Abstract
As research in the biological sciences continues to advance at a rapid pace, it is increasingly important that the data be captured, standardized, organized and made accessible to the scientific community. This is the job of a biocurator. Here we describe the process of biocuration from our perspective as FlyBase curators.
Keywords: biocuration, ontology, model organism database, FlyBase, career
Introduction
If a discovery is made and no one knows about it, is it really a discovery? In recent years, scientists have been routinely using online databases to gather information, keep abreast of the latest findings, and publish and/or publicize their data. Whether it is to search the literature, identify genes with similar phenotypes, determine a gene structure, compare sequences across species, or obtain stocks or reagents, biologists use databases to obtain data quickly and accurately. The types of biological databases available online include model organism databases (MODs) such as FlyBase, WormBase and Mouse Genome Informatics (and at least 20 others); sequence banks like GenBank, UniProt and the Protein Data Bank; and bibliographic databases like NCBI PubMed. In addition, there are hundreds of small, often lab-driven, databases designed around a distinct topic, such as BioGRID, a repository for interaction datasets; FlyExpress, an expression pattern search engine; and Homophila, a human disease-associated Drosophila gene database. The amount and variety of data found in these databases is enormous, and its availability depends upon the work of biocurators. The field of biocuration is emerging and developing in response to the growing quantity of scientific data and the need for scientists to rapidly search and analyze that data. Here we describe the processes and challenges of biocuration from the perspective of a curator at FlyBase: What is biocuration? How is it done? What is it like to do it?
Biocuration: Its Charge and Challenges
In order for the increasingly complex torrent of novel biological data to be understood to its full potential, scientists need easy access to the data in a standardized and organized form. This is the job of a biocurator: to identify and collect data; to analyze and extract the findings; and to incorporate, represent and display the data in formats useful for different types of audiences. For a model organism database like FlyBase, in practical terms this means identifying the relevant literature, translating experimental results into a standardized, searchable language (through the use of controlled vocabularies), and working with database and website developers to ensure usability.
As research methods evolve, so must biocuration adapt. Not only is the amount of published curatable data growing each year, but the variety of data types is also increasing. In fact, this increase in the variety of data is proving to be the greatest challenge. It is relatively straightforward to continue to curate the types of data that are well understood and for which a data handling pipeline already exists. It is much harder to understand, create the database structures for, and develop curation protocols for new types of data. An example is the modENCODE project, which is seeking to create an encyclopedia of DNA elements for the model organisms Drosophila melanogaster and Caenorhabditis elegans (www.modencode.org). While the data produced by modENCODE will benefit the fly (and worm) communities greatly, the incorporation of their data into FlyBase will be a major challenge over the next few years. Initial modENCODE data is already forcing us to review our definitions of genomic elements such as transcription start sites and even the transcripts themselves.
While biocuration is becoming more and more complex, in most cases there has been no comparable increase in funding. Thus, we need to do significantly more with (relatively) less. All databases are exploring ways of automating some curation tasks, but biocuration, as it currently exists, depends on the expertise of scientists. Some databases are able to make use of text-mining, or other computational methods of extracting data from the literature. However, for some MODs, and especially for FlyBase, attempts to use various text-mining tools as an aid to curation have hit significant stumbling blocks. Even the seemingly simple task of identifying the genes mentioned in a research paper is quite a challenge when so many gene names are common English words (for example the fly genes ‘or’, ‘a’, ‘white’, ‘for’).
The challenge of data extraction is not solely a computational issue. Even human curators have difficulty with this task. Accurately, consistently and quickly capturing details of genetic objects (such as genes, alleles and recombinant constructs) and experimental results requires immense attention to detail by biocurators, as well as by authors. When insufficient information is given in a paper to determine, for example, exactly which allele of a gene is being studied, the process of curating a paper is delayed, and the data may never be included in the database. For this reason, biocurators are currently lobbying journals to require authors to use unique database identifiers (such as FlyBase gene ID numbers) in their papers. In addition, advice is provided for authors to help make their papers more curatable (http://flybase.org/static_pages/docs/author-suggestions.html), thus making curation faster, and the chance of errors lower.
