Abstract
Identifying the targets of immune response after allogeneic hematopoietic cell transplantation (HCT) promises to provide relevant immune therapy candidate proteins. We used protein microarrays to serologically identify nucleolar and spindle-associated protein 1 (NuSAP1) and chromatin assembly factor 1, subunit B (p60; CHAF1b) as targets of new antibody responses that developed after allogeneic HCT. Western blots and enzyme-linked immunosorbent assays (ELISA) validated their post-HCT recognition and enabled ELISA testing of 120 other patients with various malignancies who underwent allo-HCT. CHAF1b-specific antibodies were predominantly detected in patients with acute myeloid leukemia (AML), whereas NuSAP1-specific antibodies were exclusively detected in patients with AML 1 year after transplantation (P < .001). Complete genomic exon sequencing failed to identify a nonsynonymous single nucleotide polymorphism (SNP) for NuSAP1 and CHAF1b between the donor and recipient cells. Expression profiles and reverse transcriptase–polymerase chain reaction (RT-PCR) showed NuSAP1 was predominately expressed in the bone marrow CD34+CD90+ hematopoietic stem cells, leukemic cell lines, and B lymphoblasts compared with other tissues or cells. Thus, NuSAP1 is recognized as an immunogenic antigen in 65% of patients with AML following allogeneic HCT and suggests a tumor antigen role.
Introduction
After HLA-identical allogeneic hematopoietic cell transplantation (HCT), hematologic malignancies are cured via graft-versus-leukemia (GVL) effects that target minor histocompatibility antigens (mHAs) and tumor antigens. Besides the beneficial GVL response, allogeneic immune responses frequently damage normal recipient tissues, causing graft-versus-host disease (GVHD). A more extensive characterization of mHAs responsible for GVL and GVHD will lead to an ability to augment GVL response and improve GVHD monitoring and guide immune suppression. Thus far, allogeneic immune responses after HCT have predominantly been characterized as T-cell responses,1–5 providing limited numbers of mHAs.6 Alternatively, this study shows B-cell responses after allogeneic HCT can be characterized as specific new antibody responses from peripheral blood using high-throughput protein microarray technology.
Previous work demonstrated allogeneic antibodies develop against multiple mHAs encoded on the Y chromosome, called H-Y antigens, after male patients undergo HCT using female hematopoietic grafts.7–9 Sex-mismatched HCT studies suggest allogeneic B-cell responses against ubiquitously expressed H-Y antigens may play a role in both GVHD and GVL. Furthermore, rituximab treatment specifically targeting CD20 on B cells provides therapeutic benefit for many patients with chronic GVHD (cGVHD).10–14 This study extends allogeneic B-cell analysis beyond H-Y antigens to test for novel antibody (Ab) development against 5056 human proteins.
A patient with acute myeloid leukemia (AML) who relapsed twice and remained with persistent disease underwent myeloablative HLA-identical unrelated donor allogeneic HCT and had blood prospectively collected through 18 months after transplantation. The patient remains disease-free 2.5 years after transplantation, suggesting a benefit from GVL responses. Two novel targets, nucleolar and spindle-associated protein 1 (NuSAP1) and chromatin assembly factor 1, subunit B (p60; CHAF1b), were identified serologically using protein microarrays as antigens newly recognized 1 year after transplantation but absent in the donor or pretransplantation plasma. Subsequent enzyme-linked immunosorbent assay (ELISA) testing of 120 HCT patient samples collected 1 year after transplantation from patients with different malignancies showed Ab against NuSAP1 and CHAF1b predominately developed in patients with AML. Gene expression profiles showed NuSAP1 and CHAF1b were highly expressed in CD34+CD90+ hematopoietic stem cells (HSCs), leukemic cell lines, and AML primary tumors. Together, exclusive development of NuSAP1 in AML and high expression of NuSAP1-specific Ab in 24 of 37 patients with AML suggests NuSAP1 is a clinically relevant AML tumor antigen.
Methods
Patients
Plasma samples were obtained from a 40-year-old female patient with AML before transplantation, after transplantation (1, 2, 11, 12, 14, 16, and 18 months), and from the respective male donor after allogeneic HCT. She underwent myeloablative conditioning therapy using cyclophosphamide and total body irradiation with her AML French-American-British (FAB) classification as M4 and with PR3 10% blasts at the time of transplantation. Her cytogenetics profile was normal and her AML did not arise from multilineage dysplasia. She developed extensive cGVHD 12 months after HCT with 75% skin erythema, fasciitis of 50% skin, oral pharyngeal moderate ulceration, and liver function abnormalities with a maximum aspartate aminotransferase (AST) of 4.59 μkat/L (275 U/L) and an alanine aminotransferase (ALT) of 2.51 μkat/L (150 U/L).
Plasma samples were also obtained from 120 patients (Table 1) 1 year after transplantation and from 70 healthy persons matched for age and sex for validation studies using quantitative IgG ELISA for CHAF1b and NuSAP1. The samples were cryopreserved at −80°C until further use. A total of 6 patients were followed longitudinally over time; their characteristics are given in Table 2. Approval was obtained from the Stanford institutional review board for these studies, and individual informed consent for further studies was obtained from all patients and donors in accordance with the Declaration of Helsinki.
Table 1.
Patient characteristics
| HCT patient characteristics | n = 120 |
|---|---|
| Median patient age, y (range) | 49 (19-66) |
| Median donor age, y (range) | 48 (15-74) |
| Primary disease | |
| Acute lymphoblastic leukemia | 9 (8%) |
| Acute myelogenous leukemia | 37 (31%) |
| Chronic lymphocytic leukemia | 8 (7%) |
| Chronic myelogenous leukemia | 10 (8%) |
| Hodgkin disease | 2 (1%) |
| Myelodysplastic syndrome | 8 (7%) |
| Myelofibrosis | 3 (2%) |
| Multiple myeloma | 18 (15%) |
| Mantle cell lymphoma | 8 (7%) |
| Non-Hodgkin lymphoma | 13 (11%) |
| Severe aplastic anemia | 4 (3%) |
| Conditioning | |
| Myeloablative | 52 (43%) |
| Nonmyeloablative | 68 (57%) |
| Donor | |
| Related–HLA identical | 92 (77%) |
| Unrelated–HLA identical | 25 (21%) |
| Unrelated–HLA mismatch | 3 (2%) |
| Graft | |
| Bone marrow | 8 (7%) |
| Peripheral blood stem cell | 112 (93%) |
Patient characteristics of 120 patients with various leukemic diseases are described. These patients were screened for NuSAP1 and CHAF1b antibodies using quantitative IgG ELISA.
Table 2.
