Abstract
While automatic spike sorting has been investigated for decades, little attention has been allotted to consistent evaluation criteria that will automatically determine whether a cluster of spikes represents the activity of a single cell or a multiunit. Consequently, the main tool for evaluation has remained visual inspection by a human. This paper quantifies the visual inspection process. The results are well-defined criteria for evaluation, which are mainly based on visual features of the spike waveform, and an automatic adaptive algorithm that learns the classification by a given human and can apply similar visual characteristics for classification of new data. To evaluate the suggested criteria, we recorded the activity of 1652 units (single cells and multiunits) from the cerebrum of 12 human patients undergoing evaluation for epilepsy surgery requiring implantation of chronic intracranial depth electrodes. The proposed method performed similar to human classifiers and obtained significantly higher accuracy than two existing methods (three variants of each). Evaluation on two synthetic datasets is also provided. The criteria are suggested as a standard for evaluation of the quality of separation that will allow comparison between different studies. The proposed algorithm is suitable for real-time operation and as such may allow brain–computer interfaces to treat single cells differently than multiunits.
1. Introduction
Voltage on chronically implanted extracellular electrodes, such as depth electrodes or electrode arrays, can be generated by the electrical activity of one or more neurons per electrode. In the latter cases, it is usually desirable to separate the spikes of each single neuron from the rest of the spikes. This is commonly obtained by signal-processing methods, known as spike detection and sorting, whose ideal goal is to isolate the spikes and relate each of them to the original neuron that created it (Lewicki 1998, Herbst et al 2008, Benitez and Nenadic 2008, Fletcher et al 2008, Ding and Yuan 2008, Bar-Hillel et al 2006, Shoham et al 2003, Zhang et al 2004, Zouridakis and Tam 2000, Yang and Shamma 1988). In some cases, however, relating the spikes to single cells is impossible. In these cases, one may group together, for each electrode, all spikes that cannot be separated into their generating single neurons and call these groups multiple unitary potentials, or multiunits.
Notwithstanding, many of the automatic spike sorting algorithms report their classification as clusters of spikes, with no distinction between single neurons and multiunits, and rely on a manual decision of their type (single cell or multiunit). In addition, there are no clear, objective and agreed-upon criteria for making this type of decision. Vargas-Irwin and Donoghue (2007, p 1) state that ‘a stereotypical spike shape is generally the major or even sole characteristic used to verify that a set of waveforms is attributable to a single neuron’. Stark and Abeles (2007), for example, determined well-isolated single units ‘by the homogeneity of spike waveforms, separation of the projections of spike waveforms onto principal components during spike sorting, and clear refractory periods in interspike interval histograms’. Suner et al (2005) and Kim et al (2008) were based on distinguishing waveforms with the naked eye and on a quantitative evaluation using the signal-to-noise ratio (SNR). The spike shape and the interspike interval (ISI) distribution were used only in relative measures of reliability: their change over time between days of recording for each electrode. Kim et al (2006) define multiunits as the remainder of unclassifiable spike activities following classification by mixture-of-t-distributions (Shoham et al 2003). Few groups assessed the quality of clustering by comparing the within-cluster variability with that of the noise (Pouzat et al 2002, Schmitzer-Torberta et al 2005, Joshua et al 2007).
The lack of uniform criteria for the evaluation of the quality of separation (i.e. single neuron or multiunit) causes multiple flaws in processing of neural data. First, different studies may employ different criteria for evaluation of the quality of separation. Second, even with the same general criteria, the results may be inconsistent between experimenters, because the final decision is still manual. Third, when recording from multiple channels, manual decisions may be time consuming. Fourth, decisions based on manual revision of the data are inappropriate for automatic processing systems such as brain–computer interfaces (BCI; also called brain–machine interfaces (BMI) or neuroprostheses). The goal of this paper is to define criteria for evaluation of the quality of separation between clusters of spikes in a quantitative way that will allow an automatic implementation.
The quality of separation is affected by variation in the spike waveform or ISI distribution which is, in turn, affected by a large number of factors such as systematic reduction in the spike amplitude during a high-frequency burst (Harris et al 2001), systematic drift in the electrode position (Snider and Bonds 1998), systematic changes to single-unit spike waveforms as a function of the ISI (i.e. dependence on the firing history) (Fee et al 1996b, Kaneko et al 2007) and hardware-level filters (Quiroga 2009). However, instead of modeling the exact phenomena affecting the quality of separation (Quirk and Wilson 1999), we define measures of evaluation that mimic a human making a manual decision. Human classifiers rely on visual inspection as their main evaluation tool (see for example Vargas-Irwin and Donoghue (2007), Rutishauser et al (2006), Tankus et al (2009), Stark and Abeles (2007), Suner et al (2005), Kim et al (2008)). We therefore bypass the need to characterize spike generation precisely, using a computer vision approach, that quantifies the visual properties of the waveforms of a cluster of spikes in a manner similar to the one used by a human classifier. This novel approach yields the multiple contributions of this paper. (1) We define a new ‘visual’ measure of the shape of a cluster of spikes. (2) Using this feature along with a requirement for a refractory period, we define an algorithm for evaluation of the quality of separation. (3) We suggest a methodology for automatic tuning of classification thresholds using linear discriminant analysis.
2. Materials and methods
2.1. Patients and electrophysiology
Subjects were 12 patients (15–42 years old (mean: 26.4 years; SD = 8.3), 5 right-handed females, 2 left-handed females, 4 right-handed males, 1 left-handed male) with pharmacologically resistant epilepsy. As non-invasive monitoring did not yield concordant data corresponding to a single resectable focus, the subjects were implanted with chronic depth electrodes for 1–2 weeks to determine the seizure focus for potential surgical treatment (Fried et al 1999, Engel et al 2005, Wilson 2004). Each patient had between 7 and 12 electrodes (surgeries were performed by IF). The electrodes were implanted based on clinical criteria only. Each of these clinical electrodes terminated with a set of nine 40 μm platinum–iridium microwires. Signals from these microwires were recorded at either ~28 kHz or ~30 kHz and bandpass filtered in the range 0.3–3~ kHz using a 64-channel acquisition system (Neuralynx, Tucson, AZ, USA). Spikes of individual cells were isolated based on their wavelet coefficients and the distribution of interspike intervals using the method suggested by Quian-Quiroga et al (2004) (original code courtesy of the original authors). This method was employed in multiple research studies (Mukamel et al 2005, Gelbard-Sagiv et al 2008, Quian-Quiroga et al 2005). Magnetic resonance images (MRI) following placement of the electrodes, or post-placement computed tomography scans, coregistered to preoperative MRI scans, were used to verify the anatomical location of the electrodes. Participation in this study was completely voluntary. All patients provided informed consent. All studies conformed with the guidelines of the Medical Institutional Review Board at the University of California, Los Angeles.
