Abstract
Little is known about clinical and virologic manifestations of rhinovirus (HRV) and coronavirus (HCoV) infections after hematopoietic cell transplantation (HCT). We performed surveillance for 1 year and describe the natural history of these infections during the first 100 days after allogeneic HCT, when symptom surveys and upper respiratory samples were collected weekly. Samples were tested using RT-PCR for HRVs and HCoVs (OC43, 229E, HKU1, and NL63). Among 215 patients, 64 (30%) patients had 67 infections. Day 100 cumulative incidence estimate was 22.3% for HRV and 11.1% for HCoV. Median duration of viral shedding was 3 weeks; prolonged shedding of at least 3 months occurred in 6 of 45 patients with HRV and 3 of 22 with HCoV. Six patients with HRV and 9 with HCoV were asymptomatic. HRV infection was associated with rhinorrhea, congestion, postnasal drip, sputum, and cough; HCoV infection was not associated with respiratory symptoms or hepatic dysfunction. Lower respiratory infection developed in 2 patients with HRV before day 100, and 1 each with HRV and HCoV after day 100. HRV and HCoV infections are common in the first 100 days after HCT, viral shedding lasts more than 3 weeks in half, and lower respiratory infection is rare.
Introduction
The human rhinoviruses (HRVs) and coronaviruses (HCoVs) are well known to be the predominant causes of the common cold in immunocompetent persons. The main site of viral replication is in epithelial cells of the upper respiratory tract, but these viruses can cause pneumonia and bronchiolitis in infants and young children and pneumonia among older adults.1–3 Minimal data exist regarding the incidence and clinical relevance of these virus infections among hematopoietic cell transplantation (HCT) recipients, a population in which many other respiratory viruses, such as respiratory syncytial virus (RSV), influenza viruses, parainfluenza viruses (PIVs), adenoviruses (HAdVs), and human metapneumovirus (HMPV), cause high rates of lower respiratory tract disease and death.4–10
Among HCT recipients, HRVs have been described as a cause of serious lower respiratory tract infections, frequently in association with other pathogens.11–13 A recent case report describes 2 HCT recipients with HRV as the probable etiologic agent for fatal lower respiratory tract infection.14 Several cases of HCoV-related pneumonia have been reported in immunocompromised adult and pediatric patients treated for hematologic malignancies,15,16 and 2 cases of coronavirus pneumonia have been described after HCT.17,18 These data suggest that HRVs and HCoVs may play an important role as single agents or as copathogens, although prospective studies of these viruses among HCT recipients are lacking.
We conducted a prospective surveillance study of allogeneic HCT recipients, in which weekly upper respiratory samples and symptom surveys were collected during the first 100 days after HCT regardless of the presence of symptoms. We analyzed the samples for human HRVs and HCoVs using reverse transcription polymerase chain reaction (RT-PCR).
Methods
Patients and clinical data
This study was part of a prospective longitudinal surveillance study of respiratory virus infections among HCT recipients followed for 1 year after HCT, with weekly virologic surveillance in the first 100 days. Participants underwent allogeneic HCT at the Fred Hutchinson Cancer Research Center between December 2005 and April 2008. Study patients completed a baseline information form on study entry and a weekly standardized symptom survey, including 15 symptoms, for the entire year of the study period. Clinical chart review was performed to determine additional symptom detail when necessary. The study protocol was reviewed and approved by the Fred Hutchinson Cancer Research Center Institutional Review Board, and all participants provided written informed consent in accordance with the Declaration of Helsinki.
Virologic methods
Weekly nasal wash and oropharyngeal swab samples were collected from each patient beginning 1 to 2 weeks before transplantation until 100 days after HCT, and then every 1 to 3 months or with the onset of new respiratory symptoms. Nasal washes were collected with a total of 10 mL of saline (5 mL/nostril) for adults and 5 mL (2.5 mL/nostril) for children and combined with the oropharyngeal swab into one sterile vial. Nasopharyngeal swabs were collected if the patient's clinical condition precluded collection of a nasal wash.
Patients who develop respiratory symptoms in our center routinely undergo chest radiograph; bronchoscopy is performed if lower respiratory tract infection is present. During the study period, bronchoalveolar lavage (BAL) samples were routinely evaluated for respiratory viruses by RT-PCR and often by culture using rhesus monkey kidney, Buffalo green monkey kidney, HL (transformed HeLa), human foreskin fibroblasts, and A-549 cell lines. Although viral culture of nasal washes and BAL samples is no longer routine practice at our center, these results are also captured in our study database and included in the analysis. BAL samples were also routinely submitted to pathology and to the laboratory for bacterial, fungal, and acid-fast bacilli cultures, Aspergillus galactomannan assay, and detection of cytomegalovirus (CMV) and herpes simplex virus.
Respiratory samples were tested by real-time RT-PCR using previously validated primers and TaqMan probes for the 4 non-SARS types of HCoV (OC43, 229E, HKU1, and NL63).15 Samples were tested for HRV by 2 real-time RT-PCR assays using primers and TaqMan probes targeting the highly conserved 5′ noncoding region. HRV RT-PCR assay “A” included 3 forward primers (F1 [GACCTGGCAGATGAGGCTAGA], F3 [GGAGTGTAGCTTAGGCTGATGAGTCT], and F4 [CTTGCAGATGAGGCTGGACAT]), 3 reverse primers (R1 [CAGCCACGCAGGCTAGAAC], R2 [CTAGCGTCCTGTGGTTTTGGA], and R4 [GCGGCTCTTCACACCTTGTC]), and 2 FAM-labeled MGB probes (P1 [CCCCACTGGCGACAG] and P3 [CCCCACTGGTAACAGT]). HRV RT-PCR assay “B” included 2 forward primers (F2 [CCCTCACTAGTCTGGTCGATGAG] and F4), 2 reverse primers (R1 and R3 [GGAGTGTAGCTTAGGCTGATGAGTCT]), and a FAM-labeled MGB probe (P2 [TCCCCACGGGCGAC]). Samples positive by either or both assays were considered positive for HRV. In January 2008, new HRV primers and probes were introduced19 that led to increased rhinovirus detection by 10% to 15% when tested against known culture-positive isolates.