Patient characteristics for 6 AML patients followed longitudinally after transplantation
| Patient | Patient sex | Donor sex | Patient age | FAB | Conditioning regimen | Graft | Donor | cGVHD |
|---|---|---|---|---|---|---|---|---|
| 1 | M | F | 44 | M2 | FTBI/CY | PB | URD | Y |
| 2 | M | F | 65 | M2 | TLI/ATG | PB | URD | Y |
| 3 | M | F | 64 | M2 | TLI/ATG | PB | URD | Y |
| 4 | M | M | 40 | M5 | FTBI/VP16 | PB | SIBL | Y |
| 5 | M | M | 57 | M6 | TLI/ATG | PB | URD | Y |
| 6 | M | M | 56 | NOS | BU/VP16/CY | PB | SIBL | Y |
Six AML patients were followed longitudinally for detection and development of H-Y, NuSAP1, and CHAF1b antibodies and IgG levels.
FTBI indicates fractionated total body irradiation; CY, cyclophosphamide; TLI, total lymphoid irradiation; ATG, anti-thymoglobulin; PB, peripheral blood; URD, unrelated donor; and SIBL, sibling.
Peripheral blood mononuclear cells (PBMNCs) from 12 newly diagnosed patients with AML and 5 healthy persons were also obtained for reverse transcriptase–polymerase chain reaction (RT-PCR) studies. All 12 AML samples had greater than 95% blasts, which were isolated from leukapheresis products (Table 3).
Table 3.
Patient characteristics for 12 AML patients for RT-PCR
| Sample | Age/sex | Primary/secondary | De novo/relapsed | Cytogenetics | Flt-3 ITD | % CD34 | FAB | Notes |
|---|---|---|---|---|---|---|---|---|
| 1 | 53/F | Primary | Relapsed | 46, XX, t(5;15;12;12) | Unknown | 50 | M5 | |
| 2 | 56/M | Secondary | De novo | 45, XY, t(8;9)(q24;q22),-21 | Negative | 52 | NS | Secondary to chronic eosinophilic leukemia |
| 3 | 47/M | Primary | De novo | Normal | Negative | 68 | M5 | |
| 4 | 40/F | Primary | De novo | Normal | Normal | 90 | M5 | |
| 5 | 77/F | Primary | De novo | 46, XX, der(7)t(1;7)(q25;q31.2) | Negative | 87 | M4 | FLT3 TKD positive |
| 6 | 59/M | Primary | De novo | Normal | Positive | 18 | NS | |
| 7 | 64/M | Primary | De novo | Normal | Positive | 3 | M1 | |
| 8 | 39/M | Primary | Relapsed | Normal | Positive | 27 | M4 | NPM positive |
| 9 | 61/F | Primary | De novo | 46, XX, t(10;11)(p11.2–12;q23) | Negative | 8 | M5 | MLL positive |
| 10 | 39/F | Primary | De novo | Normal | Positive | 1 | M4 | NPM positive |
| 11 | 57/F | Primary | De novo | Normal | Negative | 4 | M5 | NPM positive |
| 12 | 46/M | Primary | De novo | Failed to grow | Negative | 98 | M5 | FISH negative for monosomy 5,7 and trisomy 8 |
Twelve patients were screened by RT-PCR for NuSAP1 and CHAF1b gene expression analysis. The patient characteristics are described.
RT-PCR indicates reverse transcription–polymerase chain reaction; TKD, tyrosine kinase domain; NPM, nucleophosmin; FAB, French-American-British; MLL, mixed lineage leukemia; and FISH, fluorescence in situ hybridization.
Plasma profiling using protein arrays
Protein microarrays were obtained (ProtoArrays Version 3.0) displaying 5056 full-length human proteins, derived from the Ultimate ORF collection, printed in duplicate with N-terminal GST epitopes expressed in baculovirus and affinity-purified under native conditions, maintaining their cellular enzymatic activities/native conformations. The arrays also contained control spots consisting of buffer, empty spots, and human IgG printed in 4 different concentrations (IgG[1] to IgG[4]). The arrays were incubated with blocking buffer (50mM HEPES [pH 7.5], 200mM NaCl, 0.08% Triton X-100, 25% glycerol, 20mM reduced glutathione, 1.0mM DTT, 10M NaOH (pH of blocking buffer, 7.5-8.0), and 1% bovine serum albumin [BSA]) for 1 hour followed by application of the plasma sample diluted 1:150 for 90 minutes. After washing the array 4 times for 10 minutes each with PBST buffer (1× PBS, 1% BSA, and 0.1% Tween 20), secondary antibody was added (goat anti–human Alexa 647, diluted 1 μg/mL in PBST buffer) for 90 minutes. After washing the slides with PBST buffer, the arrays were dried and fluorescence intensity was detected using a microarray scanner (GenePix 4000B). All incubations were carried out on a rotating platform at 4°C.
Data acquisition
The slides were scanned using a GenePix scanner (GenePix 4000B) at a photomultiplier (PMT) gain of 60% with a laser power of 90% and a focus point of 0 μm. The .gal file was obtained from the ProtoArray central portal on the Invitrogen website by submitting the barcode of each ProtoArray. Data were obtained using GenePix software (Version 5 Microarray Image Analysis) and was normalized and converted to log scale (“Statistical methods”) and represented by plotting the pretransplantation normalized values on the x-axis versus the posttransplantation normalized values on the y-axis. The data discussed in this publication have been deposited in the National Center for Biotechnology Information's (NCBI's) Gene Expression Omnibus (GEO)15 and are accessible through GEO Series accession number GSE15255.16
Statistical methods
All intensities above 500 were considered positive based on the average intensity readings for negative controls plus 3 SDs. Data obtained from protein microarrays were analyzed as follows.
(1) Normalization using R.
Results were normalized using algorithms constructed in R for intraslide variation (comparing duplicate spot results) and interslide variation using the intensity reading obtained for spotted human IgG3 (details in supplemental information, available on the Blood website; see the Supplemental Materials link at the top of the online article). The normalized posttransplantation fluorescent readings were subtracted from the pretransplantation readings and ranked in descending order with the highest difference as the most significant hit. The top-ranked proteins were then compared with the donor array normalized fluorescent intensities, and only those proteins absent in the donor were called significant hits.
(2) Prospector Analyzer (Invitrogen).
(a) This software uses the Chebyshev inequality P value, which is derived by testing the null hypothesis. The pretransplantation, 1-month, and 2-month samples were compared versus 11-, 12-, 14-, 16-, and 18-month samples to obtain significant protein hits. (b) The software also calculates the z score for each printed spot's fluorescent intensity. The z score indicates the deviation of each protein's antibody reading from its distribution mean (SD). Once the z scores were obtained, the delta between the pretransplation and posttransplantion z scores was calculated and compared with the donor z scores to identify significant targets or hits.
(3) SAM analysis using the t test.
Normalized signal intensity values were used for significance analysis of microarrays (SAM) analysis. The samples were grouped as pretransplantation, 1-month, and 2-month samples versus 11-, 12-, 14-, 16-, and 18-month samples.
Optical density values obtained after ELISA for NuSAP1 and CHAF1b were compared using the Fisher exact t test for conditioning regimens, acute GVHD and cGVHD, related/unrelated donor, and relapse. Antibodies against NuSAP1 and CHAF1b in patients with AML versus all other hematologic diseases were analyzed using the Kruskal-Wallis χ2 test, Wilcox test, and Fisher exact t test. Patient and donor age and IgG levels were treated as continuous, multivariate data and compared in Table 4 using the Kruskal-Wallis χ2 test.