We recorded a total of 1652 units in the human cerebrum (temporal lobe: 790 units; frontal lobe: 739; occipital lobe: 63; parietal lobe: 60; see the supplementary data available at stacks.iop.org/JNE/6/4/056001/mmedia), of which 568 (34.4%) were single units and 1084 (65.6%) were multiunits according to manual classification (see section 2.3). The ratio of single cells to multiunits of about 1:2 respectively is common in our setup (see, for example, Quian-Quiroga et al (2005)), which suggests that the manual classification in the current study is similar in nature to other studies with the same recording setup and patient population.
The neural activity was recorded during 32 sessions, each on a different day. Units recorded from within different sessions are treated as different in this study. The total recording time in a session was between 10.7 and 50.6 min (average = 28.6, SD = 10.6). Within each session, activities of between 25 and 87 units were recorded (average = 51.6, SD = 16.3). In each session, we recorded between 0.39 and 1.36 units per microwire (average = 0.81, SD = 0.25).
2.2. The task
The patients performed two tasks. In 20 sessions, the task was to move a cursor in a simple 2D maze using an external device for control. This paradigm has been described by Tankus et al (2009). The other 12 sessions employed the center-out task described by Moran and Schwartz (1999), with the following adaptations to our setup. First, the patients used a joystick to control the cursor. Second, the hold times at the center of the screen and the peripheral targets were longer (between 2000–3000 ms and 200–400 ms, respectively), with maximal time allowed for the completion of a trial of 5000 ms.
2.3. Manual classification of single cells versus multiunits
The manual classification between single units and multiunits was done visually based on the following criteria (Tankus et al 2009, Gelbard-Sagiv et al 2008, Quiroga et al 2008): (1) the spike shape and its variance; (2) the ratio between the spike peak value and the noise level; (3) the interspike interval (ISI) distribution of each cluster and (4) the presence of a refractory period for the single units (that is, less than 1% of the inter-spike intervals are shorter than 3 ms). The recorded units were spike-sorted as they were recorded (over a period of 45 months), not in one continuous session (or on multiple consecutive dates).
2.4. Visual feature and algorithm for evaluation of the quality of separation
For every recording channel, the process of spike sorting, whether manual or automatic, results in a set of clusters of spikes. Each cluster is then inspected to decide whether it represents a single cell or a multiunit. This inspection, which we shall name evaluation of the quality of separation, is based on two main properties of the recorded signal: the spike waveform and the ISI distribution. The proposed algorithm, which we name single-unit–multi-unit (SUMU), first considers the ISI distribution for the requirement of a refractory period of single cells. A cluster is declared multiunit if the percent of ISIs shorter than 3 ms exceeds 1%. Otherwise, classification is performed according to the waveforms.
2.4.1. Evaluation of the waveform
The waveforms of a well-isolated cell are expected to present low variation around its main rise in voltage6. Our main visual feature will therefore evaluate the variation of the distribution of spike waveforms around the mean waveform of the cluster. For this, we first need to detect the main rise. While the end of the main rise is assumed to be supplied by the spike-sorting algorithm (due to alignment), finding its beginning accurately is challenging because of the high variability of characteristic waveforms of different neurons. To cope with the high variability, we first estimate upper and lower bounds for this starting point based on the local change in voltage (i.e. the derivative of the waveform). We then refine the detection based on the maximal curvature of the waveform between the two bounds. Once we detected the main rise, we can estimate the variability around it. The measure we define takes into account not only the standard deviation on the main rise but also the rate of rise.
The rest of this subsection details the detection of the main rise in voltage and the measure of deviation around it. To allow for both simple reading and mathematical insight, the latter appears in parentheses in the following.
Let us examine a cluster of suspected spikes for which we computed estimators m(t) and s(t) for the mean and standard deviation of the voltage at time t, respectively. Let be the relative sampling times of spike waveforms.
Detecting the main voltage rise
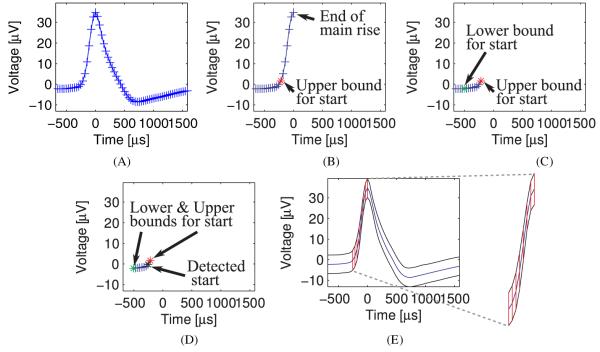
We next define a procedure for detection of the main rise in voltage and demonstrate it step-by-step in figure 1.
Figure 1.
The steps of detection of the main rise in voltage and the deviation around it for a single unit in the left vACC of case 3. (A) The mean waveform of the cluster. The plus (+) signs denote the mean waveform at sampling points. (B) The end of the main rise is at time 0 due to alignment by the spike-sorting algorithm. The upper bound for the start of the main rise is therefore at negative times only. See the text for definition of the upper and lower bounds and the estimated beginning of the main rise in voltage. (C) The lower bound, detected between the first sampling point and the upper bound. (D) The estimated beginning of main rise in voltage (based on curvature), between the lower and upper bounds. (E) The main rise in voltage. The area between the mean waveform and ± 1 standard deviation is marked on the main rise. This area is enlarged on the right-hand side of the figure. The marked area, weighed with respect to the height of the main rise, is a natural visual feature that a human classifier uses to evaluate deviation from the mean waveform (see the text for the exact definition of the feature).
The end of the main rise is assumed to have been computed by the spike-sorting algorithm, which usually aligns the detected spikes by their peak. Let us assume that the spike-sorting algorithm aligns all suspected spikes at the j th sample point and defines it as time 0 (that is, ). We therefore need to detect only the beginning of the main rise (which should be one of the sampling points: t1, . . . , tj−1 < 0) (figure 1(B)). We first estimate an upper bound for it, defined as the first point in time when the voltage change between successive points exceeded a certain (high) threshold (i.e. minimal i for which m(ti) − m(ti−1) > Thresholdhigh, where i ∈ {2, . . . , j − 1}; we used ) (figure 1(B)). If this bound is not established, we reject the cluster, as it is probably not representing a cluster of spikes. We then estimate a lower bound for the beginning of the rise by the last point in time preceding the upper bound, in which the change in voltage crosses a very low positive threshold. (Let iupper be the sampling point of the upper bound. Then, the lower bound ilow is the maximal i for which m(ti−1) − m(ti−2) ≤ Thresholdlow and m(ti) − m(ti−1) > Thresholdlow, where i ∈ {3, . . . , iupper}; we used .) (Figure 1(C).) We need to take the last point with such a change because the time period preceding the main rise may demonstrate small fluctuations in voltage. The use of an upper bound is aimed to prevent such fluctuations on the main rise itself from ‘shortening’ the main rise. In order to further refine the detection, we compute the extrinsic curvature of the mean waveform (m(ti)) between the lower and upper bounds (i ∈ {ilow, . . . , ihigh}) and estimate the beginning of the main rise in voltage as the point of maximal curvature (figure 1(D)). (We denote this estimate by icurv.)
