the single-pass sequence or the EST may represent another very closely related ALMT gene that is expressed in floral spikes. The second EST encoded a more distantly related ALMT (GenBank accession number BE194233). The clone containing this EST was used to obtain 1771 nucleotides of genomic DNA by sequencing amplified fragments (HvALMT2; GenBank accession numbers FJ200746, FJ200747, and FJ200748). The 1213 bp of coding sequence was found to be only 57% identical to TaALMT1 and 58% identical to HvALMT1 over the same region (data not shown). In contrast, HvALMT1 and TaALMT1 were 78% identical to one another over this same region. Since HvALMT1 showed the strongest similarity to TaALMT1, this gene was investigated further.
Expression of HvALMT1
The expression of HvALMT1 was measured in two cultivars of barley that differ in Al3+ resistance to determine if exposure to Al3+ induces gene expression as found for ALMT genes from some other species (Hoekenga et al., 2006; Ligaba et al., 2006; Fontecha et al., 2007; Collins et al., 2008). Dayton was the Al3+-resistant genotype and Golden Promise the Al3+-sensitive genotype. Plants were grown in hydroponic culture with solutions containing 2.5 μM Al3+ as this concentration differentiates the two cultivars based on root growth (data not shown). Since organic anion efflux from roots of P-deficient plants has been reported to play a role in the P nutrition of some species (Ryan et al., 2001), the response of HvALMT1 expression to P deficiency was also determined.
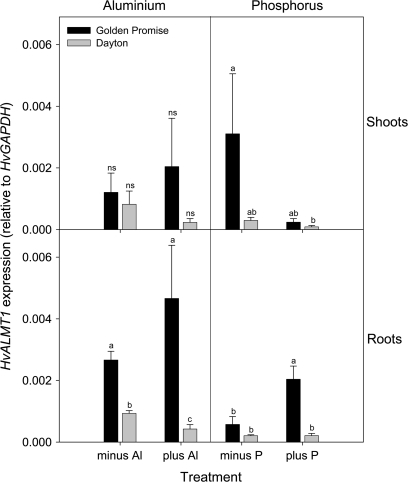
qRTPCR indicated that HvALMT1 was expressed at a low level relative to the reference gene HvGAPDH in both shoots and roots and that there were no consistent effects of either Al3+ treatment or P deficiency (Fig. 2). Expression in the Al3+-resistant cultivar Dayton was generally lower than in the sensitive cultivar Golden Promise. Since an EST on the NCBI database from floral spikes may represent HvALMT1, expression was also examined in this tissue. A pooled sample of floral spikes collected near anthesis (5 d before and after anthesis) was compared with a sample collected 10 to 31 d post-anthesis. Using HvGAPDH as the internal reference gene, expression of HvALMT1 increased from 0.0045±0.0008 (n=3) around anthesis to 0.016±0.007 (n=3) for samples collected after anthesis.
Fig. 2.
The effect of Al3+ exposure or P deficiency on HvALMT1 expression in shoots and roots of barley. HvALMT1 expression was determined using qRTPCR in plants of an Al3+-sensitive (Golden Promise) or Al3+-resistant (Dayton) cultivar grown for 4 d in hydroponics containing no added Al3+ (minus Al) or 2.5 μM Al3+ (plus Al); or for the P experiment in 13-day-old plants after 6 d of growth in hydroponics containing no added P (minus P) or 100 μM P (plus P). The expression level is shown relative to the internal reference gene GAPDH (mean ± SEM; n=3 or 4). For each panel, columns designated with different letters are significantly different (P <0.05; ns=not significant).
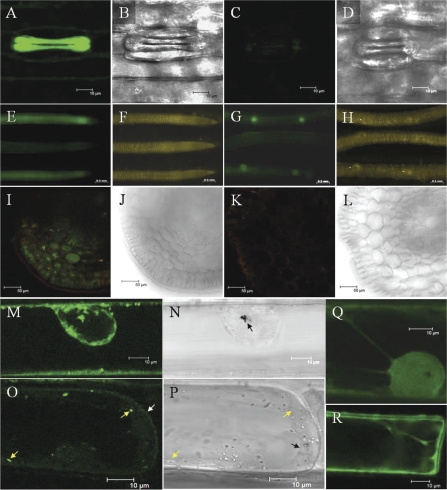
To determine the tissue specificity of HvALMT1 expression, a promoter region consisting of 2906 nucleotides upstream of the HvALMT1 start site (referred to as the HvALMT1 promoter; GenBank accession number FJ195911) was cloned, fused to the GFP reporter gene, and used to generate transgenic barley. In the leaves of dark-grown plants strong GFP fluorescence was observed specifically in the guard cells of stomata but not in adjacent subsidiary or epidermal cells (Fig. 3A). GFP fluorescence could not be reliably detected in other cells within the leaves of light-grown or dark-grown plants above the background derived from the autofluorescence of chlorophyll and its precursors. Fluorescence was observed in roots of barley grown in hydroponics, particularly in a region behind the root tip, in the buds of emerging lateral roots, and in newly emerged lateral roots (Fig. 3E, G). A transverse section through the root showed fluorescence in all cell layers (Fig. 3I). The high background autofluorescence made it difficult to detect GFP fluorescence reliably in the floral spikes. To establish whether ABA regulated HvALMT1 expression, leaves were sprayed with an aqueous solution that contained ABA, and GFP fluorescence was quantified in guard cells. The intensity of GFP fluorescence in stomatal guard cells of ABA-treated leaves incubated over 3 d did not differ from that of control leaves (Supplementary Fig. S3 at JXB online), indicating that ABA was unlikely to regulate HvALMT1 expression directly.