Table 4.
Only AML patients develop NuSAP1 Ab and CHAF1b antibody after HCT
| Yes | No | P | |
|---|---|---|---|
| NuSAP1 antibody | n = 24 | n = 96 | |
| Median patient age, y | 46 | 50 | .88 |
| Median donor age, y | 40 | 50 | .44 |
| Conditioning | .36 | ||
| Myeloablative | 13 | 40 | |
| Nonmyeloablative | 11 | 56 | |
| Donor | .16 | ||
| Related | 16 | 78 | |
| Unrelated | 8 | 18 | |
| IgG levels | 907 mg/dL | 643 mg/dL | < .001 |
| Acute GVHD | 1.00 | ||
| 0-I | 20 | 81 | |
| II-IV (15%) | 4 | 15 | |
| Chronic GVHD | .64 | ||
| Yes (66%) | 17 | 62 | |
| No | 7 | 34 | |
| Relapse | .007 | ||
| Yes (28%) | 1 | 29 | |
| No | 23 | 67 | |
| Disease | < .001 | ||
| AML | 24 | 13 | |
| Other diseases | 0 | 83 | |
| CHAF1b antibody | n = 10 | n = 110 | |
| Median patient age, y | 40 | 50 | .44 |
| Median donor age, y | 48 | 48 | .70 |
| Conditioning | .75 | ||
| Myeloablative | 5 | 48 | |
| Nonmyeloablative | 5 | 62 | |
| Donor | .052 | ||
| Related | 5 | 87 | |
| Unrelated | 5 | 23 | |
| IgG levels | 850 mg/dL | 682 mg/dL | < .001 |
| Acute GVHD | 1.00 | ||
| 0-I | 9 | 93 | |
| II-IV (15%) | 1 | 17 | |
| Chronic GVHD | .49 | ||
| Yes (66%) | 8 | 71 | |
| No | 2 | 39 | |
| Relapse | 1.00 | ||
| Yes (28%) | 2 | 29 | |
| No | 8 | 81 | |
| Disease | .001 | ||
| AML | 8 | 29 | |
| Other diseases | 2 | 81 |
One hundred twenty patients were screened for antibodies against NuSAP1 and CHAF1b. The patients were segregated into 2 groups. The first group shows patients with antibody and the second group was detected with no antibody responses. These 2 groups are correlated with variables as shown.
Protein purification
Full-length CHAF1b and NuSAP1 clones were obtained from the Ultimate ORF clone collection and expressed using Escherichia coli expression system with Gateway technology. The Ultimate ORF clones were transferred from pENTR 221 entry vector to pDEST-15 expression vector with an LR recombination reaction using the Gateway LR Clonase enzyme mix. Transformed BL21-AI One Shot cells were induced for protein production with L-arabinose. Cells were lysed, and protein expression was confirmed in the soluble portion of the lysate using Western blotting with anti-GST mAb. The recombinant proteins were then expressed and purified from BL21-AI–transformed cells using B-PER GST Fusion Protein Purification Kits. Because all patients had tested negative for HIV, HIV-p24 protein was used as a negative control and was also expressed in E coli and purified in a similar fashion. Insect-expressed CHAF1b and NuSAP1 were obtained from Invitrogen.
Western blotting
E coli–derived or baculovirus-infected insect cell–derived purified proteins (0.5 μg/lane) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto Hybond C+ membranes. Proteins were detected with either anti-GST (1:1000) or patient plasma diluted 1:500. After washing, the membranes were incubated with secondary antibody goat anti–human IgG conjugated to horseradish peroxidase. The membranes were washed and visualized by enhanced chemiluminescence (ECL).
ELISA for antibodies to recombinant proteins
Baculovirus-infected insect-derived purified proteins were diluted to 2.0 μg/mL, and 50 μL/well of each protein was coated per well in 96-well ELISA plates. The plates were washed with 1× Tris-buffered saline with 0.1% Tween-20 (TBST) and blocked with 2% nonfat milk for 1 hour. Anti-GST (1:1000) or patient plasma samples (1:50) were incubated overnight at 4°C. Antibodies were detected with goat anti–human IgG conjugated to alkaline phosphatase. Absorbance was measured at 450 to 550 nm.
Flow cytometric analysis and sorting of the leukocyte subsets
Briefly, 10 × 106 cells from G-CSF–mobilized leukapheresis products or bone marrow fine-needle aspirates from consenting allogeneic donors were stained with the respective leukocyte subset Abs for 30 minutes. Cells were sorted with a Cytopeia InFlux high-speed cell sorter equipped with 488 nm Coherent Sapphire and 635 nm Coherent Radius lasers. Separated subsets were reanalyzed for purity greater than 98%. CD34+CD90+ HSC cells were sorted from purified CD34+ cells (Isolex 300i) obtained from mobilized peripheral leukapheresis products. The anti-human monoclonal antibodies used were all purchased from Becton Dickinson: phycoerythrin (PE)– or allophycocyanin (APC)–labeled CD3 (clone SK7), fluorescein isothiocyanate (FITC)–labeled anti-CD4 (clone RPA-T4), APC- or phycoerythrin-Cy7 (PE-Cy7)–labeled anti-CD19 (clone SJ25C1), PE-Cy7–labeled anti-CD8 (clone RPA-T8), APC-, PerCP-, or FITC-labeled anti-CD45 (clone 2D1), allophycocyanin-Cy7–labeled anti-CD14 (clone MphiP9), PE-labeled anti-CD34 (clone 8G12), and APC-labeled anti-CD90 (Thy1; clone 5E10).
RT-PCR analysis of CHAF1b and NuSAP1 mRNA expression
A total of 1 × 106 cells sorted after flow cytometry were lysed using TRIzol reagent and stored at −80°C until further use. Total RNA was isolated as per the manufacturer's instructions (Qiagen) and was treated with DNAse. RNA concentration and purity were measured spectrophotometrically and used for quantification of target mRNAs. A total of 20 ng of RNA was reverse-transcribed to cDNA using Superscript II Reverse Transcriptase and random primers (0.2 μg/reaction). Primers were designed for CHAF1b and NuSAP1 (CHAF1b forward: TTCAGTCAGAGACGCCTGGA; CHAF1b reverse: GCTTTAGCTCTGGGGGACTG; NUSAP1 forward: AAACTTACAAACAACCCCATCTCC; NUSAP1 reverse GTTTCTTCGGTTGCTCTTCCTTT). The RT-PCR analysis was performed with SYBR green PCR core reagents in a 7900HT Fast Real-Time PCR Cycler, and the data were analyzed using Applied Biosystems Sequence Detection Software Version 2.3. Relative message levels were calculated with a comparative threshold cycle (Ct) method by which message levels were normalized to endogenous actin message levels.