A measure of deviation around the main rise
Once we isolated the main rise, we would like to test whether the deviation around it is low. Inspection of the standard deviation alone may however turn out to be misleading, because the faster the rise, the higher standard deviation one can tolerate, due to errors on the time axis: the vertical distance between the waveforms of two identical spikes shifted by one sample duration will be larger for a fast-rising spike than for a slow-rising one. Consequently, the threshold should be relative to the rate of rise, a good approximation of which is the change in voltage (neglecting the differences in the duration of the main rise). In other words, we are interested in the standard deviation of each point on the main rise, divided by the change of voltage at that point (s(ti)/(m(ti) − m(ti−1)) where i ∈ {icurv, . . . , j}). To average this measure over the whole period of the main rise, we sum the standard deviations over that period and divide it by the vertical length of the rise (a = m(tj) − m(ticurv)). (The average relative standard deviation along the main rise is therefore: b/a.) This is equivalent to dividing the area circumscribed by the average waveform and the average waveform plus 1 standard deviation by the vertical length of the average waveform (see figure 1(E)). Clusters having the average measure lower than a threshold (b/a < Thresholddeviation; we employed for the real data and for simulated data) are declared single units whereas the rest of the clusters are declared multiunits. Figure 2 provides two more examples of detection of the main rise in voltage and the deviation along it.
Figure 2.
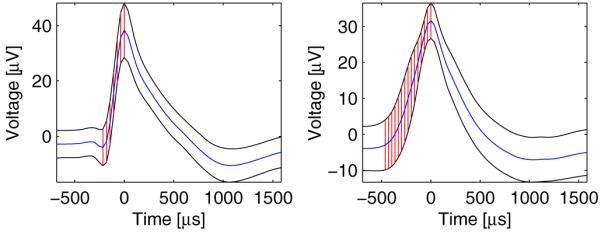
Detection of the main rise and the area circumscribed by the average waveform ±1 standard deviation in two recorded units. (Left) A single cell in the left dACC of case 3 with a small ‘valley’ at the beginning of the main rise. Despite the ‘valley’, the maximal curvature estimate is accurate and lies exactly at the beginning of the main rise. (Right) A multiunit in the left SMA proper of case 9. As expected from a multiunit, the marked area is larger than that on the left or figure 1(E).
2.4.2. Theoretical analysis of the visual feature
We analyzed the visual feature (b/a) under the assumptions that the main rise is linear, the distribution around it is normal and samples are pairwise uncorrelated (figure 3 (left)). We compared the main rise of a multiunit composed of such linear single units with an ‘equivalent’ single unit (i.e. a single unit with the same average main rise and the same average variance). Appendix A shows that in the general case, b/a is higher for the multiunit than for the ‘equivalent’ single unit.
Figure 3.
Linear main rise and relation to SNR. Center: the mean waveform (±1 standard deviation) of a single neuron in the left SMA of case 9. b/a = 2.18. Left: an example of a linear main rise (produced by linear interpolation of the main rise of the neuron in the center). Circles mark sampling points on the main rise. Right: lower peak with identical standard deviation (produced by scaling the mean amplitude of the neuron in the center by a factor of 0.25, without any change to the standard deviation). The SNR is obviously lower. b/a = 8.70 > 3, so the scaled unit would be classified as a multiunit. Visual inspection agrees with this classification.
Relation to SNR
The proposed feature b/a is inversely related to SNR, because b is the total standard deviation and a is the peak height (the signal). When comparing units with identical standard deviations (b), the method will classify units with smaller peaks (i.e. smaller a) as multiunits. In this case, 4 smaller peaks indeed imply lower SNR, as demonstrated in figure 3 (right).
Waveform alignment
The suggested way of detecting the main rise relies on the alignment of individual spikes at their peak (section 2.4.1). Appendix B explores this dependence by comparing b/a of a well-aligned single unit with that of a misaligned one under the assumption of a linear rise with a Gaussian distribution of spikes around it. It shows that under reasonable conditions (mainly that the variability in the spike shape prior to the rise is limited), b/a of the misaligned cluster is greater than that of the well-aligned one, implying that better alignment of spikes (e.g. Lewicki (1994), Wehr et al (1999)) would improve the suggested criterion.
2.5. Linear discriminant analysis—automatic learning of the threshold
A different methodology for classification of a cluster as a single cell or a multiunit is based on automatic learning of the threshold of the visual feature using linear discriminant analysis (LDA). This variant of SUMU, which we name SUMU-LDA, examines all possible divisions of the training set by a threshold on the visual feature, and finds the division with the highest accuracy with respect to single cell versus multiunit classification. The corresponding threshold is then used for predicting the quality of classification in the test set.
Linear discriminant analysis
Let {v1, v2, . . . , vk} be the unique values of the aforementioned visual feature in the training set, and let {g1, g2, . . . , gk} ∈ {0, 1} indicate whether each respective cluster is a single cell (1) or a multiunit (0). Let vs1 < vs2 < ⋯ < vsk be the sorted sequence of values (s1, s2, . . . , sk ∈ {1, 2, . . . , k). For each threshold , we define the prediction
The classification accuracy for threshold Thl is the percentage of elements i for which . The threshold Thl with maximal accuracy is denoted: ThLDA. Each cluster in the test set with visual feature v is classified as a single cell if v < ThLDA and as a multiunit otherwise.
2.6. Comparison with other methods
As mentioned in the introduction (section 1), most studies rely on visual inspection of the clusters to determine the quality of separation. One of the few studies suggesting clear criteria that can be implemented as an automatic method is that of Rutishauser et al (2006) with a method called online sorting (osort). We selected osort for comparison due to the similar setup for which it was developed (human epilepsy patients implanted with depth electrodes) and the availability of the original code. Osort uses three criteria to evaluate the quality of separation.
The fraction of ISIs shorter than 3 ms (specified in % of all ISIs). Osort requires this fraction to be less than 3% of all ISIs.
The absence of peaks in the power spectrum in the range 20–100 Hz. A peak is defined as a frequency whose power is more than 5 standard deviations away from the mean.
An approximately zero autocorrelation for small (<3 ms) time lags. The threshold for this criterion was defined neither in the paper (Rutishauser et al 2006) nor in the code. We therefore defined the threshold as autocorrelation <2, based on figure 7(C) of the original paper.