Fig. 3.
Localization of GFP fluorescence in plants transformed with either a HvALMT1 promoter::GFP or a HvALMT1 protein::GFP reporter fusion. (A and B) The stomata of barley transformed with a HvALMT1 promoter::GFP fusion. (C and D) The stomata of wild-type barley. In E–H the upper and lower roots are of two lines of barley independently transformed with the HvALMT1 promoter::GFP fusion while the middle root is that of wild-type barley. (I and J) A transverse section through the root of barley transformed with the HvALMT1 promoter::GFP fusion. (K and L) A transverse section through the root of wild-type barley. I and K used the same image acquisition and manipulation settings. (M and O) The subcellular localization of fluorescence in leek epidermal cells transiently expressing a fusion of GFP with the C-terminus of HvALMT1. (N and P) The corresponding brightfield images. GFP fluorescence from the control (pPSUbiGFP) is shown in Q and R. The nucleus is visible within M, N, and Q. The gold particles used for the transformation of cells are marked in N by the arrow. Within O the position of the plasma membrane is marked by the white arrow and the position of two vesicles with GFP fluorescence is marked by the yellow arrows. In P the positions of the same two vesicles are marked with yellow arrows and the tonoplast is marked by the black arrow.
HvALMT1 localizes to the plasma membrane and internal vesicles
The GFP reporter molecule was fused to the C-terminus of the full-length HvALMT1 protein (HvALMT1::GFP) to enable the subcellular localization of HvALMT1 to be determined from transient expression in leek. The control included in these experiments was GFP under the control of the ubiquitin promoter. Fluorescence from the HvALMT1::GFP fusion was clearly observed in small particles or vesicles in the cytoplasm of ∼0.8–1.8 μm in diameter that moved with the streaming cytoplasm (Fig. 3M, O, and Supplementary GFP movies at JXB online). A number of these vesicles were commonly seen to congregate around the nucleus (Fig. 3M). In areas where the vacuole did not force the cytoplasm up against the periphery of the cell, a faint fluorescent signal was also detected at the plasma membrane (Fig. 3O). The nucleus did not fluoresce and the localization differed from that of the GFP control which fluoresced strongly in the nucleus (Fig. 3Q, R). Fluorescence was not observed at the tonoplast or on many other vesicles observed in the cytoplasm. The pattern of fluorescence described was observed in a range of cells with various levels of fluorescence intensity, indicating that it was not an artefact due to high level expression of the transgene.
Electrophysiology of HvALMT1 in Xenopus laevis oocytes
The transport properties of HvALMT1 were examined by expressing it in Xenopus oocytes. By convention, inward currents are negative currents representing cation influx or anion efflux, and outward currents are positive currents representing cation efflux or anion influx. Considering the components of the bathing solution used in these experiments, an inward current could be mediated by the influx of Ca2+ or the efflux of anions from within the oocyte. Similarly, an outward current could be mediated by the efflux of cations from the oocyte or by the uptake of anions from the bathing solution.
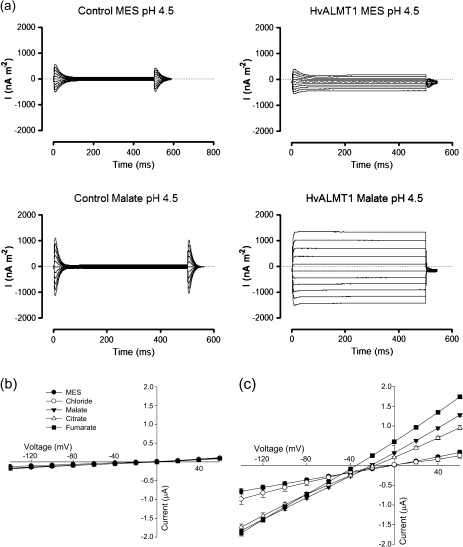
Expression of HvALMT1 in oocytes conferred an increase in both inward and outward currents not observed in the water-injected control oocytes (Fig. 4a). The currents activated rapidly in response to the voltage changes and remained relatively stable during the 500 ms pulses. The HvALMT1-dependent increase in current density was small with MES in the bathing solution but significantly larger when MES was replaced by malate. Current–voltage curves for these two anions as well as for chloride, fumarate, and citrate are shown in Fig. 4b and c. The magnitudes of the inward and outward currents were between 2- and 5-fold greater with malate, fumarate, or citrate in the bathing solution compared with chloride or MES (Fig. 4). Reversal potentials were approximately −20 mV with malate and citrate in the bathing solution and −35 mV with fumarate. In contrast, reversal potentials near zero were measured for control oocytes and for oocytes expressing HvALMT1 and bathed in chloride or MES. The equilibrium potential for Ca2+ is very positive in these solutions so the negative reversal potentials measured in oocytes expressing HvALMT1 indicate that Ca2+ fluxes did not contribute significantly to the currents.