Results
NuSAP1 and CHAF1b were identified as new antibody targets after allo-HCT
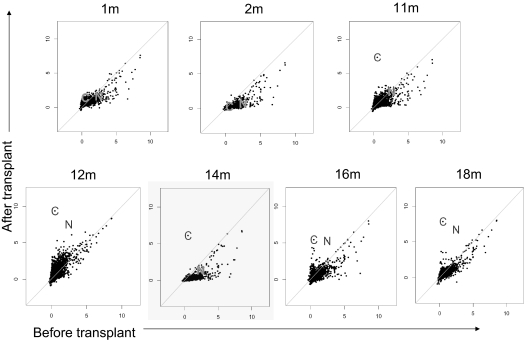
Plasma was collected from a patient with AML before transplantation, after transplantation (1, 2, 11, 12, 14, 16, and 18 months), and from her unrelated donor. Plasma was incubated with protein microarrays that displayed 5056 full-length human proteins with N-terminal GST epitopes expressed in baculovirus-infected insect cells and affinity-purified under native conditions.17 Because proteins were printed in duplicate, the florescent intensity readings for antibodies against each protein were averaged, logged, and plotted in Figure 1. Longitudinal analysis of patient antibody responses identified new antibodies detected 1 year after transplantation. For the first and second months, all pretransplantation readings for proteins were either equal to or greater than any posttransplantation readings. New antibody responses developed 11 months after transplantation and peaked with highest signal intensities at 12 months. At 1 year after HCT, the patient developed cGVHD, and 2 proteins were targeted with high allo-Ab signals: CHAF1b and NuSAP1 (indicated by “C” and “N,” respectively, in subsequent figures). The patient was treated for cGVHD with high-dose steroids and both total IgG as well as antigen-specific Ab binding diminished except for CHAF1b. Further, at 16 months, after tapering of steroids, the 12-month Ab profile recurred, with CHAF1b and NuSAP1 again the strongest Ab targets. The donor had low antibody responses against CHAF1b (signal intensity, 0.64) and no responses against NuSAP1, proving these are not adoptive immune responses but ones that develop newly after allogeneic HCT (supplemental Table 1).
Figure 1.
Allogeneic antibodies develop 1 year after HCT. Plasma samples were obtained from a patient with AML 1, 2, 11, 12, 14, 16, and 18 months after transplantation and before transplantation, and from the donor. Raw data from protein microarrays were normalized and plotted, with posttransplantation results plotted on the y-axis against the pretransplantation measurements on the x-axis. The 2 proteins identified as targets were CHAF1b and NuSAP1 (labeled “C” and “N,” respectively) and were first detected 11 months after transplantation. For the diagonal lines, the x-axis denotes the pretransplantation log-normalized fluorescent intensity and the y-axis denotes the posttransplantation log-normalized fluorescent intensity.
SAM software analysis also identified NuSAP1 and CHAF1b as the most significant Ab response after HCT as shown in supplemental Figure 1. t test results comparing normalized antibody results collected pretransplantation, 1 month, and 2 months after transplantation versus 11, 12, 14, 16, and 18 months after transplantation identified NuSAP1 and CHAF1b with 30- and 128-fold change, respectively (false discovery q value, 0). We also analyzed the data using company-specific software (Prospector Analyzer). Again, CHAF1b and NuSAP1 were the only 2 significant hits between the 2 groups of early and late transplantation time points (P = .018). Thus, 3 independent analyses identified CHAF1b and NuSAP1 as the most significant new Ab targets after allogeneic HCT (supplemental Figure 1).
CHAF1b and NuSAP1 Ab recognition was validated by Western blot and ELISA
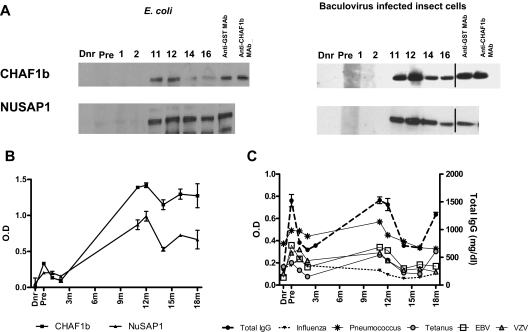
To confirm CHAF1b and NuSAP1 antibody recognition by independent methods, the proteins were expressed in eukaryotic and prokaryotic systems, and patient sera was tested by western blots and quantitative ELISA. GST affinity purification following insect cells and E coli protein expression yielded full-length highly purified NuSAP1 (55 kDa) and CHAF1b (60 kDa) with an expected 25-kDa GST addition. Anti-GST and anti-CHAF1b mAb detection confirmed protein purity (Figure 2A). Western blot detection of patient plasma reproduced protein microarray results. There was no recognition of either CHAF1b or NuSAP1 by donor plasma, the pretransplantation plasma, 1- or 2-month patient plasma, but specific recognition for both proteins developed 11 months after HCT and persisted thereafter.
Figure 2.
Validation of targets (CHAF1b and NuSAP1) by Western Blot and ELISA. Using protein microarrays, CHAF1b and NuSAP1 were identified as target proteins recognized by new antibodies that first developed 1 year after transplantation and were absent in the donor plasma or before transplantation in the patient, corroborating protein microarray results. CHAF1b and NuSAP1 were made using the E coli expression system as well as baculovirus-infected insect cells. (A) 0.5 μg protein was loaded in each well. Each lane was cut and incubated with the patient's pretransplantation (Pre) and posttransplantation plasma samples, along with the patient's donor plasma (Dnr). The probed lanes were realigned and detected by anti–human IgG Ab. Vertical lines have been inserted to indicate a repositioned gel lane. Positive controls were anti-GST mAb and anti-CHAF1b mAb. (B) Affinity purified NuSAP1 and CHAF1b obtained from E coli– and baculovirus-infected insect cells were quantitatively detected by patient plasma using IgG ELISA. There were no antibodies detected in the sera of donor, pretransplantation, or 1 month or 2 months posttransplantation, but antibodies against NuSAP1 and CHAF1b were detected 11 months after transplantation and later. (C) Antibodies against infectious antigens influenza, pneumococcus, tetanus, EBV, and VZV were detected in the donor, pretransplantation, and 1, 2, 11, 12, 14, 16, and 18 months after transplantation using IgG ELISA. Total IgG was also measured in triplicate for the same time points and is plotted as milligrams per deciliter. Error bars indicate SE of measurement.
ELISAs were developed using insect cell–expressed N-terminal GST fusion proteins. Figure 2B shows quantitative NuSAP1 and CHAF1b ELISAs fail to detect Ab in donor, pretransplantation, or 1- or 2-month samples, but ELISA quantification 11 months after transplantation and beyond reflect both Western blot qualitative Ab detection and protein microarray quantification. To assess whether NuSAP1 and CHAF1b Ab development reflected a specific alloimmune response or resulted from general Ab augmentation, we measured Ab specific for Epstein-Barr virus (EBV), varicella zoster virus (VZV), pneumococcus, influenza A, tetanus, and total IgG concentration (Figure 2C). Figure 2B and C show that antibody development against CHAF1b and NuSAP1 developed concurrently with antimicrobial Ab responses, suggesting broad immune reconstitution occurred 11 and 12 months after HCT. Total IgG levels and specific antimicrobial responses decrease dramatically after steroid therapy beginning at 12 months.