In order to match the first criterion with that of our manual classification, we examined two variants of this criterion. The first keeps the original requirement of the fraction of ISIs shorter than 3 ms to be <3% of all ISIs; we will call it osort3. The second requires that this fraction is <1% of all ISIs; we will call it osort1. In addition, we checked a variant of osort1 in which small autocorrelations are defined as those <1.5; it will be marked osort C. We also examined other thresholds on the autocorrelation: 0.25, 0.5 and 1, but in these cases all recorded units were reported as multiunits, so the performance was identical to the naive algorithm (to be defined in section 3.1).
Current studies which focus on the problem of quantification of the separation quality are mainly based on the distance between the spike cluster and the noise cluster (Pouzat et al 2002, Schmitzer-Torberta et al 2005, Joshua et al 2007). We therefore compared our results also with one of the latest methods with this approach, the isolation score measure by Joshua et al (2007). We used the code provided with the original paper and adopted the same parameters with the only change being our different sampling frequency. We split the neural recording into 60 s segments and averaged the resultant isolation scores as described by Joshua et al. We considered a unit to be a single unit if its isolation score was ≥0.75, based on the suggestion of Joshua et al to exclude recording periods with an isolation score of 0.7–0.8. We also tested the thresholds of 0.50 and 0.10. Thresholds lower than 0.10 did not change the results significantly and are therefore omitted. We shall denote the three variants of the algorithm of Joshua et al by: isol75, isol50 and isol10, respectively.
2.7. Synthetic databases
In addition to the aforementioned recordings in the human, we also compared the performance of the two algorithms on two synthetic databases: the one by Quian-Quiroga et al (2004) and the other by Rutishauser et al (2006). For each database, we applied the spike-sorting method suggested in the respective paper (WaveClus and osort, respectively) and tested the quality of separation of each using three methods: the proposed one, osort (all three variants) and isolation score (three variants). A cluster defined by the relevant spike-sorting method is considered a single cell if more than 90% of its spikes belong to the same cluster according to the ground truth provided with the database.
Note that even though the two databases provide the ground truth spike train, the input to the methods of evaluation of the quality of separation may not be exactly identical to the original papers. This stems from the fact that the database provided with osort (Rutishauser et al 2006) contains ground truth spike trains, but not the added noise. We therefore added noise using the original simulator, with the same standard deviations as in the original paper, but still the datasets may not be exactly identical. The discrepancy with WaveClus is due to the fact that the superparamagnetic clustering is stochastic.
2.8. Runtime
Runtimes of the different algorithms were evaluated on the real dataset of the 1652 units from the human cerebrum. All runtime tests were performed on the same dedicated Dell XPS M1530 laptop (Duo-core 2.4 GHz CPUs with 4 Gb RAM). All implementations were in MATLAB 7.2 (Mathworks, Natick, MA) and used the original code distributed by the original authors of the methods (with the required minor code additions to match different data formats). Runtimes are reported for one MATLAB process running on one CPU (i.e. no parallel MATLAB processes).
3. Results
Whenever relevant, results are reported as average ± standard error.
3.1. Recordings in the human
Table 1 compares the accuracy in classification of recorded units into single cell versus multiunits of the proposed algorithms, SUMU and SUMU-LDA, the three variants of osort and the three variants of the isolation score measure, with respect to manual classification (see section 2.3). SUMU and SUMU-LDA result in a significantly higher accuracy (92.0% and 91.4%, respectively) than all other methods (p < 0.0001; one-sided paired-sample t-test, for all variants of the two methods) (compare also to 66.3% in a naive algorithm that labels all data as multiunits7). Its false-positive rate is an order of magnitude less than that of osort, and its false-negative rate is an order of magnitude lower than that of the isolation score method.
Table 1.
Comparison of SUMU with three variants of osort: osort1, osort3, osort_C and three variants of the isolation score: iso75, isol50, isol10 (see section 2.6 for definitions of the variants) in evaluating the quality of separation on real data from the human cerebrum. Values in the table are average percentages (± standard error) per session, except for SUMU-LDA, for which percentages are average (±SE) in a 12-fold cross-validation (numbers in parentheses refer to the training set)
| Algorithm | SUMU | osort1 | osort3 | osort_C |
|---|---|---|---|---|
| Accuracy | 92.0* ± 1.1 | 54.3 ± 2.7 | 37.8 ± 1.7 | 54.3 ± 2.7 |
| False-positive rate | 4.5 ± 1.1 | 45.5 ± 2.7 | 62.2 ± 1.7 | 45.5 ± 2.7 |
| False-negative rate | 3.5 ± 0.5 | 0.3 ± 0.1 | 0.0 ± 0.0 | 0.3 ± 0.1 |
| Algorithm | SUMU-LDA | isol75 | isol50 | isol10 |
|---|---|---|---|---|
| Accuracy | 91.4* ± 1.3 (91.5 ± 0.1) | 66.8* ± 1.5 | 67.5* ± 1.4 | 69.7* ± 1.5 |
| False-positive rate | 4.5 ± 1.4 (4.4 ± 0.1) | 0.0 ± 0.0 | 0.1 ± 0.1 | 2.6 ± 0.7 |
| False-negative rate | 4.1 ± 0.7 (4.1 ± 0.1) | 33.3 ± 1.5 | 32.4 ± 1.4 | 27.7 ± 1.2 |
Asterisks denote significant accuracy over the naive algorithm (see the text for details). False-positive rate is the percentage of units that are multiunits but were marked as single cells by the algorithm. False-negative rate is the percentage of units that are actually single cells but were marked as multiunits by the algorithm. The accuracy of SUMU is the highest.
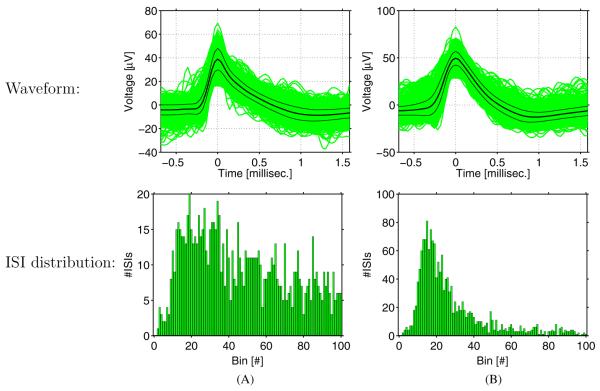
Figure 4 demonstrates classification of two clusters of spikes recorded from different regions in the cerebrum of two patients by various methods. The vast majority of the errors of the isolation score method were false negatives (figure 4(A)), while almost all errors of osort were due to false positives (figure 4(B)) (cf table 1).
Figure 4.