Samples were also tested for RSV, influenza virus types A and B, PIV types 1 through 4, HMPV, and HAdV using qualitative RT-PCR with the use of jellyfish DNA (EXO) to assess extraction and assay inhibition and a low positive control for each respiratory virus.15,20–23 Each assay reliably detected 10 viral copies per reaction, providing a sensitivity of 1000 copies/mL (10 μL of specimen added per reaction).20–23
For testing by a panel of multiplexed qualitative assays, RT-PCR reaction strips were prepared containing specific primer and probe sets for 8 multiplexed reactions, including 5 reactions in which EXO was one of the targets. The specific primers and probes were added to each well, dried, and one strip was used for each patient or control RNA sample. A single RT-PCR master mix (AgPath; Applied Biosystems) was added to every well, and samples were amplified without standard curves. The reaction mixes using rehydrated, multiplexed primers and probes in the 8-well panels had the same sensitivity and specificity as did the individually run quantitative reactions. Results were reported as positive (amplification plot crossed the threshold at < 40 cycles) or negative (amplification plot did not cross the threshold) for each target in each well. Samples with average EXO threshold cycle values greater than 35.5 were considered to be unsatisfactory and sample processing and/or amplification was repeated.
Samples testing positive for HCoV by the qualitative assay were subsequently tested by quantitative RT-PCR using a standard curve of known numbers of HCoV subtype OC43 RNA transcripts ranging from 10 to 107 copies per reaction. HCoV+ samples were also retested using subtype-specific PCR assays.15 No quantitative assay for HRVs was available because of the inability to reliably quantify the viral load of more than 100 unique HRV subtypes compared with a single standard curve. All PCR methods were performed according to College of American Pathologist standards, and the laboratories passed proficiency testing in viral diagnostics.
Definitions
Subjects were required to have submitted at least one respiratory sample within the first 100 days after transplantation. Asymptomatic infection was defined as detectable virus in the patient's upper respiratory sample without documented symptoms of respiratory illness on the patient symptom survey.9 An upper respiratory tract infection was defined as detection of respiratory virus from the upper respiratory tract (nose, throat) in conjunction with any reported respiratory symptoms. A lower respiratory tract infection (LRI) was defined as virus detection in a BAL or lung biopsy from patients with new pulmonary infiltrates on chest radiograph or computed tomography.
Total duration of virus shedding for a given HRV or HCoV infection was calculated using all available results from 2 weeks before and up to 1 year after HCT. During the first 100 days, to account for possible missed weekly samples and weeks when the virus RNA was below the PCR assay's limit of detection, total duration of virus shedding for a given HRV or HCoV infection was calculated from the first positive to the final positive sample, allowing no more than 4 weeks or 2 negative samples between any 2 consecutive positive samples. After day 100, duration of shedding was calculated to the final positive sample, allowing no more than 8 weeks or 2 negative samples between any 2 consecutive positive samples. Consecutive detections that failed to meet these criteria were considered separate infection episodes. If any other respiratory virus was detected during the duration of HRV or HCoV shedding, a subject was defined to have virus codetection. Hospital-acquired infections were defined as occurring 7 days or more after admission to the inpatient wards; the remainder were defined as community-acquired infections.
The patient's underlying disease stage was categorized as low, intermediate, or high risk, as previously described.24 Donor match status was determined according to donor-recipient human leukocyte antigen-A (HLA-A), HLA-B, and HLA-DR status. Acute graft-versus-host disease (GVHD) was graded using standard criteria based on stages of organ involvement and categorized as acute GVHD grades 0 or 1 and grades 2 to 4.25
Statistical analysis
The probability of acquiring at least one HCoV or HRV infection was estimated using cumulative incidence curves. Patient records were censored at 14 days past the time of last eligible respiratory sample, death, or day 100 after transplantation, whichever occurred first. Death before day 100 and within 14 days of a patient's last sample was treated as a competing risk for infection. Cox regression models were fit to evaluate potential risk factors for acquisition of HCoV and HRV infections. We also assessed whether HCoV and HRV infections in the first 100 days after HCT were predictors of 1-year survival using a Cox regression model with the viral infections as time-dependent covariates.