Antibodies against NuSAP1 and CHAF1b develop in patients with AML after HCT
To determine the frequency and intensity of NuSAP1 and CHAF1b antibody development after allogeneic HCT, plasma was collected 1 to 2 years after allo-HCT from 120 patients with a variety of malignancies (Table 1). These patients were screened by quantitative IgG ELISA for NuSAP1 and CHAF1b Ab. The threshold used for NuSAP1 and CHAF1b seropositivity was 3 SDs above the mean seropositivity of 70 healthy persons.
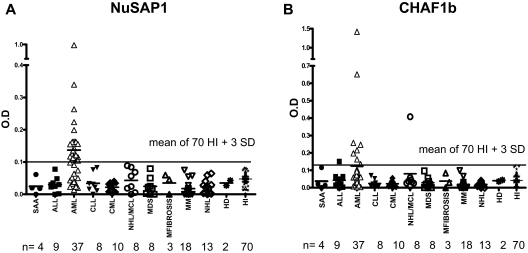
NuSAP1 Abs were exclusively detected in 24 (65%) of 37 patients with AML after allo-HCT compared with no patients with acute lymphoblastic leukemia (ALL), chronic lymphocytic leukemia (CLL), chronic myelogenous leukemia (CML), mantle cell leukemia (MCL), multiple myeloma (MM), and non-Hodgkin lymphoma (NHL; Wilcox test, P < .001; Table 4; Figure 3A). Abs against CHAF1b were detected in 8 (21%) of 37 patients but was associated with AML compared with other malignancies (Wilcox test, P < .001; Table 4; Figure 3B). Univariate analysis of NuSAP1 or CHAF1b seropositivity did not associate with GVHD risk factors (donor or patient age, related or unrelated donor, myeloablative or nonmyeloablative conditioning intensity), nor did NuSAP1 or CHAF1b Ab associate with acute or chronic GVHD development, but NuSAP1 Ab development was significant for disease relapse (P = .007) and increased IgG levels (P < .001). We used multivariate logistic regression to explain the development of NuSAP1 Ab using total IgG and AML status across 120 patients studied. AML disease was highly significant (P < .001) in the multivariate model, but total IgG levels did not associate with NuSAP1 Ab development (P = .7). Further exploring the univariate association between total IgG and NuSAP1 Ab development, we found patients with AML had higher total IgG levels compared with other hematologic malignancies. We suspect the lower IgG levels measured in ALL and NHL result from their preceding prolonged chemotherapy and prednisone treatment. Our further analysis of total IgG levels of 24 patients with AML with NuSAP1 Ab development and 13 patients without NuSAP1 Ab showed there was no difference between the 2 AML groups. This confirms the multivariate analysis that there is an association of NuSAP1 Ab development with AML, but no association with total IgG among all the 120 patients 1 year after transplantation for NuSAP1 Ab. To demonstrate immune competence in NuSAP1-seronegative patients, we tested for the development of allogeneic Ab in these 36 HCT male patients with female donors. A total of 17 (47%) of 36 developed H-Y antibodies, confirming their ability to develop new antibody responses despite remaining NuSAP1 seronegative.
Figure 3.
Only patients with AML develop NuSAP1 Ab and CHAF1b Ab after HCT. (A-B) NuSAP1 and CHAF1b are predominant in patients with AML compared with other diseases/leukemia as identified by quantitative IgG ELISA. OD readings that tested positive were higher than the mean of 70 healthy persons plus 3 SDs (solid line across the graphs).
Pretransplantation plasma samples from 17 of these 37 patients with AML were available for testing, and none were seropositive for NuSAP1 (supplemental Figure 2). Only 4 pretransplantation samples from an additional 33 patients with AML tested NuSAP1 seropositive. Overall, only 4 (8%) of 50 patients with AML tested NuSAP1 seropositive before transplantation compared with 24 (65%) of 37 patients with AML after transplantation (P < .001). This frequent and exclusive development of NuSAP1 Ab in patients with AML suggests NuSAP1 is an AML-specific tumor antigen resulting from new Ab development after allogeneic HCT.
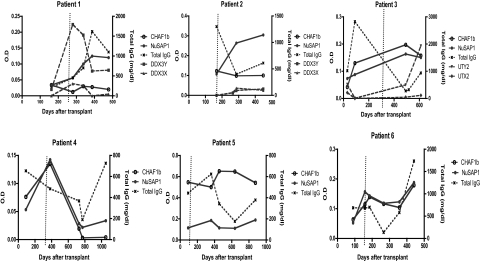
To determine when NuSAP1 Ab develops, longitudinal samples of 6 patients with AML (Table 2) who developed NuSAP1 Ab were tested by ELISA. All 6 remained seronegative through the first 150 days, and their NuSAP1 Ab peaked between 350 to 450 days after HCT. To determine whether NuSAP1 Ab development reflected general Ab augmentation, Figure 4 compares NuSAP1 and CHAF1b, with total IgG measurements. NuSAP1 Ab development does not develop in association with increase in total IgG. NuSAP1 Ab developed in patients 2, 3, and 4 when IgG was decreasing. A total of 3 male patients with female donors (patients 1, 2, and 3) were tested for the development of allogeneic H-Y Ab.7 H-Y Ab against DDX3Y developed in patient 1 while patient 3 developed UTY Ab, but their development was temporally distinct from NuSAP1 Ab. In contrast, Ab against CHAF1b did develop concurrently with NuSAP1 in 3 patients (3, 4, and 6), suggesting their Ab development may share a common eliciting event such as AML tumor lysis. Furthermore, no autoimmune Ab developed against 19 autoantigens (supplemental Figure 3), suggesting NuSAP1 and CHAF1b Ab developed in an antigen-specific manner. In summary, the development of NuSAP1 and CHAF1b Ab in these 6 patients does not reflect augmentation of total antibody quantity.
Figure 4.
Development of H-Y, NuSAP1, and CHAF1b antibodies and IgG levels. A total of 6 patients were followed longitudinally after transplantation. Antibody intensities against NuSAP1, CHAF1b, H-Y (namely DDX3Y/X and UTY/X for patients 1-3, female donors with male recipients), and total IgG levels are plotted. The dotted vertical line represents cGVHD development. As observed, NuSAP1 antibodies peak between days 350 to 450. NuSAP1 Ab development does not correlate with total IgG levels, indicating that NuSAP1 Ab responses are specific and not due to general Ab augmentation.
NuSAP1 and CHAF1b amino acid sequences are identical in the donor and recipient
We hypothesized that NuSAP1 and CHAF1b immunogenicity resulted from immune responses targeting disparate polymorphic amino acids (ie, expression of an allele in the recipient that is absent in the donor cells). We exon-sequenced both CHAF1b and NuSAP1 and failed to detect any nonsynonymous SNPs (nsSNPs) in the 13 CHAF1b and 11 NuSAP1 exons. Interestingly, in the last exon 11 of NuSAP1, the recipient's stop codon was TAG compared with the donor's TAA stop codon. Although both are stop codons for the protein NuSAP1, read-through by selenium incorporation can result from TAG codon and would add 4 amino acids, making this protein immunogenic in the recipient to the donor cells. We have no experimental evidence this occurred in our patient. In addition, the patient's new Ab response recognized the full-length NuSAP1 presented on the protein microarray.