Examples of spike clusters from the human medial temporal lobe and their classification into single cells or multiunits by the various methods. Top row: waveforms of all spikes, centered at the peak. The mean and mean ±1 standard deviation are marked (black). Bottom row: ISI distribution histogram (bin size: 1 ms). (A) A cluster of spikes from the right parahippocampal gyrus (PHG) of case 10 (average firing rate: 1.50 Hz), classified as a single unit by manual inspection as well as SUMU and osort. All isolation score variants rendered it a multiunit. (B) A unit from the left anterior hippocampus (AH) of case 11 (average firing rate: 1.34 Hz) classified as a multiunit by a human, due to high variability of the waveforms (especially near the beginning of the main rise in voltage). SUMU and isolation score (all variants) agreed with that classification. In contrast, osort (all variants), whose only input is the ISI distribution, classified the unit as a single neuron, because of the resemblance of its ISI distribution to that of a single unit.
In order to estimate the contribution of the waveform-based feature (section 2.4.1) to classification by SUMU and SUMU-LDA, we repeated the classification using this feature only (i.e. not constraining the refractory period). The classification accuracy in this case was 89.1% ± 1.5% and 88.5% ± 2.0%, respectively. The waveform-based feature explains over 80% of the variation in classification using the two features (r2 = 0.81, r2 = 0.86, respectively), which demonstrates the importance of this feature to classification. Notwithstanding, the improvement in accuracy when adding the constraint on the refractory period is significant (p < 0.0005, p = 0.011, respectively; paired-sample t-test).
3.2. Synthetic databases
We employed the database provided with osort (Rutishauser et al 2006) in the comparison of all methods (table 2). The spike-sorting algorithm (Rutishauser et al 2006) identified 44 units, of which 22 (50%) were single units. No method performed significantly better than the naive algorithm (defined in section 3.1) on this database (one-sided paired-sample t-test: p > 0.1 for all methods). Isol50 and SUMU equate in their accuracy (~80%) and so are Isol75 and SUMU-LDA (~75%). The differences between the four algorithms (SUMU, SUMU-LDA, isol75 and isol50) were not significant (p > 0.5; paired-sample t-tests for comparisons without SUMU-LDA; two-sample t-tests of SUMU-LDA versus other methods). SUMU, SUMU-LDA and isol50 are the only methods significantly above the 50% chance level in a one-sided t-test (p < 0.05).
Table 2.
Comparison of SUMU, SUMU-LDA, the three variants of osort: osort1, osort3, osort_C, and the three variants of the isolation score: isol75, isol50, isol10 in evaluating the quality of separation on synthetic data provided with osort (Rutishauser et al 2006). The table is organized similarly to table 1. No classification is significantly higher than the naive algorithm
| Algorithm | SUMU | osort1 | osort3 | osort_C |
|---|---|---|---|---|
| Accuracy | 80.8 ± 6.1 | 57.1 ± 13.2 | 53.1 ± 15.9 | 57.1 ± 13.2 |
| False-positive rate | 12.0 ± 8.9 | 42.9 ± 13.2 | 46.9 ± 15.9 | 42.9 ± 13.2 |
| False-negative rate | 7.2 ± 3.9 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Algorithm | SUMU-LDA | isol75 | isol50 | isol10 |
|---|---|---|---|---|
| Accuracy | 75.0 ± 7.3 (79.5 ± 0.6) | 75.7 ± 10.0 | 80.8 ± 6.1 | 67.7 ± 8.8 |
| False-positive rate | 15.3 ± 5.9 (13.6 ± 0.5) | 5.9 ± 5.9 | 16.5 ± 8.7 | 32.4 ± 8.8 |
| False-negative rate | 9.7 ± 5.2 (6.8 ± 0.3) | 18.4 ± 5.9 | 2.8 ± 2.8 | 0.0 ± 0.0 |
The database provided with WaveClus (Quian-Quiroga et al 2004) contains 60 s of simulated data. WaveClus managed to correctly separate the spike trains into single cells in 59/60 = 98.3% of the simulated units. SUMU, SUMU-LDA and osort (all three variants) denoted all units (60/60 = 100%) as single cells, yielding an accuracy of 59/60 = 98.3%. The accuracy of isolation score was lower than the 50% chance level (isol75: 10/60 = 16.7%; isol50: 13/60 = 21.7%; isol10: 20/60 = 33.3%). For all variants, all errors were false negatives.
When comparing classification by the feature of the waveforms alone versus its combination with the refractory period constraint on the two synthetic databases, there was no change in the results (i.e. the refractory period added no information). This emphasizes that classification is based on the visual feature.
3.2.1. Bursting units
In addition to the aforementioned 60 units, the WaveClus database provides one channel with three simulated bursting cells. WaveClus correctly separated the spikes into single units in 2/3 = 66.7% of the units. The accuracy of SUMU, osort (all three variants), isol10 and Isol75 was 66.7%, at the chance level (SUMU, osort (all three variants), isol10: one false positive; isol75: one false negative). Isol50 and SUMU-LDA (3-fold cross-validation) gained an accuracy of 33.3%.
3.3. Runtime
The total amount of time required to compute SUMU was 381 s, SUMU-LDA: 385 s (12-fold cross-validation, including feature computation, training and testing), each variant of osort: 1099 s and each variant of the isolation score: 111 023 s. Computing the isolation score lasted over 30 h because of the direct processing of the raw spike train, in contrast with the other methods whose inputs are isolated spikes.
4. Discussion
This study defined quantitative measures for evaluation of the quality of separation of a cluster of spikes (single cell or multiunit) and analyzed the conditions under which the main feature classifies between single cells and multiunits. Based mainly on the spike waveform, we were able to correctly match the classification of a human observer in 92.0% of the 1652 units recorded from inside the human cerebrum. The error rate (8.0%) is much lower than the variability in the number of neurons and spikes detected by human classifiers on synthetic data and tetrode data, which may be as high as 23% of false positives and 30% of false negatives (Wood et al 2004, Harris et al 2000). The type I and type II error rates can both exceed 50% for single wire recordings (Harris et al 2000). Harris et al (2000) also estimated the optimal performance for a given set of feature vectors of any clustering system, manual or automatic, based on intracellular recordings. They found the optimal error rate to lie between 0 and 8%, which indicates that our prediction rate is close to the optimal clustering rate.
It is important to note, that in contrast with spike-sorting algorithms, which should consider the interrelationships between the different clusters formed on the same electrode (as described, for example, by Rutishauser et al (2006, p 216)), evaluation criteria only examine the final product of the spike sorter. As such, they are only intended to mark whether a given cluster is a single or a multiunit. They cannot and need not compensate for sorting errors.
The literature has only a few studies aimed directly at evaluation of the quality of separation. One approach which quantifies the certainty with which spikes are classified is Bayesian clustering (see Lewicki (1998) for a review). This type of method relates a probability to each cluster which may be used for evaluating the quality of separation. However, this probability relies on the specific statistical model employed by the spike-sorting algorithm. An alternative Bayesian approach is to represent uncertainty in the assignment of waveforms to units and incorporate this uncertainty into the analysis (Wood and Black 2008); this approach bypasses the need to classify units into single cells or multiunits but requires a different type of analysis than that of most studies today.