Logistic regression was used to analyze the relationship between occurrence of each symptom during the first 100 days after HCT and virus detection within the past week or past month (for wheezing and cough). Virus detection within the last week was defined as a positive sample at either the current or the last study contact. If the time lag from the last contact was greater than 14 days and the current test was negative for virus, then virus within the last week was defined as negative. Virus detection within the last month was defined as a positive sample at either the current or any of the last 4 study contacts, as long as those contacts occurred within 45 days of the current visit. Logistic regression was also used to analyze the incidence of secondary pathogenic bacterial or fungal infections in the lungs or bloodstream as a function of current and past detection of HRV or HCoV detection within the past week. Blood or lung samples from posttransplantation days before any viral testing, and from 2 weeks or more after discontinuation of viral testing, were excluded from the analysis. Otherwise, all blood and lung samples were used as possible outcome indicators. Differences in log-transformed hepatic transaminases and bilirubin levels between values corresponding to a positive virus sample within the last week and those with a negative sample were evaluated via linear regression. Adjustment to all models was made for the following covariates: detection of other respiratory viruses within the last week, day relative to transplantation, age at transplantation, pretransplantation seasonal allergies (for symptom analysis only), conditioning regimen (myeloablative vs nonmyelablative), donor type, and acute GVHD. Measurements based on multiple patient contacts were entered into logistic and linear regression analyses as repeated measures, with adjustment for possible correlation between values within a subject using generalized estimating equations.26
P values from regression models were obtained from the Wald test, and no adjustments were made for multiple comparisons. Two-sided P values less than .05 were considered statistically significant. Statistical analyses were performed using SAS, Version 9.0 (SAS Institute Inc).
Results
Incidence of HRV and HCoV after HCT, respiratory virus codetections, secondary infections, and duration of viral shedding
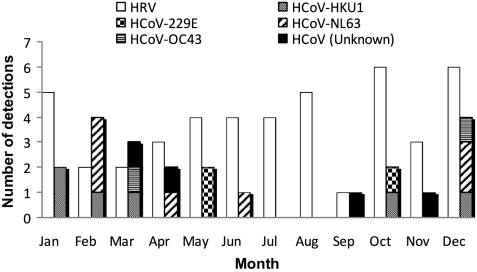
Of 215 patients who underwent allogeneic HCT, 64 (30%) were infected with HRV and/or HCoV in the first 100 days. HRVs and HCoVs were detected from 45 (21%) and 22 (10%) patients, respectively. Clinical characteristics of the patients with HRV and HCoV detection were similar to the characteristics of the other 151 patients in the study population (Table 1). HCoV-NL63 accounted for the majority of the HCoV infections (7, or 32%); 5 cases of HCoV-HKU1, 3 cases of HCoV-229E, and 3 cases of HCoV-OC43 were also documented. In 4 cases, HCoV type could not be identified because the samples were negative on retesting. Three patients had codetection of both HRV and HCoV. Among those patients with HRV infection, 18 had a concomitant viral culture performed on 24 clinical nasal wash samples that were also tested by PCR. Of those samples, 3 (12%) were positive for rhinovirus by culture and 15 (62%), including the 3 culture-positive samples, were positive by PCR. Six (9%) patients, 5 with HRV infection, first had HCoV or HRV detected in the month before transplantation. All 6 patients reported concurrent respiratory symptoms, and 3 with HRV exhibited prolonged virus shedding from 14 to 30 weeks.
Table 1.
Characteristics of 215 HCT recipients in study cohort
| Characteristic | RhV or HCoV (n = 64), n (%) | No RhV or HCoV (n = 151), n (%) | All patients (n = 215), n (%) |
|---|---|---|---|
| Median age, y (range) | 50.2 (0.8-72.2) | 51.6 (0.6-72.2) | 51.2 (0.6-72.2) |
| No. of patients age < 12 y | 4 (6) | 5 (2) | 9 (4) |
| Sex | |||
| Female | 27 (42) | 58 (38) | 85 (40) |
| Male | 37 (58) | 93 (62) | 130 (60) |
| Underlying disease risk | |||
| Low | 6 (10) | 7 (5) | 13 (6) |
| Intermediate | 39 (60) | 103 (68) | 142 (66) |
| High | 19 (30) | 41 (27) | 60 (28) |
| Stem cell source | |||
| Bone marrow | 7 (11) | 21 (14) | 28 (14) |
| Peripheral blood | 53 (83) | 120 (79) | 173 (80) |
| Cord blood | 4 (6) | 10 (7) | 14 (7) |
| CMV serostatus | |||
| D+/R+ | 17 (27) | 35 (23) | 52 (24) |
| D+/R− | 8 (13) | 7 (4) | 15 (7) |
| D−/R+ | 20 (31) | 57 (38) | 77 (36) |
| D−/R− | 19 (29) | 52 (35) | 71 (33) |
| Donor match | |||
| Related-matched | 22 (34) | 52 (34) | 74 (34) |
| Related-mismatched | 4 (6) | 6 (4) | 10 (5) |
| Unrelated-matched | 28 (44) | 66 (44) | 94 (44) |
| Unrelated-mismatched | 10 (16) | 27 (18) | 37 (17) |
| Acute graft-versus-host disease | |||
| Grade 0 or 1 | 15 (23) | 43 (28) | 58 (27) |
| Grade 2-4 | 49 (77) | 108 (72) | 157 (73) |
| Transplantation type | |||
| Myeloablative | 32 (50) | 70 (46) | 102 (47) |
| Nonmyeloablative | 32 (50) | 81 (54) | 113 (53) |
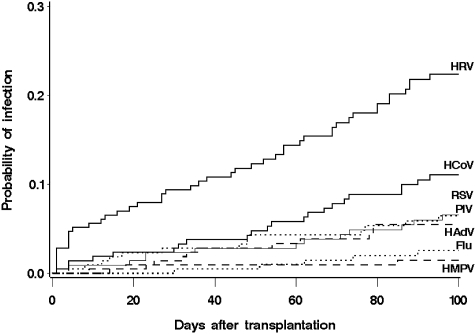
The cumulative incidence estimate at day 100 for HRV was 22.3% (95% confidence interval [CI], 16.5%-28.1%) and for HCoV infection was 11.1% (95% CI, 6.7%-15.4%; Figure 1). Cumulative incidences for PIV, RSV, HMPV, influenza, and adenovirus are also shown. Excluding the 6 patients positive before HCT, median time of first detection was 44 days (range, 2-93 days) and 53 days (range, 2-93 days), for HRV and HCoV, respectively. Five (7%) patients acquired these viruses during their hospitalization: 1 with HCoV and 4 with HRV detection. Seasonal outbreaks were more common in the winter for HCoV (13 of 22 cases were first detected in December through March), and there was no apparent seasonality for HRV detection (Figure 2). Univariate analysis of potential risk factors showed no significant association of patient age, gender, underlying disease risk, stem cell source, CMV serostatus, donor type, acute GVHD, conditioning regimen, or engraftment status with acquisition of HCoV and HRV infection (data not shown).