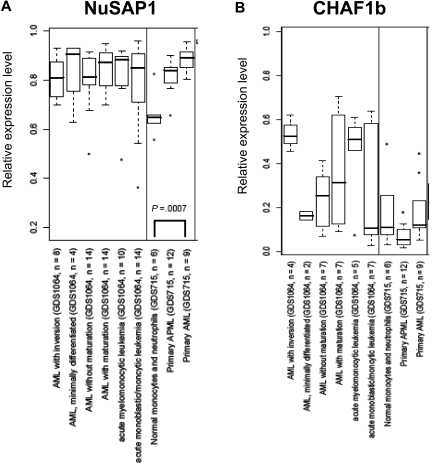
NuSAP1 gene expression is in the top quartile of AML gene repertoire
An alternative cause of NuSAP1 and CHAF1b immunogenicity hypothesized could be their aberrant expression or increased expression in AML cells. Data from GEO18 were analyzed for CHAF1b and NuSAP1 gene expression relative to normal neutrophils. In Figure 5A and B, the x-axis indicates a separate group/subset of samples from 5 different GEO datasets that were analyzed because they included large numbers of well-characterized AML samples studied by experienced DNA microarray laboratories. NuSAP1 is statistically higher in AML samples than in the healthy samples within each experiment. However, no correlation was found for CHAF1b and patients with AML. This overexpression of NuSAP1 in AML further indicates that NuSAP1 could be a potential tumor-associated antigen compared with CHAF1b.
Figure 5.
NuSAP1 gene expression is in the top quartile of AML gene repertoire. Data obtained from 5 independent experiments in GEO is graphed as a box and whisker plot for NuSAP1 (A) and CHAF1b (B). The x-axis indicates a separate group/subset of samples from each GEO dataset (GDS). The GDS accession number for each is also shown. The y-axis indicates the normalized level of expression of the respective mRNA. These GDSs were picked because they were among the largest (in terms of number of arrays) experiments related to AML.
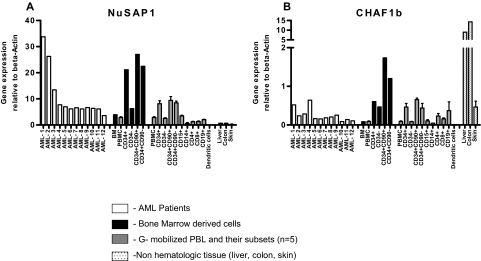
NuSAP1 is expressed predominantly in the peripheral blood of patients with AML by RT-PCR
To directly confirm whether NuSAP1 or CHAF1b were expressed in patients with AML, RT-PCR was standardized using primers specific for NuSAP1 and CHAF1b with plasmids from previous transfections in E coli. The gene expression results of the 2 proteins were normalized to an internal control β-actin and are represented as fold change (Figure 6A-B). PBMNCs from 12 patients with AML with greater than 95% blasts demonstrated high heterogeneous NuSAP1 gene expression. In contrast, all 12 AML samples demonstrated low gene expression of CHAF1b (Figure 6A-B). After sorting 5 healthy donor GM-CSF–mobilized PBMNCs into T and B cells, we found no relative difference between CD3, CD4, or CD8 T cells; CD19 B cells; CD15 granulocytes; CD14 monocytes; or monocyte-derived dendritic cells for either gene expression. Both Nusap1 and CHAF1b gene expression was relatively increased in undifferentiated progenitor cells, specifically CD34+CD90+ and CD34+CD90− cell populations. Increased expression of NuSAP1 and CHAF1b in CD34+ progenitor blood cells is similarly reported in virtual online gene expression studies.19–21 Skin, liver, and colon are frequently affected by GVHD and were also analyzed for NuSAP1 and CHAF1b. NuSAP1 was not detected in these GVHD tissues. In contrast, CHAF1b was highly expressed, especially in liver and colon. In conclusion, the high NuSAP1 gene expression in AML cells and its frequent and exclusive antibody development after allo-HCT suggests NuSAP1 may be an AML tumor–associated antigen.
Figure 6.
NuSAP1 and CHAF1b are highly expressed in CD34+CD90+HSCs as demonstrated by RT-PCR quantification. One bone marrow biopsy was sorted by flow cytometry into CD34+, CD34−, CD34+CD90+, and CD34+CD90− subpopulations and screened for NuSAP1 (A) and CHAF1b (B) expression. A total of 12 AML PBMNC samples and GM-CSF–mobilized peripheral blood samples (n = 5) were screened, and these samples were further sorted into CD34+, CD34−, CD34+CD90+ and CD34+CD90−, CD15, CD14, CD4, CD8, CD19, and dendritic cell subpopulations. DNA was obtained from all subpopulations and reverse-transcribed to cDNA using OligodT primers. cDNA was screened for NuSAP1 and CHAF1b expression using the respective specific primers using RT-PCR. Results obtained by RT-PCR were plotted relative to the positive control gene β-actin. Error bars indicate SE of measurement.
Discussion
Reports demonstrate the identification of mHAs or tumor antigens using serologic expression methodologies like SEREX22,23 or chimeric antigen receptors of tumor-specific T lymphocytes coupled with phage display library24 after transplantations. We hypothesized that novel mHAs could be serologically identified as targets of antibody responses that develop after allogeneic transplantations and are absent before transplantation. More than 70 000 nsSNPs encode polymorphic amino acid residues in human proteins that are potential mHAs, and their systemic analysis as possible mHAs requires a high-throughput approach like protein microarrays. To determine whether targets of new antibody responses can be detected using protein microarray technology, we longitudinally followed a single patient with cGVHD for new Ab development after allogeneic HCT.
With the advantage of screening 5056 proteins simultaneously using high-density protein microarrays, we serologically identified 2 proteins, NuSAP1 and CHAF1b, as specific Ab targets that were high in the posttransplantation plasma compared with pretransplantation or donor plasma in a patient with AML and developed 1 year after HCT. We then determined if the antibody responses against NuSAP1 and CHAF1b after allogeneic HCT correlated with disease type. Of the 120 patients with various hematologic diseases, NuSAP1 antibodies were frequently and exclusively detected in 24 (65%) of 37 patients with AML, suggesting NuSAP1 development required AML exposure and may be a GVL antigen. In support of this GVL role, NuSAP1 Ab development correlated with decreased disease relapse (P = .007).