The proposed evaluation criteria, SUMU, matched the manual classification on the real dataset to a significantly higher degree than the other methods. This may be due to the two methods (SUMU and manual classification) being based on visual features of the spike waveforms and the ISI distribution histogram. The criteria of osort, on the other hand, rely solely on ISI distribution8, which may explain its low accuracy. The lower accuracy of the isolation score measure may stem from its development for recordings with higher SNR obtained by real-time isolation of the signals and selection of units by an experimenter advancing the electrode to the target neurons and monitoring spike-sorting quality online. The fact that decreasing the threshold on the isolation score resulted in higher accuracy (table 1) implies that single units have low isolation scores and are therefore hard to distinguish from multiunits in the same score range. This is typical in chronically implanted electrodes.
For real data, our comparison employed manual classification by human classifiers as its reference, due to the lack of the ground truth number of recorded units, or the ground truth classification of the clusters into single cells and multiunits. Manual classification may however disagree with the ground truth in some cases. For synthetic data, the ground truth is known by design, but the synthetic data themselves are only an approximation of the real spiking process of interest. Thus, the gap of knowledge of the ground truth is inherent in the type of extracellular recordings we use. To reduce this uncertainty, we tested the proposed methods on both a large real dataset from the human and two different synthetic datasets.
The low standard errors on the real database by all methods in table 1 is indicative of the consistency of the manual classification of different sessions, and thus of its suitability for testing the criteria. While our testing data were recorded from the human cerebrum, the algorithms SUMU and SUMU-LDA are not specific to the human and may thus be applied to extracellular recordings from other species as well.
Comparison of the criteria on the synthetic database provided with osort (Rutishauser et al 2006) shows that SUMU, SUMU-LDA, isol50 and isol75 equate with each other. The low accuracy of osort, which relies solely on ISI distribution, is probably due to the small amount of data (only 100 s of simulated spike trains) yielding inaccurate estimation of the distribution. The isolation scores seem to be less sensitive to the amount of available data, probably due to its intensive processing of the raw signal directly (see discussion of the runtime below) and the relatively higher SNR of this dataset. The database provided with WaveClus (Quian-Quiroga et al 2004), when spike-sorted by the WaveClus method, resulted in an almost perfect classification, which dooms all methods to perform not significantly better than the naive one. This phenomenon demonstrates the weakness of the database as a benchmark for evaluation of the quality of separation, rather than a flaw in the evaluation methods themselves. We chose to include the database in the analysis anyway because of its use as a benchmark in the literature (Quian-Quiroga et al 2004, Herbst et al 2008).
Bursting units are a challenge for both spike-sorting and quality of separation algorithms, because the waveforms for spikes in a burst are typically different from each other. SUMU and SUMU-LDA performed similarly to the other methods, but still at the chance level, due to the weakness of the database as a benchmark (as mentioned above). Interestingly, SUMU, osort (all three variants) and isol10 marked all units as single units, which is contrary to the intuition that clusters with different waveforms due to bursting will be incorrectly denoted multiunits (i.e. misclassification was due to false positives rather than the expected false negatives).
Runtime
SUMU had the lowest runtime of all algorithms examined and is suitable for real-time applications such as BCI. The runtime of the isolation score is two orders of magnitude longer than that of the SUMU, SUMU-LDA and osort algorithms, and is thus inappropriate for real-time implementation.
Generality, limitations and future work
Fundamentally, spike-sorting methods try to use a minimal set of assumptions (e.g. refractory period) about the properties of neural coding, so as not to introduce biases into subsequent stages of the analysis. The proposed methods follow this approach while modeling the assumptions in a rigorous mathematical manner. This yields a smaller set of assumptions than is usually used (implicitly) during visual inspection of the waveform, the ISI distribution histogram and maybe other features such as wavelet coefficients.
Moreover, the proposed features can be combined with other methods for evaluation of the quality of separation so as to reduce the set of assumptions (e.g. omit the constraint for a refractory period) or model noise too. An example is the combination of the proposed visual feature of the waveform with the notion of cluster distances (see Fee et al (1996a, 1996b)), whose set of assumptions is quite small. This is a subject for future research.
5. Conclusions
We have developed two measures of the quality of separation of a cluster of spikes, SUMU and SUMU-LDA, that can be computed automatically and in real time with performance resembling that of a human classifier on real data recorded from the human cerebrum. As more and more studies are geared toward automatic spike sorting that will serve in brain–computer interfaces (Kipke et al 2008), the role of the man-in-the-loop need be replaced by computer algorithms. The proposed algorithm therefore mimics the behavior of humans in order to maintain similar performance and utilizes the main tool that humans use for manual spike sorting: visual inspection.
The proposed methods outperformed existing state-of-the-art techniques on real data in both accuracy and runtime, and equated with the best of them on synthetic databases provided by other authors.
Our criteria may serve two purposes. On the one hand, they are suggested as a standard that will allow comparison of the quality of separation between different studies. On the other hand, they can serve in motor BCIs to automatically distinguish between well-isolated cells and multiunits. Rather than intermixing the two types of units (Hochberg et al 2006, Velliste et al 2008), a motor BCI will be able to weigh each unit according to its quality of separation.
Acknowledgments
The authors are grateful to the anonymous second referee for insightful comments. The authors thank Rodrigo Quian-Quiroga and Ueli Rutishauser for the code and data of WaveClus and osort, respectively, and Mati Joshua for the code of the isolation score method. The authors also thank Roy Mukamel for his assistance. This research was supported in part by grant no. 2497 from the PVA Research Foundation and by NINDS grant.
Appendix A. b/a for a linear multiunit and its ‘equivalent’ single unit
Let us assume that the main rise of all single cells is linear with normal distribution and it has the same length. We assume no overlap between spikes. As a consequence, for each single cell, two constants, c and d, exist such that the measured voltage on the main rise at sampling time ti (i = 1, . . . , n) is an uncorrelated random variable . For a multiunit composed of l such neurons, there exist constants cj and dj such that if the spike belongs to the jth neuron (j = 1, . . . , l), its voltage measure is . The probability of a spike of the jth neuron is . By the partition theorem, the expected value of Vi is
Therefore,
| (A.1) |
By the law of total variance,
So, by definition,
| (A.2) |
A single unit with an equivalent linear rise: and variance results in
| (A.3) |
| (A.4) |
and therefore,
Equality occurs when all neurons composing the multiunit have an identical main rise (Var(cjti + dj) = 0), which is indeed indistinguishable from the equivalent single unit. In all other cases, the multiunit obtains higher values of the feature b/a than the ‘equivalent’ single unit.