Fig. 4.
Electrophysiology of Xenopus oocytes expressing HvALMT1. (a) Family of current curves measured in Xenopus laevis oocytes injected with water (control) or HvALMT1 cRNA in response to 500 ms voltage pulses between –140 mV and 60 mV in 20 mV steps. The data were collected with either 10 mM MES–BTP or 10 mM malate–BTP in the bathing solution (pH 4.5). Current–voltage curves generated from the currents measured in control oocytes (b) or HvALMT1-expressing oocytes (c) with different anions in the bathing solution. Currents were recorded from oocytes incubated in MES–BTP (MES), chloride–BTP (Chloride), malate–BTP (Malate), citrate–BTP (Citrate), or fumarate–BTP (Fumarate) at pH 4.5. Error bars show the SEM, n=6.
The activity of several other members of the ALMT family is increased several-fold by exposure to Al3+. Tests were carried out to determine whether oocytes expressing HvALMT1 responded to Al3+ treatment in a similar way. When Al3+ was added to the bathing solution (pH 4.5) a relatively small increase of ∼40% at –140 mV was observed in the inward current which was absent from cells treated with La3+ (Supplementary Fig. S4 at JXB online). These increases were observed after 30 s of Al3+ treatment, and no further increases occurred over the following 6 min that the cells were monitored (data not shown).
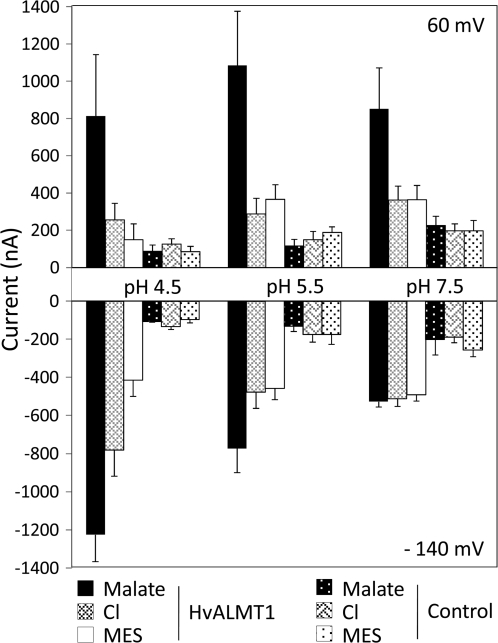
To examine the dependency of these currents on external pH (pHext), currents were measured in control oocytes and HvALMT1-expressing oocytes at pH 4.5, 5.5, and 7.5 (Fig. 5; Supplementary Fig. 5 at JXB online). With malate in the bathing solution, the magnitude of inward current in oocytes expressing HvALMT1 decreased at higher pHext from approximately –1200 nA at pHext4.5 to approximately –500 nA at pHext7.5 and a holding voltage of –140 mV (Fig. 5). The inward currents measured with chloride in the bathing solution also decreased at higher pH but only between pHext 4.5 and 5.5, whereas the currents with MES in the bathing solution were independent of pHext. The magnitudes of outward currents in oocytes bathed in malate, chloride, or MES were less dependent on pHext but they were dependent on the anion in the bathing solution such that fumarate>malate>citrate>Cl–=MES (Fig. 4c).
Fig. 5.
Effect of pH on currents in Xenopus oocytes expressing HvALMT1. Xenopus laevis oocytes injected with HvALMT1 cRNA or water as a control were incubated overnight in 10 mM malate–BTP. The currents across the oocyte membrane were recorded using the two-electrode voltage clamp technique, while the oocytes were bathed in either 10 mM MES–BTP (MES), 10 mM chloride–BTP (Cl), or 10 mM malate–BTP (malate) at external pH 4.5, 5.5, or 7.5. Shown is the current across the membrane of oocytes clamped at 60 mV and –140 mV. Error bars show the SEM (n=3).