Both NuSAP1 and CHAF1b are intracellular proteins with increased expression in rapidly proliferating cells.25,26 NuSAP1 expression peaks at G2-M cell-cycle transition, and associates with microtubule formation while localizing to nucleoli during interphase.27 Recent studies support NuSAP1 as a transcriptional target gene of c-Myc, and also suggests a novel mechanism by which c-Myc promotes proliferation by stabilizing the mitotic spindle in fast-dividing cells via NuSAP1 and a nucleolar RNA methyltransferase Misu.25,28 Our virtual gene expression analysis of published data and RT-PCR analysis of primary AML samples determined NuSAP1 has increased expression in AML, suggesting a role in AML cell proliferation, and may lead to antibody induction via DNA or RNA association, as many autoantibody targets have their immunogenicity ascribed to DNA/RNA-dependent antigen presentation.29,30
CHAF1 is a heterotrimeric protein composed of 3 subunits, CHAF1a, CHAF1b, and CHAF1c, comprising p150, p60, and p48, subunits respectively.31 It was shown that deletion of CHAF1b abolished chromatin assembly,32 and was shown to be a marker for cellular proliferation distinguishing cells from the quiescent state.26 CHAF1b is highly expressed in cGVHD target tissues (liver and colon), but our serologic testing of 120 allogeneic HCT patients did not support an association with cGVHD development. Virtual gene expression studies and our RT-PCR studies show CHAF1b expression is increased in progenitor HSCs, but a GVH role for CHAF1b after allo-HCT remains speculative, requiring further studies.
We hypothesized new Ab development against NuSAP1 and CHAF1b might be due to nsSNPs between the donor and the patient, resulting in single amino acid disparities. However, complete exon sequencing revealed no differences between the donor and the recipient. For NuSAP1, the donor and the patient were identical for all codons except the last exon 11, where the recipient had a TAG amino acid SNP and the donor had a TAA. Although TAA and TAG are stop codons, due to the change in the reading code, the stop codon could be translated to incorporate amino acids like tryptophan, glutamine, or selenocysteine, thus causing disparity between the donor and recipient cells, eliciting an immune response.33,34 However, we have no evidence supporting TAG-dependent read-through or the incorporation of additional amino acid residues. In fact, the protein microarray and our ELISA presented full-length NUSAP1. Thus, our measured NuSAP1 antibody responses targeted the full-length protein.
NuSAP1 and CHAF1b were identified as new Ab targets despite their intracellular localization.27,31,32 Recent literature has identified that HSCs express leukemia-associated antigens, correlated their expression with stage of the disease for CML, and also showed that mHAs were ubiquitously present in the CD34+ population.35–37 Our data are in concordance with the literature where intracellular targets, especially on HSCs, are targeted. It could be as proposed by Jordan and Kubler that nuclear antigens may be found on the cell surface when damage occurs to cells and intracellular antigens are exposed and stick to the cell surface, or they could be isoforms of cell nucleus–related proteins, or just are found in response to physiologic stress responses.29 Furthermore, it could be hypothesized that CHAF1b- and NuSAP1-increased expression in rapidly proliferating cells results in “accidental exposure” on the AML cell surface. However, neither CHAF1b monoclonal Ab or NuSAP1 Ab detected cell-surface staining on primary AML cells, GM-CSF–mobilized PBMNCs, or bone marrow CD34+CD90+-sorted cells (data not shown).
The relatively exclusive development of CHAF1b and NuSAP1 Ab in patients with AML (with their expression absent before transplantation) supports a tumor antigen role due to aberrant or increased expression in patients with AML, stimulating new Ab responses after transplantation. In support of this, our virtual gene expression profiles showed CHAF1b and NuSAP1 are predominately expressed in leukemic cell lines and B lymphoblasts compared with other tissues/cells (Symatlas). Directly testing this, our RT-PCR results showed NuSAP1 is highly expressed in primary AML cells. Thus, we propose NuSAP1 as a novel tumor-associated antigen since antibodies against NuSAP1 were exclusively and frequently detected in 24 (65%) of 37 patients with AML after allogeneic HCT. The specificity of NuSAP1 Ab development in patients with AML suggests a tumor antigen etiology and not generalized dysregulated immune reconstitution. Most important, NuSAP1 antibody development was associated with persistent disease remission (P = .007), again supporting a GVL role. Our observation that patients with NuSAP1 Ab had a greater total IgG quantity (9.07 g/L) than seronegative patients (6.43 g/L; P < .001) suggests NuSAP1-seropositive patients have improved immune reconstitution, and that this improved immune reconstitution may contribute to improved disease remission.
Our study demonstrates global assessment of alloimmunity after HCT can be achieved by combining the advantages of antibodies and protein microarray technology. The advantages of using antibodies for screening are that new Abs can discriminate mHAs, can freely diffuse throughout the body, making blood testing appropriate, and can be cryopreserved, allowing extensive sample analysis. Protein microarray technology advantages include concurrent screening across thousands of proteins, multiplexed data acquisition, and analysis. The proteins printed can reflect varied tissue organs, and no cell culture or viral mitogens are required. Protein microarrays are quantitative, reproducible, and rapid. Protein microarray technology was used to identify differentially expressed proteins in ovarian cancer, and further validated their ovarian antigen tumor antigen discovery by demonstrating increased antigen expression via tissue microarrays.38 The significance of antibody development against NuSAP1 and CHAF1b are limited by testing blood samples collected 1 year after HCT because this requires 1 year of survival, preventing the analysis of all patients dying from disease relapse during the first year after transplantation. Nonetheless, NuSAP1 Ab development may predict for decreased disease relapse and longer clinical follow-up of this 120-patient cohort, especially for the ongoing studies for the 37 patients with AML. Like other technologies, protein microarray is a nascent technology which has limitations. These include the potential for Ab cross-reactivity, absence of alternate splice variants, purity of proteins printed, and varying protein concentrations, leading to false-positive results. Thus, validation of targets by various other technologies is important. Our study shows serologic screening with protein microarrays for new Ab responses can identify disease-specific, clinically relevant tumor antigens.
Acknowledgments
We acknowledge Dr Ravi Majeti for providing 11 newly diagnosed AML blood samples, Dr Howard Chang for providing skin cDNA samples, and Dr Edgar Engelman for providing dendritic cells obtained from monocytes. We also thank Dr Ruby Wang and Ms Linda Elder for their help in assimilating patient information; the staff of Stanford Cellular Therapeutics and Transplantation Laboratory for their technical assistance in procuring patient samples; and the Stanford Genome Technology Center for genotyping services, especially Ms Julie Wilhelmy and Ms Sujatha Krishnakumar. D.B.M. is an American Society of Hematology (ASH) Translational Research Scholar and American Society for Blood and Marrow Transplantation (ASBMT) Young Investigator.
This study was supported by National Institutes of Health (NIH) research grants T32 A107290-21 and PO1 CA049605.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.P.W. and D.B.M. designed and performed research, and collected and analyzed data; P.P.W., M.C., A.J.B., and D.B.M. interpreted data and performed statistical analysis; R.J.A. and M.M. contributed vital new reagents and analytical tools; and P.P.W. and D.B.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Miklos, Division of Blood and Marrow Transplantation, 269 West Campus Dr, CCSR 2205, Stanford University Medical Center, Stanford, CA 94305-5623; e-mail: dmiklos@stanford.edu.