Figure 1.
Cumulative incidence of first infection episodes of HRV, HCoV, and other respiratory viruses (RSV, PIV, HMPV, influenza, adenovirus) after transplantation in 215 HCT recipients.
Figure 2.
Incident detections of HRV and all subtypes of HCoV by month (n = 67 incident detections).
Including the 3 patients with HRV/HCoV codetection, we observed codetection with one or more respiratory viruses in 12 (19%) of the 64 patients. Additional HRV codetections included 2 patients with RSV, 1 with PIV2, 2 with HMPV, 1 with HAdV, and 1 with both RSV and PIV2. HCoV codetections included a patient with influenza A and a patient with both influenza A and HMPV. In a multivariable regression model, neither HRV nor HCoV infection in the last week was associated with secondary pulmonary or bloodstream infections caused by pathogenic bacteria or fungi during the first 100 days after HCT (data not shown). There were too few positive secondary viral infections to analyze this outcome, and there was only one case of sinusitis in the cohort, in a patient with concomitant HRV and HCoV infection. This patient had one colony of Streptococcus pneumoniae isolated in sinus culture; no fungal organisms were isolated.
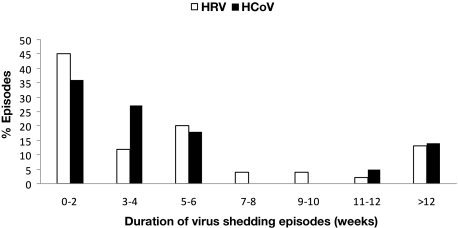
The median duration of shedding was 3 weeks (range, 0-49 weeks) for HRV and 3 weeks (range, 0-22 weeks) for HCoV. Excluding 21 patients who were positive only once (16 with RHV and 5 with HCoV), the median duration of viral shedding was 5 weeks (range, 1-49 weeks) and 4 weeks (range, 1-22 weeks), for HRV and HCoV, respectively. The majority of virus detections had a duration of viral shedding of less than a month, 25 (56%) for HRV, and 14 (63%) for HCoV (Figure 3). Six (13%) patients with HRV detection and 3 (14%) patients with HCoV had a prolonged shedding duration for more than 12 weeks; 3 with HRV first had virus detected before transplantation. Among these 9 patients, 4 with HRV and 2 with HCoV had viral detection for more than 5 months. One patient with HRV was positive the entire year. All patients with HCoV had the same subtype of virus during the shedding period, and the 3 patients with prolonged shedding had HCoV-NL63.
Figure 3.
Duration of HRV (white) and HCoV (black) shedding for 45 and 22 infection episodes, respectively.
Neither virus was significantly associated with death in the first year after HCT when included in a model together and adjusted for age, donor/recipient CMV serostatus, and donor type: HCoV hazard ratio 0.8 (95% CI, 0.3-2.0) and HRV hazard ratio 0.5 (95% CI, 0.2-1.1).
Asymptomatic infection
The median number of weekly sample collections with concurrent symptom surveys between day 0 and day 100 for each patient was 13 (range, 2-15). Among patients with HRV detections, 6 (13%) of 45 patients had no reported respiratory symptoms on symptom survey during the duration of viral shedding. Subject ages ranged from 39 to 63 years; 3 received an unrelated transplant and 4 had nonmyeloablative conditioning. All were diagnosed as outpatients a median of 70 days (range, 21-88 days) after HCT. HRV was detected only once in 5 of these patients; in the sixth patient, viral shedding lasted for 5 weeks. All follow-up samples from the 6 patients were negative. Two additional patients with more than 10 positive samples reported symptoms only once during the duration of viral shedding.
Among patients with HCoV detections, 9 (41%) of 22 were asymptomatic during the time virus was detected. Subject ages ranged from 47 to 68 years; 8 received an unrelated transplant and 4 had nonmyeloablative conditioning. All were diagnosed in the outpatient department a median of 60 days (range, 48-93 days) after HCT. For 5 (55%) patients, the virus was detected only once, and for the other 4 patients the median duration of viral shedding was 4 weeks (range, 2-6 weeks). All had provided follow-up samples that were negative. None of the 3 patients with HRV and HCoV codetection was asymptomatic.