[14C]Malate transport by oocytes expressing HvALMT1
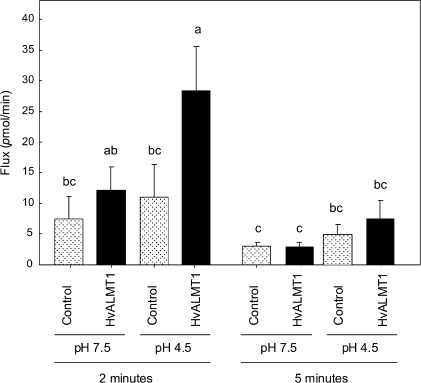
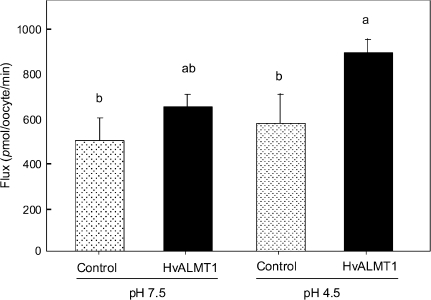
Malate efflux from oocytes was also monitored after they were injected with [14C]malate. In these experiments the malate efflux after 2 min was ∼2.5-fold greater in oocytes expressing HvALMT1 (pHext 4.5) than in control oocytes injected with water (Fig. 6). This difference was not apparent 5 min after injection or at time points up to 30 min later (data not shown for later time points). Furthermore, the efflux was not significantly different from that of control oocytes when the external pH was 7.5 rather than 4.5, which is consistent with the electrophysiological data. Malate uptake by the oocytes was monitored by bathing the oocytes in [14C]malate and measuring the radioactivity taken up. Although the difference was smaller than observed for efflux, the rate of malate uptake was significantly greater in oocytes expressing HvALMT1 than in oocytes injected with H2O at pHext 4.5, but not at pHext 7.5 (Fig. 7).
Fig. 6.
Efflux of radioactively labelled malate from Xenopus oocytes expressing HvALMT1. Oocytes were pre-loaded with [14C]malate and the radioactivity of the bathing solution was measured. Oocytes were injected with either HvALMT1 cRNA or water as a control and incubated at external pH 7.5 or pH 4.5. Shown is the mean efflux of radioactive malate from the oocyte 2 min and 5 min after injection. Error bars show the SEM (n=5), and columns with different letters are significantly different (P <0.05; n=5).
Fig. 7.
Uptake of radioactively labelled malate by Xenopus oocytes. Oocytes injected with HvALMT1 cRNA or water as a control were incubated in [14C]malate at external pH 7.5 or pH 4.5 for 30 min and then digested in acid to determine the rate of malate uptake. Error bars show the SEM (n=5), and columns with different letters are significantly different (P <0.05; n=5).
Discussion
In this work HvALMT1 from barley, which was identified as the closest homologue to the well characterized TaALMT1 gene from wheat, was cloned and characterized. Despite its strong similarity it is evident that HvALMT1 functions differently from TaALMT1. Xenopus oocytes expressing HvALMT1 induced larger inward and outward currents than water-injected controls when bathed in malate, fumarate, or citrate. The ability of HvALMT1 to transport malate in oocytes was verified by radiotracer experiments that showed greater efflux and uptake of [14C]malate compared with controls. These data support the hypothesis that HvALMT1 is a transport protein that facilitates the movement of malate and other ions across oocyte membranes. Pineros et al. (2008) similarly found that expression of TaALMT1 in oocytes conferred inward and outward currents of a similar magnitude to those described here for HvALMT1. Interestingly, Kovermann et al. (2007) found fumarate- and malate-dependent inward currents in vacuoles isolated from tobacco leaves heterologously expressing AtALMT9. Pineros et al. (2008b) found that oocytes expressing ZmALMT1 showed citrate-dependent currents although ZmALMT1 appeared to have low transport capacity for organic anions compared with inorganic anions. HvALMT1 was weakly activated by Al3+, showing a small increase in inward current of a similar magnitude to that observed for ZmALMT1 (Pineros et al., 2008b), which contrasts with the much larger activation observed for TaALMT1 (Sasaki et al., 2004; Pineros et al., 2008a). In view of this weak activation it is concluded that HvALMT1 is likely to function in the plant independently of Al3+. Seven members of the ALMT family have now been functionally characterized in oocytes, with four strongly activated by Al3+ and three, including HvALMT1, showing little or no activation by Al3+. To identify amino acid residues that potentially interact with Al3+ to activate the proteins, all seven sequences were aligned, and amino acids shared by the Al3+-activated proteins but absent from the others were looked for. Although no identical amino acids were apparent, seven amino acids with strong similarity to one another were present only in the Al3+-activated proteins (Supplementary Fig. S6 at JXB online). These residues are located in the C-terminus hydrophilic region thought to be located in the extracellular region (Motada et al. 2007). Interestingly, one of these amino acids is a serine located at position 384 of TaALMT1, a residue required for activity and that is potentially phosphorylated (Ligaba, 2009). However, the function of this serine appears not to be conserved since the B. napus proteins possess alanine at the equivalent position yet are still functional and Al3+ activated (Ligaba et al., 2006).