Appendix B. b/a of a misaligned single unit
Under the assumptions of appendix A, let us examine a single unit whose spikes are misaligned. We assume that misalignment occurs by choosing a wrong sample to be the peak, but the length of the main rise remains unchanged (n sample points), so that the whole main rise shifts either to the left or to the right. Let X be the random variable denoting the offset of the main rise to the left (so negative = shift to the right; 0 = no shift). We consider offsets in the range X = j ∈ [−n, n]. Note that for X = n and X = −n, the misaligned main rise contains no elements of the true main rise.
The probability of misalignment by j sample points to the right is denoted P(X = −j) = p−j. In this case, the first j sample points of the misaligned rise are not part of the true main rise, so
where lm is the voltage at the mth sample point prior to the true main rise, relative to the first sample point of the true rise (i.e. ct1 + d). In figure 3 (left) lm is the vertical distance between the leftmost circle and the mth sample before the main rise. For the rest of the points, the main rise is shifted:
The probability of correct alignment is P(X = 0) = p0:
Similarly, for shifts to the left, with probability P(X = j) = pj, the voltage is
where km is the voltage at the mth sample point following the true main rise, relative to the last sample point of the true rise (i.e. ctn + d). In figure 3 (left) km is the vertical distance between the rightmost circle and the mth sample after the main rise. ∀ m, km > 0 because ctn + d is the maximal voltage.
By the partition theorem,
where Δt is the sampling interval (assumed constant), and . Because the slope c > 0, also αm > 0.
The misaligned numerator thus becomes
Substituting asu (equation (A.3)) yields
If we assume that αn > α1 due to the decay that follows the peak, all terms in the expression in brackets are negative or zero, except maybe for those involving βj . Now, due to decay after the peak and the relatively ‘flat’ waveform before the rise, it is reasonable to assume that αn − α1 ≥ |βn − β1|. If we also assume pn = p−n, the last two in brackets are negative or zero. From the same reason, it makes sense to assume that the expected value of αj is larger than that of βj , so . Under constraints on the variation of the voltage before the rise and following it: ama ≤ asu.
By the law of total variance,
From equation (A.4), it follows that
Consequently,
Thus, under the aforementioned assumptions, the feature b/a is higher for the misaligned cluster than for the well-aligned one.
Footnotes
W.L.O.G we assume the peak of the mean waveform is positive. For negatively peaked waveforms, the same procedure is applied to minus the waveform.
We selected the naive algorithm to report ‘multiunit’ rather than ‘single unit’ based on the posterior distribution of recorded data. As such, it has knowledge which a truly naive algorithm lacks (i.e. the analysis is more strict than a real naive algorithm).
Note that the spike-sorting algorithm proposed by Rutishauser et al (2006) does indeed refer also to spike waveforms; only the three criteria that allow a sufficient classification of clusters as single unit or not rely only on the ISI.
References
- Bar-Hillel A, Spiro A, Stark E. Spike sorting: Bayesian clustering of non-stationary data. J. Neurosci. Methods. 2006;157:303–16. doi: 10.1016/j.jneumeth.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Benitez R, Nenadic Z. Robust unsupervised detection of action potentials with probabilistic models. IEEE Trans. Biomed. Eng. 2008;55:1344–54. doi: 10.1109/TBME.2007.912433. [DOI] [PubMed] [Google Scholar]
- Ding W, Yuan J. Spike sorting based on multi-class support vector machine with superposition resolution. Med. Biol. Eng. Comput. 2008;46:139–45. doi: 10.1007/s11517-007-0248-0. [DOI] [PubMed] [Google Scholar]
- Engel AK, Moll CK, Fried I, Ojemann GA. Invasive recordings from the human brain: clinical insights and beyond. Nat. Rev. Neurosci. 2005;6:35–47. doi: 10.1038/nrn1585. [DOI] [PubMed] [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Automatic sorting of multiple unit neuronal signals in the presence of anisotropic and non-Gaussian variability. J. Neurosci. Methods. 1996a;69:175–88. doi: 10.1016/S0165-0270(96)00050-7. [DOI] [PubMed] [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Variability of extracellular spike waveforms of cortical neurons. J. Neurophysiol. 1996b;76:3823–33. doi: 10.1152/jn.1996.76.6.3823. [DOI] [PubMed] [Google Scholar]
- Fletcher M, Liang B, Smith L, Knowles A, Jackson T, Jessop M, Austin J. Neural network based pattern matching and spike detection tools and services—in the CARMEN neuroinformatics project. Neural Netw. 2008;21:1076–84. doi: 10.1016/j.neunet.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Fried I, Wilson CL, Maidment NT, Engel J, Behnke E, Fields TA, MacDonald KA, Morrow JW, Ackerson L. Cerebral microdialysis combined with single neuron and electroencephalographic recording in neurosurgical patients. J. Neurosurg. 1999;91:697–705. doi: 10.3171/jns.1999.91.4.0697. [DOI] [PubMed] [Google Scholar]
- Gelbard-Sagiv H, Mukamel R, Harel M, Malach R, Fried I. Internally generated reactivation of single neurons in human hippocampus during free recall. Science. 2008;322:96–101. doi: 10.1126/science.1164685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J. Neurophysiol. 2000;84:401–14. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Harris KD, Hirase H, Leinekugel X, Henze DA, Buzsáki G. Temporal interaction between single spikes and complex spike bursts in hippocampal pyramidal cells. Neuron. 2001;32:141–9. doi: 10.1016/s0896-6273(01)00447-0. [DOI] [PubMed] [Google Scholar]
- Herbst JA, Gammeter S, Ferrero D, Hahnloser RHR. Spike sorting with hidden Markov models. J. Neurosci. Methods. 2008;174:126–34. doi: 10.1016/j.jneumeth.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Hochberg LR, Serruya MD, Friehs GM, Mukand JA, Saleh M, Caplan AH, Branner A, Chen D, Penn RD, Donoghue JP. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature. 2006;442:164–71. doi: 10.1038/nature04970. [DOI] [PubMed] [Google Scholar]
- Joshua M, Elias S, Levine O, Bergman H. Quantifying the isolation quality of extracellularly recorded action potentials. J. Neurosci. Methods. 2007;163:267–82. doi: 10.1016/j.jneumeth.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Tamura H, Suzuki SS. Tracking spike-amplitude changes to improve the quality of multineuronal data analysis. IEEE Trans. Biomed. Eng. 2007;54:262–72. doi: 10.1109/TBME.2006.886934. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Manyam SC, Warren DJ, Normann RA. Electrophysiological mapping of cat primary auditory cortex with multielectrode arrays. Ann. Biomed. Eng. 2006;34:300–9. doi: 10.1007/s10439-005-9037-9. [DOI] [PubMed] [Google Scholar]