Data Identification, Curation and Representation: The FlyBase Model
FlyBase (flybase.org) is the main web portal for the Drosophila research community and serves as a searchable compendium of phenotypic, molecular and genomic data. While our curated data remains Drosophila-melanogaster-centric, we have recently incorporated gene annotations for 11 other fully sequenced Drosophila species.1
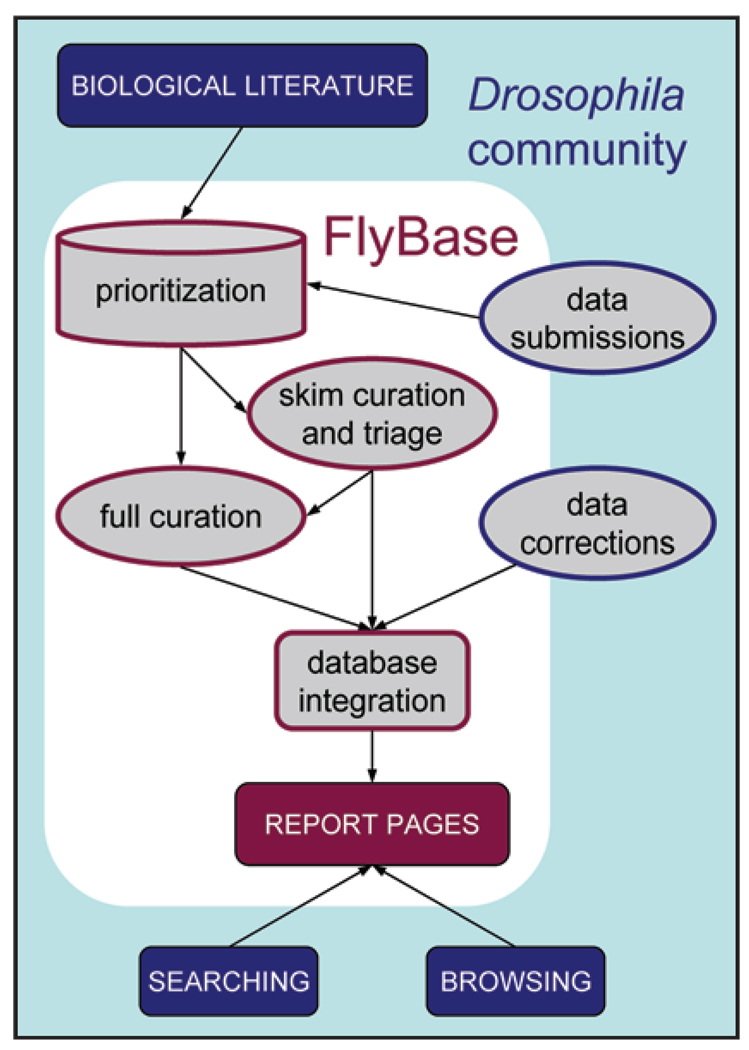
FlyBase literature curation is the process of extracting relevant data from a research paper in a formulaic manner and then assimilating it with existing data in the database (see Fig. 1). The goal of literature curation is to identify genetic, molecular and genomic objects used in experiments and capture the results of those experiments. By connecting the experimental results to the object (for example, a phenotype to an allele), we allow our users to browse and search the database either by object (e.g., find all alleles of the dpp gene) or by biological role (e.g., all alleles with phenotypes manifest in the eye).
Figure 1.
Data flow into and out of FlyBase.
The first step in curation is to identify the sources of data to curate. The main data sources are the primary literature, and increasingly, high-throughput data sets and other databases. We estimate that approximately 2,150 primary research papers are published in English on Drosophila every year, which is more than can be curated in depth by FlyBase. For every paper identified as being a Drosophila-related paper, we perform two tasks: skimming and triaging. Skimming is the process of identifying the genes (if any) that are the main focus of the paper. This allows the paper to be indexed on FlyBase such that it appears in the reference section on the relevant gene report pages. At the same time, we make an initial determination of the types of data that require full curation, a process we call triaging. We attach internal flags to papers indicating the type(s) of data contained in the paper. Examples of such flags include a newly generated allele, a gene that has been newly characterized, evidence for changes to the gene model (the genomic structure of the gene, i.e., intron/exon structure), or gene expression data. Through the processes of skimming and triaging, we simultaneously perform a very basic curation and create a framework that allows us to prioritize full curation of those papers that contain more of the information that users would like to see.
Once a paper has been skimmed and triaged, it may be targeted for further curation. Genetic literature curators extract phenotypic data from the results section (text, tables and figures) of a paper. We also use the materials and methods sections of papers to identify alleles and recombinant constructs used in a paper. Additional types of data include Gene Ontology (GO) terms, genetic interactions, gene expression patterns, activity of regulatory elements, and gene model corrections. Approximately 57 percent of all the primary research papers listed in the FlyBase bibliography have been curated for genetic data, GO, phenotypes, molecular data and genetic interactions.