The outward currents from oocytes expressing HvALMT1 were independent of external pH, while the inward current measured with malate in the bathing solution increased with decreasing pH. The efflux of [14C]malate from oocytes appeared to be similarly regulated by pH since [14C]malate efflux was greater at pH 4.5 than at pH 7.5. The relative current magnitude cannot be directly compared with the [14C]malate fluxes because the currents were measured while membrane potentials were clamped at each voltage, whereas for the [14C]malate measurements the voltage was not clamped. Furthermore 14C measurements will also include any non-electrogenic fluxes occurring. The efflux of [14C]malate decreased after 2 min, suggesting either that internal malate was depleted or that transport through HvALMT1 was rapidly stalled for some other reason such as by the flow of counter ions. When malate, citrate, or fumarate were added to the external solution at pH 4.5 the magnitude of the inward current (and hence efflux of an endogenous anion) increased when compared with Cl– and MES as well as control oocytes bathed in the same solutions. This suggests that HvALMT1 transport activity is enhanced by external organic anion supply, possibly providing a positive feedback mechanism where organic anion (e.g. malate) efflux by HvALMT1 promotes a sustained efflux of organic anions. These results indicate that HvALMT1 functions as a transport protein, but more work is required to show how the protein is regulated and to determine whether these findings reflect the function of HvALMT1 in plants. Specifically, it will be important to ascertain whether the protein is regulated by the external pH and if high concentrations of external anions enhance efflux.
The strong sequence similarity between HvALMT1 and the anion channel TaALMT1 suggests that HvALMT1 also functions as an anion channel selective for organic anions. A proportion of the HvALMT1::GFP fusion protein localized to the plasma membrane, which would lead to the efflux of organic anions from the cell. HvALMT1::GFP was also located on motile vesicles which may represent the endomembrane system that delivers the protein to the plasma membrane. Therefore, the localization of HvALMT1 to the vesicles may function to load such vesicles with organic anions. The vesicles could then be transferred to other internal organelles or their contents released to the apoplasm by exocytosis if they fuse with the plasma membrane. A similar cellular localization pattern between the plasma membrane and motile vesicles was reported for a K+ transporter (KAT1)::GFP fusion in the guard cells of faba bean (Vicia faba; Hurst et al., 2004). ABA triggers the retrieval of the KAT1 proteins from the plasma membrane via the movement of vesicles and the transporters are returned to the plasma membrane after ABA is withdrawn (Sutter et al., 2007). The dual subcellular localization of KAT1 represents a mechanism whereby the KAT1 protein is transported to the plasma membrane via exocytotic vesicles during the period of guard cell expansion (stomatal opening). The model suggested that during stomatal closure (guard cell contraction), transporter-rich areas of the plasma membrane are recovered by endocytotic vesicles, providing a mechanism for changing the area of plasma membrane surrounding the guard cell and the activity (number) of transporters within the membrane during expansion and contraction (Hurst et al., 2004; Meckel et al., 2004; Sutter et al., 2007).
The HvALMT1 promoter::GFP fusion fluoresced strongly in guard cells of stomata, suggesting that HvALMT1 is involved in stomatal function, perhaps even regulating stomatal aperture. Anion channels are essential for regulating turgor pressure, and malate transport is known to be pivotal in this regulation (Roelfsema and Hedrich, 2005). HvALMT1 may be one of the pathways for malate efflux from guard cells either by functioning as an anion channel at the plasma membrane or via exocytosis by first loading vesicles with organic anions. The density of anion channels in the plasma membrane may even be regulated by a similar mechanism to that of the KAT1 transporters discussed above, with HvALMT1 being similarly transported to, and retrieved from, the plasma membrane via motile vesicles.
Patch clamping of guard cell protoplasts from faba bean has identified two anion conductances that are involved in the control of the stomatal aperture: a rapid (R-type) conductance and a slow (S-type) conductance (Schroeder and Keller, 1992). The R-type channel activates rapidly and is voltage dependent such that conductance increases at membrane potentials more positive than –80 mV. In contrast, the S-type channel is active over a wider range of membrane potentials including those more negative than –80 mV. The conductance of HvALMT1 is unknown, and nor is it clear how rapidly the protein is activated, but the current–voltage curves generated by expression in oocytes show some similarity with the S-type channel.