Upper respiratory tract infection and LRI
Thirty-nine patients with HRV reported upper respiratory symptoms at the time of HRV detection. Bronchoscopy was performed in 3 (7%) of these patients before day 100, concurrent with HRV detection in the upper respiratory tract. Two patients were found to have progression of disease to LRI with BAL samples positive by PCR and negative by culture. These patients have been described previously; both died of respiratory failure with HRV as the most probable etiologic agent.14 After day 100, one additional patient in this cohort had a BAL sample positive for HRV. This patient was a 12-month-old boy transplanted for severe combined immunodeficiency who was positive for both HRV and HMPV before transplantation with recurrent intermittent positive nasal wash samples for HMPV, HRV, and PIV type 3 (after 7 months) for more than a year after HCT. BAL on day 274 was positive only for HRV. He had been intubated for respiratory distress and had no changes on chest computed tomography. Repeat BAL on day 350 was negative for all respiratory viruses, and he subsequently recovered from his respiratory illness.
Thirteen patients with HCoV infection reported upper respiratory symptoms; none of these patients had clinical evidence of LRI and hence underwent bronchoscopy before day 100. However, after day 100, one patient with HCoV-NL63 detection had a BAL sample positive for HCoV. This was a 41-year-old woman who received a nonmyeloablative haploidentical transplant for refractory/relapsed Hodgkin disease. On day 30 after HCT, her nasal wash was positive for both HCoV and influenza A; oseltamivir therapy was initiated. A repeat bronchoscopy on day 85 revealed persistence of both influenza A and HCoV as well as HMPV. On day 101, she was readmitted with hypoxia, worsening dyspnea, and progressive infiltrates on chest computed tomography. Repeat BAL was positive for HCoV, influenza A, and HMPV on day 101 and positive for influenza A and HMPV on day 110. She died of respiratory failure on day 112. Limited lung autopsy showed diffuse alveolar damage, and all viral cultures from the autopsy were negative.
HRV and HCoV detection associated with symptoms and hepatic values
The mean number of respiratory symptoms present with HRV and HCoV detection were 4 (range, 0-11) and 2 (range, 0-9), respectively. For patients with positive detection, the time lag between current and previous visits ranged from 4 to 17 days, with 90% of those values between 5 and 9 days. In a multivariable regression model, a positive HRV sample within the last week was significantly associated with rhinorrhea, sinus congestion, postnasal drip, sputum production, and cough (Table 2). HCoV infection was not significantly associated with any respiratory symptoms. HRV within the last month was also a significant predictor of cough (odds ratio = 2.0; 95% CI, 1.2-3.5; P = .01) but not of wheezing (odds ratio = 1.7; 95% CI, 0.7-4.6; P = .26). These data were also reanalyzed for patients with at least 2 positive samples (ie, patients with only one positive sample were classified as negative), and still HCoV infection within the last week was not significantly associated with any respiratory symptoms. HRV within the last week remained significantly associated with rhinorrhea, sinus congestion, postnasal drip, and cough.
Table 2.
Respiratory and systemic symptoms associated with coronavirus or rhinovirus detection at time of virus detection or within the prior week
| Symptom | Coronavirus |
Rhinovirus |
||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Rhinorrhea | 1.4 (0.7-2.6) | .33 | 2.3 (1.3-4.1) | .004 |
| Sinus congestion | 0.9 (0.4-2.2) | .83 | 3.4 (1.8-6.9) | < .001 |
| Postnasal drip | 0.7 (0.3-1.6) | .38 | 2.4 (1.3-4.8) | .009 |
| Shortness of breath | 0.9 (0.3-2.8) | .79 | 1.5 (0.7-3.3) | .32 |
| Sputum | 0.9 (0.4-2.0) | .74 | 1.9 (1.1-3.3) | .03 |
| Pharyngitis | 0.8 (0.2-2.6) | .70 | 0.8 (0.4-1.7) | .61 |
| Sneezing | 0.8 (0.2-2.4) | .63 | 0.9 (0.5-1.8) | .88 |
| Cough | 1.9 (0.9-4.1) | .12 | 2.3 (1.3-4.2) | .006 |
| Wheezing* | — | — | 2.0 (0.8-5.1) | .16 |
| Fever | 0.8 (0.3-1.8) | .58 | 0.7 (0.4-1.2) | .22 |
| Headache | 0.9 (0.5-1.9) | .85 | 1.0 (0.6-1.7) | .99 |
| Myalgias | 0.3 (0.1-0.8) | .02 | 0.6 (0.3-1.3) | .22 |
| Diarrhea | 1.2 (0.6-2.5) | .55 | 0.7 (0.4-1.3) | .28 |
Results are for 195 of 215 patients. Nine patients with no symptom survey data and 11 patients with no data regarding seasonal allergies were excluded from these analyses. Each result was adjusted for the following covariates: other respiratory viruses within the last week, day relative to transplantation, age at transplantation, pretransplantation allergies, conditioning regimen, donor type, and grades 2 to 4 acute graft-versus-host disease.
— indicates not applicable.
There were too few occurrences of coronavirus to include as a predictor for wheezing. Ear pain and watery eyes were too rare for analysis.
All 6 patients with HRV and 3 patients with HCoV prolonged shedding for more than 12 weeks were symptomatic during most periods of viral detection. The most common symptoms among the patients with HRV were rhinorrhea (concurrent with 64% of all samples with HRV detected; range for individual patients, 4%-100%), cough (45%; range, 7%-100%), postnasal drip (43%; range, 0%-100%), and sinus congestion (28%; range, 7%-33%). The most frequently reported symptoms among patients with HCoV prolonged shedding were cough (63%; range, 25%-100%), rhinorrhea (56%; range, 28%-100%), sputum production (30%; range, 0%-85%), and postnasal drip (26%; range, 0%-64%). Notably, wheezing was not a frequent symptom among prolonged shedders (8% for HRV, 0% for HCoV).