- Kim SP, Simeral JD, Hochberg LR, Donoghue JP, Black MJ. Neural control of computer cursor velocity by decoding motor cortical spiking activity in humans with tetraplegia. J. Neural Eng. 2008;5:455–76. doi: 10.1088/1741-2560/5/4/010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipke DR, Shain W, Buzsaki G, Fetz E, Henderson JM, Hetke JF, Schalk G. Advanced neurotechnologies for chronic neural interfaces: new horizons and clinical opportunities. J. Neurosci. 2008;28:11830–8. doi: 10.1523/JNEUROSCI.3879-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki MS. Bayesian modeling and classification of neural signals. Neural Comput. 1994;6:1005–30. [Google Scholar]
- Lewicki MS. A review of methods for spike sorting: the detection and classification of neural action potentials. Netw. Comput. Neural Syst. 1998;9:R53–78. [PubMed] [Google Scholar]
- Moran D, Schwartz A. Motor cortical representation of speed and direction during reaching. J. Neurophysiol. 1999;82:2676–92. doi: 10.1152/jn.1999.82.5.2676. [DOI] [PubMed] [Google Scholar]
- Mukamel R, Gelbard H, Arieli A, Hasson U, Fried I, Malach R. Coupling between neuronal firing, field potentials, and fMRI in human auditory cortex. Science. 2005;309:951–4. doi: 10.1126/science.1110913. [DOI] [PubMed] [Google Scholar]
- Pouzat C, Mazor O, Laurent G. Using noise signature to optimize spike-sorting and to assess neuronal classification quality. J. Neurosci. Methods. 2002;122:43–57. doi: 10.1016/s0165-0270(02)00276-5. [DOI] [PubMed] [Google Scholar]
- Quian-Quiroga R, Nadasdy Z, Ben-Shaul Y. Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput. 2004;16:1661–87. doi: 10.1162/089976604774201631. [DOI] [PubMed] [Google Scholar]
- Quian-Quiroga R, Reddy L, Kreiman G, Koch C, Fried I. Invariant visual representation by single neurons in the human brain. Nature. 2005;435:1102–7. doi: 10.1038/nature03687. [DOI] [PubMed] [Google Scholar]
- Quirk MC, Wilson MA. Interaction between spike waveform classification and temporal sequence detection. J. Neurosci. Methods. 1999;94:41–52. doi: 10.1016/s0165-0270(99)00124-7. [DOI] [PubMed] [Google Scholar]
- Quirk MC, Wilson MA. Interaction between spike waveform classification and temporal sequence detection. J. Neurosci. Methods. 2000;99:143–5. doi: 10.1016/s0165-0270(99)00124-7. erratum. [DOI] [PubMed] [Google Scholar]
- Quiroga RQ. What is the real shape of extracellular spikes? J. Neurosci. Methods. 2009;105:194–8. doi: 10.1016/j.jneumeth.2008.09.033. [DOI] [PubMed] [Google Scholar]
- Quiroga RQ, Mukamel R, Isham EA, Malach R, Fried I. Human single-neuron responses at the threshold of conscious recognition. Proc. Natl Acad. Sci. 2008;105:3599–604. doi: 10.1073/pnas.0707043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Schuman EM, Mamelak AN. Online detection and sorting of extracellularly recorded action potentials in human medial temporal lobe recordings, in vivo. J. Neurosci. Methods. 2006;154:204–24. doi: 10.1016/j.jneumeth.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Schmitzer-Torberta N, Jacksona J, Henzeb D, Harrisc K, Redish AD. Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience. 2005;131:1–11. doi: 10.1016/j.neuroscience.2004.09.066. [DOI] [PubMed] [Google Scholar]
- Shoham S, Fellows MR, Normann RA. Robust, automatic spike sorting using mixtures of multivariate t-distributions. J. Neurosci. Methods. 2003;127:111–22. doi: 10.1016/s0165-0270(03)00120-1. [DOI] [PubMed] [Google Scholar]
- Snider RK, Bonds AB. Classification of non-stationary neural signals. J. Neurosci. Methods. 1998;84:155–66. doi: 10.1016/s0165-0270(98)00110-1. [DOI] [PubMed] [Google Scholar]
- Stark E, Abeles M. Predicting movement from multiunit activity. J. Neurosci. 2007;27:8387–94. doi: 10.1523/JNEUROSCI.1321-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suner S, Fellows MR, Vargas-Irwin C, Nakata GK, Donoghue JP. Reliability of signals from a chronically implanted, silicon-based electrode array in non-human primate primary motor cortex. IEEE Trans. Neural Syst. Rehabil. Eng. 2005;13:524–41. doi: 10.1109/TNSRE.2005.857687. [DOI] [PubMed] [Google Scholar]
- Tankus A, Yeshurun Y, Flash T, Fried I. Encoding of speed and direction of movement in the human supplementary motor area. J. Neurosurg. 2009;110:1304–16. doi: 10.3171/2008.10.JNS08466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Irwin C, Donoghue JP. Automated spike sorting using density grid contour clustering and subtractive waveform decomposition. J. Neurosci. Methods. 2007;164:1–18. doi: 10.1016/j.jneumeth.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. Cortical control of a prosthetic arm for self-feeding. Nature. 2008;453:1098–101. doi: 10.1038/nature06996. [DOI] [PubMed] [Google Scholar]
- Wehr M, Pezaris JS, Sahani M. Simultaneous paired intracellular and tetrode recordings for evaluating the performance of spike sorting algorithms. Neurocomputing. 1999;26–27:1061–8. [Google Scholar]
- Wilson CL. Intracranial electrophysiological investigation of the human brain in patients with epilepsy: contributions to basic and clinical research. Exp. Neurol. 2004;187:240–5. doi: 10.1016/j.expneurol.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Wood F, Black MJ. A nonparametric Bayesian alternative to spike sorting. J. Neurosci. Methods. 2008;173:1–12. doi: 10.1016/j.jneumeth.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood F, Black MJ, Vargas-Irwin C, Fellows M, Donoghue JP. On the variability of manual spike sorting. IEEE Trans. Biomed. Eng. 2004;51:912–8. doi: 10.1109/TBME.2004.826677. [DOI] [PubMed] [Google Scholar]
- Yang X, Shamma SA. A totally automated system for the detection and classification of neural spikes. IEEE Trans. Biomed. Eng. 1988;35:806–16. doi: 10.1109/10.7287. [DOI] [PubMed] [Google Scholar]
- Zhang PM, Wu JY, Zhou Y, Liang PJ, Yuan JQ. Spike sorting based on automatic template reconstruction with a partial solution to the overlapping problem. J. Neurosci. Methods. 2004;135:55–65. doi: 10.1016/j.jneumeth.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Zouridakis G, Tam DC. Identification of reliable spike templates in multi-unit extracellular recordings using fuzzy clustering. Comput. Methods Programs Biomed. 2000;61:91–8. doi: 10.1016/s0169-2607(99)00032-2. [DOI] [PubMed] [Google Scholar]