For the guard cell anion channel GCAC1 (characterized from faba bean) the addition of malate to the external medium shifts the membrane potential at which maximum current occurs towards the resting potential of the cell (Hedrich et al., 1994). Addition of malate also increases the magnitude of the inward current (Lohse and Hedrich, 1995), indicative of a greater efflux of malate from the cell. Similarly, the addition of organic anions to the bathing solution of oocytes expressing HvALMT1 increased the current density at positive and negative membrane potentials, and shifted the reversal potential more negatively. GCAC1 activity is also affected by external pH. A decrease in the external pH decreases the inactivation of the conductance in response to membrane depolarization and also decreases the activation in response to membrane repolarization (Schulz-Lessdorf et al., 1996). This change is probably related more to the pH gradient across the plasma membrane than the apoplasmic pH per se. The present data demonstrate that conductance of HvALMT1 also increases at low external pH. In Arabidopsis the SLAC1 gene is highly expressed in guard cells of stomata and has a role in the function of the S-type channel function (Negi et al., 2008; Vahisalu et al., 2008). Although direct evidence of transport activity was not demonstrated in that study, the disruption of organic and inorganic anions in protoplasts of guard cells isolated from slac1 mutants was indicative that SLAC1 is important in anion homeostasis and hence stomatal function. More recently, two studies have identified SLAC1 as an anion channel that is regulated by ABA in a signal transduction pathway involving a kinase–phosphatase pair (Geiger et al., 2009; Lee et al., 2009). Although ABA regulates SLAC1 activity, the Genevestigator database indicates that ABA does not induce SLAC1 expression (Zimmermann et al., 2004). Similarly, the present study showed that HvALMT1 expression was not induced by ABA and it is more likely that if HvALMT1 activity is regulated by ABA then it will be via a similar signal transduction pathway. Interestingly, although SLAC1 was permeable to a range of anions, it had relatively low permeability towards malate (Geiger et al., 2009; Lee et al., 2009) and it is possible that malate efflux from guard cells is conferred by ALMT proteins acting in parallel to SLAC1.
In addition to the guard cells, HvALMT1 is expressed in the root elongation zone, lateral roots, and floral spikes, indicating that the protein might facilitate organic anion transport within these tissues perhaps to regulate turgor. Both processes are important in growing and elongating cells due to their high metabolic activity.
Several ALMT-like genes are present in the barley genome, with HvALMT1 appearing to be the most similar to the wheat Al3+-resistance gene TaALMT1. The analysis of wheat–barley addition lines showed that HvALMT1 localized to chromosome 2H and not to chromosome 4H to where a major Al3+-resistance locus has been mapped (Ma et al., 2004; Wang et al., 2007). Furthermore, the expression of HvALMT1 was not induced by Al3+ treatment nor was it expressed at a higher level in the Al3+-resistant cultivar Dayton. Taken together these data confirm that, despite its similarity to TaALMT1, HvALMT1 is not the major determinant of Al3+ resistance in barley. Instead it is now apparent that a gene located on chromosome 4H and encoding a member of the unrelated MATE family of proteins controls Al3+ resistance in barley by facilitating citrate efflux from roots (Furukawa et al., 2007; Wang et al., 2007). Fontecha et al. (2007) identified a partial (524 nucleotide) ALMT sequence generated by PCR from barley that appeared to localize to chromosome 4H. Since this sequence is identical to a PCR-amplified sequence from Phaseolus vulgaris, it is likely that it was generated from contaminated DNA. More recently, Collins et al. (2008) probed wheat–barley addition lines with ScALMT1 from rye (Secale cereale; >90% nucleotide sequence identity to TaALMT1) using Southern blots and only identified an ALMT gene that was located on chromosome 2HL, consistent with the present result. Indeed, when primers specific to TaALMT1 were used in PCR, products were amplified from samples that contained wheat DNA but not barley DNA (Fig. 1 and data not shown). TaALMT1 is located on chromosome 4D in wheat, and the absence of a barley ALMT gene on chromosome 4H with greater sequence similarity to TaALMT1 than HvALMT1 (75% identical nucleotide sequence) is puzzling. It is possible that HvALMT1 was originally located on chromosome 4H in the common ancestor of wheat and barley but was then translocated to chromosome 2H in barley subsequent to the divergence of the species. Alternatively, it is possible that a TaALMT1 orthologue present in the common ancestor of wheat and barley was lost in the barley lineage and that HvALMT1 represents a related but distinct homologue. The second alternative is more likely given the presence of AetALMT2 (GenBank accession EF424085) in Aegilops tauschii (the D-genome donor of bread wheat) which has 95% nucleotide sequence identity to HvALMT1 and two wheat ESTs (GenBank accessions BF483400 and BQ170664) with identical sequence to AetALMT1 that map to chromosome 2B (Graingenes 2.0 database; http://wheat.pw.usda.gov/) the homeologous chromosome to 2H in barley. Therefore, it is likely that AetALMT2 and other highly similar sequences in hexaploid wheat (such as those represented by the ESTs) are genes orthologous to HvALMT1 of barley.
In summary, it has been shown that HvALMT1 from barley is capable of facilitating the transport of malate and probably other anions. The gene does not confer the major Al3+ resistance of barley but its high expression in guard cells and roots suggests a role in anion transport and regulation in these tissues. Further work will study the details of HvALMT1 function in planta.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The genomic structure and hydropathy plot of HvALMT1.