No significant relationships between HRV or HCoV and mean values for alanine aminotransferase, aspartate aminotransferase, and total bilirubin were found in a multivariable linear regression model of log-transformed values, adjusted for other respiratory viruses within the last week, day relative to transplantation, age at transplantation, transplantation type, donor type, and grades 2 to 4 acute GVHD.
Symptoms and respiratory viral load
For HRV, RT-PCR threshold cycle values were used as a proxy measure of viral load. Forty-two patients had concomitant respiratory symptom data and threshold cycle values available. The median threshold cycle value corresponding to 2 or more symptoms (31.9; range, 25.0-39.4) was significantly lower than the median threshold cycle value corresponding to no or one symptom (34.8; range, 30.1-39.5, P = .04). Among patients with HCoV, 20 had concomitant symptom and quantitative viral load data available. The RT-PCR viral load level corresponding to 2 or more symptoms (median, 5.2 log10 copies/mL; range, 3.2-7.1 log10 copies/mL) was higher but not significantly different from the viral load that corresponded to no or one symptom (median, 4.9 copies/mL; range, 2.9-7.4 log10 copies/mL, P = .40). The RT-PCR viral load level among 13 patients with 2 or more samples (median, 5.7 log10 copies/mL; range, 3.6-7.1 log10 copies/mL) was higher than for those 7 patients with only one sample (median, 3.5 log10 copies/mL; range, 2.9-7.4 log10 copies/mL, P = .02). Although 10 of 13 (77%) patients with more than one sample also had at least 2 respiratory symptoms, compared with 3 of 7 (43%) with only one sample, this association was not statistically significant (P = .17).
Discussion
This prospective surveillance study of HRV and HCoV infections in a large cohort of allogeneic HCT recipients provides new insights about the incidence and clinical course of these infections early after HCT. The day 100 cumulative incidence estimates for HRV and HCoV were high at 22% and 11%, compared with our previous and current estimates for PIV, RSV, HMPV, and influenza of approximately 7% to 18%, 6% to 7%, 2% to 6%, and 3% to 4%, respectively.9 Therefore, rhinoviruses are the most common virus type detected; and coronaviruses are second, surpassing or at least comparable with PIVs in incidence. Not surprisingly, these estimates of infection exceed results from other studies in which conventional viral culture was used for viral detection.6,12,27 Even our estimates are probably low because we changed HRV assays near the end of the study period and were able to detect at least an additional 10% of HRV infections with the newer assay. Most of the patients were diagnosed in the outpatient department. Half of the HRV and HCoV infections demonstrated viral shedding for more than 3 weeks, and we observed prolonged periods of symptomatic viral shedding for more than 3 months in 6 (13%) patients with HRV and 3 (14%) patients with HCoV. This finding confirms previous reports of frequent prolonged shedding among immunosuppressed patients.13,28 We found that HCoVs exhibited a winter seasonality, consistent with other reports noting a predominance of coronavirus infections in winter months.15,29 For HRVs, however, there was no seasonal preference, also consistent with several reports using both conventional and molecular detection methods that have demonstrated HRVs to cause infections year-round.30–32
Despite the high incidence of HRV and HCoV infection among HCT recipients, the resultant morbidity and mortality were relatively low in our cohort. Asymptomatic detections were common, accounting for 13% and 41% of persons with HRV and HCoV detections, respectively. These were all detected in the outpatient department, and the implications for infection control policies, with possible transmission from asymptomatic patients, are unknown. A recent study of HRV transmission within families suggests that symptoms are necessary for HRV transmission because no asymptomatic subjects transmitted virus to their household contacts.33 During the period of this study at our center, we did not require isolation of patients who were asymptomatic with either HRV or HCoV detected in upper respiratory samples by RT-PCR. In approximately 20% of cases of HRV and HCoV infection, these viruses were detected in association with another respiratory virus, suggesting that codetection is a common occurrence. However, we found no association with HRV or HCoV infection and the incidence of concomitant or subsequent secondary bacterial or fungal infections.
We used a rigorous multivariable model to evaluate symptoms associated with HRV and HCoV infection and found that only HRV infection was associated with respiratory symptoms, including rhinorrhea, sinus congestion, postnasal drip, sputum production, and cough, whereas HCoV infection was not. These findings were not affected when the definition of infection was limited to patients who had more than one positive sample. We found no significant association between either virus and mean hepatic enzyme or bilirubin values. For HRV, the presence of 2 or more symptoms compared with 0 or one symptom was significantly associated with a lower PCR threshold cycle value, corresponding to higher levels of detectable virus. Similar results have been seen for hospitalized patients with rhinovirus13 and HCT recipients with RSV, PIV, HMPV, and influenza infection during the first 100 days after HCT,9 suggesting that the quantity of respiratory virus directly affects the clinical symptoms associated with illness. Although we did not find a significant association with HCoV viral load and symptoms, viral loads were significantly higher among those patients with 2 or more samples positive for HCoV than those with only one sample, suggesting that persistent virus positivity may be associated with higher virus quantity.