References
- 1.Rosinski KV, Fujii N, Mito JK, et al. DDX3Y encodes a class I MHC-restricted H-Y antigen that is expressed in leukemic stem cells. Blood. 2008;111(9):4817–4826. doi: 10.1182/blood-2007-06-096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terakura S, Murata M, Warren EH, et al. A single minor histocompatibility antigen encoded by UGT2B17 and presented by human leukocyte antigen-A*2902 and -B*4403. Transplantation. 2007;83(9):1242–1248. doi: 10.1097/01.tp.0000259931.72622.d1. [DOI] [PubMed] [Google Scholar]
- 3.Spierings E, Vermeulen CJ, Vogt MH, et al. Identification of HLA class II-restricted H-Y-specific T-helper epitope evoking CD4+ T-helper cells in H-Y-mismatched transplantation. Lancet. 2003;362(9384):610–615. doi: 10.1016/S0140-6736(03)14191-8. [DOI] [PubMed] [Google Scholar]
- 4.Mommaas B, Kamp J, Drijfhout JW, et al. Identification of a novel HLA-B60-restricted T cell epitope of the minor histocompatibility antigen HA-1 locus. J Immunol. 2002;169(6):3131–3136. doi: 10.4049/jimmunol.169.6.3131. [DOI] [PubMed] [Google Scholar]
- 5.Mutis T, Blokland E, Kester M, Schrama E, Goulmy E. Generation of minor histocompatibility antigen HA-1-specific cytotoxic T cells restricted by nonself HLA molecules: a potential strategy to treat relapsed leukemia after HLA-mismatched stem cell transplantation. Blood. 2002;100(2):547–552. doi: 10.1182/blood-2002-01-0024. [DOI] [PubMed] [Google Scholar]
- 6.Goulmy E. Minor histocompatibility antigens: from transplantation problems to therapy of cancer. Hum Immunol. 2006;67(6):433–438. doi: 10.1016/j.humimm.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Miklos DB, Kim HT, Zorn E, et al. Antibody response to DBY minor histocompatibility antigen is induced after allogeneic stem cell transplantation and in healthy female donors. Blood. 2004;103(1):353–359. doi: 10.1182/blood-2003-03-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zorn E, Miklos DB, Floyd BH, et al. Minor histocompatibility antigen DBY elicits a coordinated B and T cell response after allogeneic stem cell transplantation. J Exp Med. 2004;199(8):1133–1142. doi: 10.1084/jem.20031560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miklos DB, Kim HT, Miller KH, et al. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105(7):2973–2978. doi: 10.1182/blood-2004-09-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler C, Miklos D, Kim HT, et al. Rituximab for steroid-refractory chronic graft-versus-host disease. Blood. 2006;108(2):756–762. doi: 10.1182/blood-2006-01-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohty M, Marchetti N, El-Cheikh J, Faucher C, Furst S, Blaise D. Rituximab as salvage therapy for refractory chronic GVHD. Bone Marrow Transplant. 2008;41(10):909–911. doi: 10.1038/bmt.2008.12. [DOI] [PubMed] [Google Scholar]
- 12.Kebriaei P, Saliba RM, Ma C, et al. Allogeneic hematopoietic stem cell transplantation after rituximab-containing myeloablative preparative regimen for acute lymphoblastic leukemia. Bone Marrow Transplant. 2006;38(3):203–209. doi: 10.1038/sj.bmt.1705425. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Simon JA, Sanchez-Abarca I, Diez-Campelo M, Caballero D, San Miguel J. Chronic graft-versus-host disease: pathogenesis and clinical management. Drugs. 2006;66(8):1041–1057. doi: 10.2165/00003495-200666080-00002. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer M, Stanojevic S, Feuchtinger T, et al. Rituximab mediates in vitro antileukemic activity in pediatric patients after allogeneic transplantation. Bone Marrow Transplant. 2005;36(2):91–97. doi: 10.1038/sj.bmt.1705014. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database. [Accessed March 17, 2009]. Available at http://www.ncbi.nlm.nih.gov.
- 17.Mattoon D, Michaud G, Merkel J, Schweitzer B. Biomarker discovery using protein microarray technology platforms: antibody-antigen complex profiling. Expert Rev Proteomics. 2005;2(6):879–889. doi: 10.1586/14789450.2.6.879. [DOI] [PubMed] [Google Scholar]
- 18.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genomics Institute of the Novartis Research Foundation (GNF) BioGPS database. [Accessed December 7, 2008]. Available at http://biogps.gnf.org.
- 20.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brickner AG, Evans AM, Mito JK, et al. The PANE1 gene encodes a novel human minor histocompatibility antigen that is selectively expressed in B-lymphoid cells and B-CLL. Blood. 2006;107(9):3779–3786. doi: 10.1182/blood-2005-08-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krackhardt AM, Witzens M, Harig S, et al. Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood. 2002;100(6):2123–2131. doi: 10.1182/blood-2002-02-0513. [DOI] [PubMed] [Google Scholar]
- 24.Pameijer CR, Navanjo A, Meechoovet B, et al. Conversion of a tumor-binding peptide identified by phage display to a functional chimeric T cell antigen receptor. Cancer Gene Ther. 2007;14(1):91–97. doi: 10.1038/sj.cgt.7700993. [DOI] [PubMed] [Google Scholar]
- 25.Hussain S, Benavente SB, Nascimento E, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186(1):27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo SE, Theocharis SE, Klijanienko J, et al. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64(7):2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- 27.Raemaekers T, Ribbeck K, Beaudouin J, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162(6):1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28(2):446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan P, Kubler D. Autoimmune diseases: nuclear autoantigens can be found at the cell-surface. Mol Biol Rep. 1995;22(1):63–66. doi: 10.1007/BF00996307. [DOI] [PubMed] [Google Scholar]
- 30.Carl PL, Temple BR, Cohen PL. Most nuclear systemic autoantigens are extremely disordered proteins: implications for the etiology of systemic autoimmunity. Arthritis Res Ther. 2005;7(6):R1360–1374. doi: 10.1186/ar1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 32.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81(7):1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 33.Gesteland RF, Atkins JF. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura Y, Ito K. How protein reads the stop codon and terminates translation. Genes Cells. 1998;3(5):265–278. doi: 10.1046/j.1365-2443.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 35.Norde WJ, Overes IM, Maas F, et al. Myeloid leukemic progenitor cells can be specifically targeted by minor histocompatibility antigen LRH-1-reactive cytotoxic T cells. Blood. 2009;113(10):2312–2323. doi: 10.1182/blood-2008-04-153825. [DOI] [PubMed] [Google Scholar]
- 36.Barrett AJ. Understanding and harnessing the graft-versus-leukaemia effect. Br J Haematol. 2008;142(6):877–888. doi: 10.1111/j.1365-2141.2008.07260.x. [DOI] [PubMed] [Google Scholar]
- 37.Spaapen R, Mutis T. Targeting haematopoietic-specific minor histocompatibility antigens to distinguish graft-versus-tumour effects from graft-versus-host disease. Best Pract Res Clin Haematol. 2008;21(3):543–557. doi: 10.1016/j.beha.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104(44):17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]