Figure S2. Single nucleotide polymorphisms (SNPs) in HvALMT1 between four barley cultivars.
Figure S3. Effect of ABA treatment on fluorescence of guard cells expressing GFP driven by the HvALMT1 promoter.
Figure S4. Effect of Al3+ on currents in Xenopus oocytes expressing HvALMT1.
Figure S5. HvALMT1-dependent currents measured in oocytes at pH 7.5.
Figure S6. Alignments of functionally characterized ALMT proteins.
GFP Movies 1 and 2. Time-series videos of fluorescence from the HvALMT1::GFP fusion.
Table S1. Primers used for PCR.
Supplementary Material
Acknowledgments
This work was supported in part by a Postgraduate Fellowship to BDG funded by the Australian Grains Research and Development Corporation.
References
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiology. 2004;134:224–236. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP. An ALMT1 gene cluster controlling aluminium tolerance at the Alt4 locus of rye (Secale cereale L.) Genetics. 2008;179:669–682. doi: 10.1534/genetics.107.083451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao YS, Jiang LW, Ryan PR, Richardson AE. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. The Plant Journal. 2007a;51:198–210. doi: 10.1111/j.1365-313X.2007.03138.x. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Ryan PR. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Letters. 2007b;581:2255–2262. doi: 10.1016/j.febslet.2007.03.057. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. Engineering high-level aluminium tolerance in barley with the ALMT1 gene. Proceedings of the National Academy of Sciences, USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. Aluminium tolerance in wheat (Triticum aestivum L.) II. Aluminium-stimulated excretion of malic acid from root apices. Plant Physiology. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecha G, Silva-Navas J, Benito C, Mestres MA, Espino FJ, Hernández-Riquer MV, Gallego FJ. Candidate gene identification of an aluminium-activated organic acid transporter gene at the Alt4 locus for aluminium tolerance in rye (Secale cereale L.) Theoretical and Applied Genetics. 2007;114:249–260. doi: 10.1007/s00122-006-0427-7. [DOI] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF. An aluminium-activated citrate transporter in barley. Plant and Cell Physiology. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase–phosphatase pair. Proceedings of the National Academy of Sciences, USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Hedrich R, Marten I, Lohse G, Dietrich P, Winter H, Lohaus G, Heldt H. Malate-sensitive anion channels enable guard cells to sense changes in the ambient CO2concentration. The Plant Journal. 1994;6:741–748. [Google Scholar]
- Hoekenga OA, Maron LG, Pineros MA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminium tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hurst AC, Meckel T, Tayefeh S, Thiel G, Homann U. Trafficking of the plant potassium inward rectifier KAT1 in guard cell protoplasts of Vicia faba. The Plant Journal. 2004;37:391–397. doi: 10.1046/j.1365-313x.2003.01972.x. [DOI] [PubMed] [Google Scholar]
- Islam AKMR, Shepherd KW, Sparrow DHB. Isolation and characterisation of euplasmic wheat–barley chromosome addition lines. Heredity. 1981;46:161–174. [Google Scholar]
- Kovermann P, Meyer S, Hortensteiner S, Picco C, Scholz-Starke J, Ravera S, Lee Y, Martinoia E. The Arabidopsis vacuolar malate channel is a member of the ALMT family. The Plant Journal. 2007;52:1169–1180. doi: 10.1111/j.1365-313X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences, USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagudah ES, Appels R. The Nor-D3 locus of Triticum taushii: natural variation and genetic linkage to markers in chromosome 5. Genome. 1991;34:387–385. [Google Scholar]
- Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from rape encode aluminium-activated malate transporters that enhance the aluminium resistance of plant cells. Plant Physiology. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligaba A, Kochian L, Pineros MA. Phosphorylation of S384 regulates the activity of the TaALMT1 malate transporter that underlies aluminum resistance in wheat. The Plant Journal. 2009;60:411–423. doi: 10.1111/j.1365-313X.2009.03964.x. [DOI] [PubMed] [Google Scholar]
- Lohse G, Hedrich R. Anions modify the response of guard-cell anion channels to auxin. Planta. 1995;197:546–552. [Google Scholar]
- Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Takeda K. Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley. Journal of Experimental Botany. 2004;55:1335–1341. doi: 10.1093/jxb/erh152. [DOI] [PubMed] [Google Scholar]
- Meckel T, Hurst AC, Thiel G, Homann U. Endocytosis against high turgor: intact guard cells of Vicia faba constitutively endocytose fluorescently labelled plasma membrane and GFP-tagged K+-channel KAT1. The Plant Journal. 2004;39:182–193. doi: 10.1111/j.1365-313X.2004.02119.x. [DOI] [PubMed] [Google Scholar]