Progression to LRI with either HRV or HCoV was uncommon. Two (4%) of the 45 persons with HRV progressed to LRI before day 100, and these 2 patients died with HRV as the most probable cause of their respiratory failure. None of the patients in our study developed LRI attributable to HCoV during the study period, although one patient did have HCoV detected with HMPV and influenza A in a BAL on day 101. The contribution of HCoV to this patient's illness is unclear because a follow-up BAL on day 110, 2 days before her death, was positive only for HMPV and influenza A. Other authors have reported 3 cases of fatal pneumonia attributed to HCoV infection in immunosuppressed patients17,18,34; in one case, HCoV was detected on initial BAL but apparently cleared on subsequent BAL and autopsy sample.18 A recent study by Garbino et al found that HRV and HCoV were the most common respiratory viruses found in 91 of 522 BAL samples from hospitalized adults, representing 23% and 32% of positive cases.35 Of the 522 samples analyzed, 55% were from lung transplantation recipients and 7% from solid organ or HCT recipients.35 These reports and our data suggest that HCoV may occasionally cause lower respiratory tract disease in HCT recipients, but its significance as a lower respiratory tract pathogen is probably less than that of RSV, influenza, PIV, and HMPV.
Although PCR detection of viral RNA does not prove that there was active shedding of replication-competent virus, culture techniques for these viruses, particularly HCoVs, are relatively poor. Molecular methods are gaining increasing importance in clinical practice. The sensitivity of RT-PCR allows for a more complete assessment of the role these viruses play in human disease. Another limitation of our study is the lack of sequencing data for HRV to analyze whether we have observed chronic shedding of the same virus or serial infections by multiple strains. Although it is possible that a new infection may have occurred, we attempted to create definitions for shedding duration that required relatively frequent intervals for uninterrupted positive virus detections, even several months after transplantation. Using these definitions, we noted that patients with HCoV were positive for the same virus type for the entire duration of shedding. We did note that the 3 patients with HCoV infection with viral shedding for more than 3 months were all infected with HCoV-NL63. It is possible that prolonged shedding may be associated with viral subtype, but we are unable to answer this question with our relatively small sample size of HCoV infections, and we did not have subtyping available for infection with HRV.
In conclusion, we have demonstrated that HRV and HCoV infections occurred in one-third of HCT recipients during the first 100 days after transplantation. The incidence of infection was high, but the rate of pneumonia and death attributable to these viruses was low compared with other common respiratory viruses, such as RSV, PIV, influenza, and HMPV. HRV, but not HCoV, was associated with the presence of mild respiratory symptoms. In our cohort, HRV infection was the probable factor contributing to lower respiratory disease and death in 4% of patients infected with this virus. Although we did not evaluate for other conditions associated with long-term morbidity such as airflow obstruction, it seems probable that HRV, in particular, may trigger an immune response leading to airway decline. Given the frequency of infection with HRV and HCoV in this highly immunosuppressed population, further study is needed to evaluate the effect on respiratory illness and long-term sequelae of infection.
Acknowledgments
The authors thank Cheryl Callais, Christi Dahlgren, Victor Chow, Deborah Reyes, Denise Della, Srilata Remala, Noelle Gichohi, Gynevill Jolly, Jennifer Klein, Jessica Johnson, and Hilary Stempel for study coordination and assistance; Craig Silva, Chris Davis, and Gary Schoch for database services; and Nancy Wright, Terry Stevens-Ayers, Tera Matson, Sam Chatterton-Kirchmeier, Reggie Sampoleo, Rohit Shankar, and Anne Cent for laboratory expertise.
This work was supported by National Institutes of Health (grants CA 18029, HL081595, and K23HL091059, L40 AI071572, and K24HL093294). A.P.C. also received support from the MedImmune Pediatric Fellowship Grant Award and the Pediatric Infectious Diseases Society Fellowship Award funded by MedImmune.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.M. analyzed and interpreted data and wrote the manuscript; A.P.C. performed the research, collected data, analyzed and interpreted data, and wrote the manuscript; K.A.G. provided statistical expertise, analyzed data, and critically reviewed the manuscript; J.K. developed PCR assays, performed PCR testing, and critically reviewed the manuscript; J.A.E. contributed to the analysis plan and critically reviewed the manuscript; L.C. critically reviewed the manuscript and provided resources for the study; and M.B. designed and performed the research, collected data, analyzed data, and critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela Campbell, Seattle Children's Hospital, 4800 Sand Point Way NE, Mailstop R-5441, Seattle, WA 98105; e-mail: angela.campbell@seattlechildrens.org.