A major aspect of recording information in a uniform way is the use of structured controlled vocabularies (CVs). Genetic objects in FlyBase—genes, alleles, transcripts etc., are labeled with CV terms. For example, the gene twins has the label ‘protein coding gene’, while the twins transcripts have expression labels ‘oocyte’ and ‘nurse cell’. These controlled vocabularies are organized into hierarchies (called ontologies) that encode the logical relationships between terms. For example ‘protein coding gene’ is a type of gene, and ‘oocyte’ and ‘nurse cell’ are parts of ‘ovary’. Ontologies allow objects carrying related labels to be grouped. This allows the user to search for the CV term ovary, and find, in addition to the transcripts annotated with the term ovary, all the transcripts annotated with child terms of ovary (i.e., more specific terms), such as oocyte and nurse cell. By using controlled vocabularies to record information in this way we ensure consistent descriptions within FlyBase, and across databases, despite varied usage by authors, for those vocabularies that are widely used, such as GO.2 CV usage is essential for robust and accurate queries; FlyBase controlled vocabularies are integrated into our search tools QueryBuilder and TermLink.3
In addition to the large task of curating genetic and molecular data from the primary literature, FlyBase genomic curators also produce and maintain the current set of manually annotated genes for Drosophila melanogaster. Since 2002, when FlyBase collaborated with the Berkeley Drosophila Genome Project to manually annotate the euchromatic sequence of Drosophila melanogaster in its entirety,4 updates to gene models have occurred when prompted by new evidence. We compare the new evidence to our existing set of transcripts to detect which gene models require attention. While we have never completed another “tip to tail” reannotation of the genome, several thousand gene models have been updated over the past several years, and our pipeline for reexamining gene models continues to be refined. We have made changes to gene models based on new cDNA clones, proteomic data, and orthology-based gene prediction programs including a new quantitative method of identifying protein-coding regions using evolutionary signatures.5 Counts of gene model changes between releases are listed in the ‘release notes’ under the Documents menu on the website. Further discussion of our methodology for annotating genes can be found in the Gene Model Annotation Guidelines section of the FlyBase Reference Manual (http://flybase.org/static_pages/docs/refman/refman-G.html#G7).
Curators at FlyBase also work closely with software developers to help integrate large datasets into the database. For example, we have recently incorporated over 20,000 transgenic RNAi insertion lines from the Vienna Drosophila RNAi Center,6 a dataset which clearly could not have been entered manually.
The final step in curation is representing the curated data in a useful manner. Newly curated data must be integrated with the data already present and must be displayed on the website. As the types and quantity of data increase, effective and efficient presentation becomes more and more challenging. To this end, curators work closely with FlyBase developers to ensure that the data is being represented in a clear and scientifically accurate manner; we also work extensively with our developers to create and expand tools to query the database and access data. Since most of us were FlyBase users before we were curators, we attempt to view the website from a user’s perspective when we develop new tools and website features.
Biocuration as a Career
Perhaps predictably, biocuration is not easy, but its challenges are what make it interesting. At present, accurate and effective biocuration requires experienced scientists, but biocuration as a career has yet to be well defined. Recently, biocurators have begun to organize and are in the process of forming an International Society for Biocuration (ISB). The aim of the ISB will be to promote biocuration as a career.7 Anyone interested in a career in biocuration should visit www.biocurator.org and consider subscribing to the mailing list. The mailing list is currently the main avenue for spreading news to the biocurator community such as job openings, meeting details and other related items.
A day in the life of a biocurator is, in many ways, much like a day in the life of a bench scientist. Some of our work is solitary, but much of it requires discussion and collaboration with fellow curators and other project members. We participate in group meetings where we discuss problems we encounter while curating, annotating, or developing new datasets. We attend conferences such as the Annual Drosophila Research Conference where we give talks about FlyBase features and tools, and provide personalized tutorials for FlyBase users. Like laboratory work, some of our tasks are interesting, exciting and engaging while others are tedious and repetitive. Sometimes large volumes of data are moved into the database with gratifying speed, and sometimes curation tasks, especially when changes to current procedures are required, are agonizingly slow. Nevertheless, biocuration is a rewarding career. We enjoy being at the forefront of a new field, and of course, being in the privileged position of monitoring the cutting edge of scientific research across many fields. The job can be quite bipolar at times, with concentrated quiet periods, when we are curating data, punctuated with discussions on the latest advancements or helping users, either through our website help-mail, or through tutorials, talks and workshops.
Making the leap from the lab to the curation room requires that one understand the importance of the mission of biocuration. Biologists are generating more data now than at any other time in the history of science. The amount and quality of new data is a heady mix, but rapidly becomes overwhelming. Biocuration attempts to make this flood of information understandable—to catalogue it, link it, decipher it and add order. At FlyBase, we are constantly searching for ways to improve our biocuration, incorporate more data types, advance search tools or data display, reflect the continuing expansion of biology, and provide a better service to our users. It’s an exciting time in an exciting job, as the field of biocuration takes its first steps.
Acknowledgements
We would like to thank all of the FlyBase members for their critical reading of this manuscript. FlyBase is supported by the U.S. National Human Genome Research Institute, National Institutes of Health (P41 HG00739) and additional grants from the Indiana Genomics Initiative, USA and the Medical Research Council, UK (G05000293).
References
- 1.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 2.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RJ, Goodman JL, Strelets VB the FlyBase Consortium. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:588–593. doi: 10.1093/nar/gkm930. doi:10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biology. 2002;3:83. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Research. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 7.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]