- Motoda H, Sasaki T, Kano Y, Ryan PR, Delhaize E, Matsumoto H, Yamamoto Y. The membrane topology of ALMT1, an aluminum-activated malate transport protein in wheat (Triticum aestivum) Plant Signaling and Behavior. 2007;2:467–472. doi: 10.4161/psb.2.6.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Pineros MA, Cancado GMA, Kochian LV. Novel properties of the wheat aluminium tolerance organic acid transporter (TaALMT1) revealed by electrophysiological characterization in Xenopus oocytes: functional and structural implications. Plant Physiology. 2008a;147:2131–2146. doi: 10.1104/pp.108.119636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineros MA, Cancado GMA, Maron LG, Lyi SM, Menossi M, Kochian LV. Not all ALMT1-type transporters mediate aluminium-activated organic acid responses: the case of ZmALMT1—an anion-selective transporter. The Plant Journal. 2008b;53:352–367. doi: 10.1111/j.1365-313X.2007.03344.x. [DOI] [PubMed] [Google Scholar]
- Raman H, Zhang KR, Cakir M, et al. Molecular characterisation and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.) Genome. 2005;48:781–791. doi: 10.1139/g05-054. [DOI] [PubMed] [Google Scholar]
- Raskin I, Ladyman JAR. Isolation and characterization of a barley mutant with abscisic-acid-insensitive stomata. Planta. 1988;173:73–78. doi: 10.1007/BF00394490. [DOI] [PubMed] [Google Scholar]
- Roelfsema MRG, Hedrich R. In the light of stomatal opening: new insights into ‘the Watergate’. New Phytologist. 2005;167:665–691. doi: 10.1111/j.1469-8137.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiology. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ. Characterisation of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta. 1995;196:103–110. [Google Scholar]
- Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:527–560. doi: 10.1146/annurev.arplant.52.1.527. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ryan PR, Delhaize E, et al. Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminium resistance. Plant and Cell Physiology. 2006;47:1343–1354. doi: 10.1093/pcp/pcl002. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminium-activated malate transporter. The Plant Journal. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Keller BU. Two types of anion channel currents in guard cells with distinct voltage regulation. Proceedings of the National Academy of Sciences, USA. 1992;89:5025–5029. doi: 10.1073/pnas.89.11.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Lessdorf B, Lohse G, Hedrich R. GCAC1 recognises the pH gradient across the plasma membrane: a pH-sensitive and ATP-dependent anion channel links guard cell membrane potential to acid and energy metabolism. The Plant Journal. 1996;10:993–1004. [Google Scholar]
- Schunmann PHD, Richardson AE, Smith FW, Delhaize E. Characterisation of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.) Journal of Experimental Botany. 2004;55:855–865. doi: 10.1093/jxb/erh103. [DOI] [PubMed] [Google Scholar]
- Sutter JU, Sieben C, Hartel A, Eisenach C, Thiel G, Blatt MR. Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+channel and its recycling to the plasma membrane. Current Biology. 2007;17:1396–1402. doi: 10.1016/j.cub.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, Brettell R. Agrobacterium tumefaciens-mediated barley transformation. The Plant Journal. 1997;11:1369–1376. [Google Scholar]
- Upadhyaya NM, Ramm K, Gaudron J, Craig S, Wang M-B, Gupta S, Okita TW, Waterhouse PM. Can gfp replace uidA as a reporter gene to monitor transformation of cereals by biolistics or Agrobacterium? In: Larkin PJ, editor. Agriculture biotechnology; laboratory, field and market. Canberra, Australia: Under The Counter Publishing; 1998. pp. 111–113. [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkki LV, Murer H, Forster IC. Voltage clamp fluorometric measurements on a type II Na+-coupled Pi cotransporter: shedding light on substrate binding order. Journal of General Physiology. 2006;127:539–555. doi: 10.1085/jgp.200609496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Raman H, Zhou MX, Ryan PR, Delhaize E, Hebb DM, Coombes N, Mendham N. High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2007;115:265–276. doi: 10.1007/s00122-007-0562-9. [DOI] [PubMed] [Google Scholar]
- Wang M-B, Li Z-Y, Matthews PR, Upadhyaya NM, Waterhouse PM. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Horticulturae. 1998;461:401–405. [Google Scholar]
- Wang M-B, Waterhouse PM. High-efficiency silencing of a β-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Molecular Biology. 2000;43:67–82. doi: 10.1023/a:1006490331303. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H. Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1) Plant and Cell Physiology. 2005;46:812–816. doi: 10.1093/pcp/pci083. [DOI] [PubMed] [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA. A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theoretical and Applied Genetics. 2000;101:1093–1099. [Google Scholar]
- Zhang WH, Ryan PR, Sasaki T, Yamamoto Y, Sullivan W, Tyerman SD. Characterisation of the TaALMT1 protein as an Al3+-activated anion channel in transformed tobacco (Nicotiana tabacum L.) cells. Plant and Cell Physiology. 2008;49:1316–1330. doi: 10.1093/pcp/pcn107. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.