References
- 1.Heugel J, Martin ET, Kuypers J, Englund JA. Coronavirus-associated pneumonia in previously healthy children. Pediatr Infect Dis J. 2007;26(8):753–755. doi: 10.1097/INF.0b013e318054e31b. [DOI] [PubMed] [Google Scholar]
- 2.Hicks LA, Shepard CW, Britz PH, et al. Two outbreaks of severe respiratory disease in nursing homes associated with rhinovirus. J Am Geriatr Soc. 2006;54(2):284–289. doi: 10.1111/j.1532-5415.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- 3.Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185(9):1338–1341. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols WG, Guthrie KA, Corey L, Boeckh M. Influenza infections after hematopoietic stem cell transplantation: risk factors, mortality, and the effect of antiviral therapy. Clin Infect Dis. 2004;39(9):1300–1306. doi: 10.1086/425004. [DOI] [PubMed] [Google Scholar]
- 5.Nichols WG, Gooley T, Boeckh M. Community-acquired respiratory syncytial virus and parainfluenza virus infections after hematopoietic stem cell transplantation: the Fred Hutchinson Cancer Research Center experience. Biol Blood Marrow Transplant. 2001;7(suppl):11S–15S. doi: 10.1053/bbmt.2001.v7.pm11777098. [DOI] [PubMed] [Google Scholar]
- 6.Martino R, Porras RP, Rabella N, et al. Prospective study of the incidence, clinical features, and outcome of symptomatic upper and lower respiratory tract infections by respiratory viruses in adult recipients of hematopoietic stem cell transplants for hematologic malignancies. Biol Blood Marrow Transplant. 2005;11(10):781–796. doi: 10.1016/j.bbmt.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whimbey E, Champlin RE, Couch RB, et al. Community respiratory virus infections among hospitalized adult bone marrow transplant recipients. Clin Infect Dis. 1996;22(5):778–782. doi: 10.1093/clinids/22.5.778. [DOI] [PubMed] [Google Scholar]
- 8.La Rosa AM, Champlin RE, Mirza N, et al. Adenovirus infections in adult recipients of blood and marrow transplants. Clin Infect Dis. 2001;32(6):871–876. doi: 10.1086/319352. [DOI] [PubMed] [Google Scholar]
- 9.Peck AJ, Englund JA, Kuypers J, et al. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood. 2007;110(5):1681–1688. doi: 10.1182/blood-2006-12-060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. Parainfluenza virus infections after hematopoietic stem cell transplantation: risk factors, response to antiviral therapy, and effect on transplant outcome. Blood. 2001;98(3):573–578. doi: 10.1182/blood.v98.3.573. [DOI] [PubMed] [Google Scholar]
- 11.Ison MG, Hayden FG, Kaiser L, Corey L, Boeckh M. Rhinovirus infections in hematopoietic stem cell transplant recipients with pneumonia. Clin Infect Dis. 2003;36(9):1139–1143. doi: 10.1086/374340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh S, Champlin R, Couch R, et al. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin Infect Dis. 1999;29(3):528–532. doi: 10.1086/598627. [DOI] [PubMed] [Google Scholar]
- 13.Gerna G, Piralla A, Rovida F, et al. Correlation of rhinovirus load in the respiratory tract and clinical symptoms in hospitalized immunocompetent and immunocompromised patients. J Med Virol. 2009;81(8):1498–1507. doi: 10.1002/jmv.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutman JA, Peck AJ, Kuypers J, Boeckh M. Rhinovirus as a cause of fatal lower respiratory tract infection in adult stem cell transplantation patients: a report of two cases. Bone Marrow Transplant. 2007;40(8):809–811. doi: 10.1038/sj.bmt.1705827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119(1):e70–e76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 16.Pene F, Merlat A, Vabret A, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis. 2003;37(7):929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest. 1999;115(3):901–905. doi: 10.1378/chest.115.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oosterhof L, Christensen CB, Sengelov H. Fatal lower respiratory tract disease with human corona virus NL63 in an adult haematopoietic cell transplant recipient. Bone Marrow Transplant. 2009 Oct 12; doi: 10.1038/bmt.2009.292. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Holloway B, Dare RK, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46(2):533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuypers J, Wright N, Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J Clin Virol. 2004;31(2):123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuypers J, Wright N, Ferrenberg J, et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44(7):2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuypers J, Wright N, Corey L, Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33(4):299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl Infect Dis. 2009;11(4):298–303. doi: 10.1111/j.1399-3062.2009.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koc S, Leisenring W, Flowers ME, et al. Therapy for chronic graft-versus-host disease: a randomized trial comparing cyclosporine plus prednisone versus prednisone alone. Blood. 2002;100(1):48–51. doi: 10.1182/blood.v100.1.48. [DOI] [PubMed] [Google Scholar]
- 25.Flowers ME, Kansu E, Sullivan KM. Pathophysiology and treatment of graft-versus-host disease. Hematol Oncol Clin North Am. 1999;13(5):1091–1112. doi: 10.1016/s0889-8588(05)70111-8. [DOI] [PubMed] [Google Scholar]
- 26.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 27.Ljungman P, Ward KN, Crooks BN, et al. Respiratory virus infections after stem cell transplantation: a prospective study from the Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation. Bone Marrow Transplant. 2001;28(5):479–484. doi: 10.1038/sj.bmt.1703139. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser L, Aubert JD, Pache JC, et al. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med. 2006;174(12):1392–1399. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- 29.Leung TF, Li CY, Lam WY, et al. Epidemiology and clinical presentations of human coronavirus NL63 infections in Hong Kong children. J Clin Microbiol. 2009;47(11):3486–3492. doi: 10.1128/JCM.00832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.du Prel JB, Puppe W, Grondahl B, et al. Are meteorological parameters associated with acute respiratory tract infections? Clin Infect Dis. 2009;49(6):861–868. doi: 10.1086/605435. [DOI] [PubMed] [Google Scholar]
- 31.Monto AS, Sullivan KM. Acute respiratory illness in the community: frequency of illness and the agents involved. Epidemiol Infect. 1993;110(1):145–160. doi: 10.1017/s0950268800050779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigl JA, Puppe W, Meyer CU, et al. Ten years' experience with year-round active surveillance of up to 19 respiratory pathogens in children. Eur J Pediatr. 2007;166(9):957–966. doi: 10.1007/s00431-007-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypia T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197(3):382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 34.van Kraaij MG, van Elden LJ, van Loon AM, et al. Frequent detection of respiratory viruses in adult recipients of stem cell transplants with the use of real-time polymerase chain reaction, compared with viral culture. Clin Infect Dis. 2005;40(5):662–669. doi: 10.1086/427801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garbino J, Soccal PM, Aubert JD, et al. Respiratory viruses in bronchoalveolar lavage: a hospital-based cohort study in adults. Thorax. 2009;64(5):399–404. doi: 10.1136/thx.2008.105155. [DOI] [PubMed] [Google Scholar]