Abstract
Differentiation of mesenchymal stem cells into a particular lineage is tightly regulated, and malfunction of this regulation could lead to pathological consequences. Patients with osteoporosis have increased adipocyte accumulation, but the mechanisms involved remain to be defined. In this study, we aimed to investigate if micro-RNAs regulate mesenchymal progenitor cells and bone marrow stromal cell (BMSC) differentiation through modulation of Runx2, a key transcription factor for osteogenesis. We found that miR-204 and its homolog miR-211 were expressed in mesenchymal progenitor cell lines and BMSCs and their expression was induced during adipocyte differentiation, whereas Runx2 protein expression was suppressed. Retroviral overexpression of miR-204 or transfection of miR-204 oligo decreased Runx2 protein levels and miR-204 inhibition significantly elevated Runx2 protein levels, suggesting that miR-204 acts as an endogenous attenuator of Runx2 in mesenchymal progenitor cells and BMSCs. Mutations of putative miR-204 binding sites upregulated the Runx2 3′-UTR reporter activity, suggesting that miR-204/211 bind to Runx2 3′-UTR. Perturbation of miR-204 resulted in altered differentiation fate of mesenchymal progenitor cells and BMSCs: osteoblast differentiation was inhibited and adipocyte differentiation was promoted when miR-204 was overexpressed in these cells, whereasosteogenesis was upregulated and adipocyte formation was impaired when miR-204 was inhibited. Together, our data demonstrated that miR-204/211 act as important endogenous negative regulators of Runx2, which inhibit osteogenesis and promote adipogenesis of mesenchymal progenitor cells and BMSCs.
Keywords: Mesenchymal stem cells, Runx2, miR-204, Osteoblast, Adipocyte
INTRODUCTION
Mesenchymal stem cells (MSCs) have potential to undergo multilineage differentiation into multiple connective tissue cell types, such as osteoblasts, chondrocytes, adipocytes, and myoblasts [1, 2]. Differentiation of MSCs into different lineages of cells is tightly regulated, and alteration or malfunction of this regulation could result in pathological consequences [3]. For example, bone loss is often accompanied with the increase of bone marrow adiposity in aging [4]. Moreover, it was reported that adipose tissue in bone marrow is inversely related to bone formation in osteoporosis, and patients with high bone mass phenotype show inhibition of adipogenesis [5, 6]. These findings suggest a switch between the osteoblast and the adipocyte differentiation of the MSCs during the development of osteoporosis or a high bone mass phenotype.
Runx2 has been identified as a key transcription factor regulating osteogenesis and chondrogenesis [7, 8], and PPARγ was demonstrated to be an important adipogenic factor [9]. These master regulators of different lineages were expressed in low levels in undifferentiated cells, maintaining differentiation potential of MSCs [10, 11]. Runx2 and PPARγ may control the commitment of MSCs to differentiate into osteoblast/chondrocyte and adipocyte lineages. Runx2 deficiency in chondrocytes results in enhanced adipogenic differentiation [12], whereas PPARγ insufficiency increases bone mass by stimulating osteoblastogenesis [13]. In addition, a transcriptional cofactor TAZ interacts with Runx2 and activates Runx2-dependent gene transcription and promotes osteoblast differentiation, whereas its binding to PPARγ represses PPARγ-dependent gene transcription and impairs adipocyte differentiaon [14]. These findings suggest that regulation of key transcription factors, such as Runx2 or PPARγ, may affect MSC commitment and differentiation.
MicroRNAs (miRNAs or miRs) have emerged as a new dimension of gene regulation in recent years. These endogenous noncoding RNAs are ~22 nucleotides and evolutionarily conserved. They bind to the 3′-untranslated region of their target genes, including key transcription factors, receptors, and kinases, and regulate protein translation or mRNA stability, therefore tuning numerous pathways related to the development and diseases. Studies show that microRNAs have an important role in regulating stem cell self-renewal and differentiation, extensively demonstrated in embryonic stem cells and hematopoietic stem cells [15]. However, specific miRNAs and their target genes in the regulation of MSC differentiation have not been well characterized and are, therefore, poorly understood.
In this study, we found that Runx2, a master regulator of osteoblast differentiation, was regulated by specific miRNAs. MiR-204 and its homologue miR-211 were expressed in different mesenchymal progenitor cell lines and BMSCs, and their expression was induced during adipogenesis. Our gain-or loss-of-function experiments demonstrated that miR-204/211 act as endogenous attenuators of Runx2 protein expression in progenitor cells and BMSCs, through binding to multiple sites in the 3′-UTR of Runx2. Importantly, miR-204 over-expression or inhibition restrained or promoted osteoblast differentiation, respectively, demonstrated by osteoblast phenotypic assays and marker gene profiling. Also, miR-204 perturbation had significant effects on adipogenesis, and the expression of miR-204 was positively correlated with adipocyte differentiation. Our findings suggest that miR-204/211 act as Runx2 attenuators to antagonize osteoblast differentiation in progenitor cells and BMSCs.
Materials and Methods
Cell Culture, Transfection, and Infection
ST2 and C2C12 cells were cultured in α-minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum (FBS), and C3H10T1/2 cells were cultured in basal medium Eagle supplemented with 10% FBS. Human mesenchymal stem cells (hMSCs) (Lonza Group Ltd, Basel, Switzerland, http://www.lonza.com) were cultured according to the manufacturer’s instruction. COS7 cells were culture in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Bone marrow cells were isolated from long bones of 10-week-old mice. Bone marrow mesenchymal stromal cells were cultured for 5 days in α-MEM with 20% FBS and then in α-MEM with 10% FBS for 2–3 days. To induce osteoblast differentiation, after reaching 100% confluence, ST2 cells were cultured in α-MEM supplemented with 10% FBS, 300 ng/ml of BMP-2, 50 μg/ml of ascorbic acid, and10 mM β-glycerophosphate. Human mesenchy-mal stem cells were cultured in α-MEM supplemented with 10% FBS, 10 nM dexamethasone, 50 μg/ml of ascorbic acid, and 5 mM β-glycerophosphate to induce osteoblast differentiation. To induce adipocyte differentiation, ST2, C3H10T1/2, or human mesenchymal stem cells were allowed to become confluent for 1 day and then cultured in adipocyte-inducing medium (AIM, α-MEM containing 10% FBS, 1 μM dexamethasone, and 0.5 mM methylisobutylxanthine, 10 μg/ml of insulin, and 100 μM indo-methacin) for 3 days and then cultured in adipocyte-maintaining medium (AMM, α-MEM containing 10% FBS and 10 μg/ml of insulin) for 2 days. Oil droplets often formed after 3 days of treatment in C3H10T1/2 cells. ST2 cells normally need more time to develop oil droplets; thus, after the first round of AIM and AMM treatment, cells were subjected to a second round of treatment with AIM and AMM. For reporter assays, Fugene HD transfection reagents (Roche, Basel, Switzerland, http://www.roche-applied-science.com) were used to transfect plasmids in C3H10T1/2 cells. For transfection of miRNA or siRNA oligos, Dharmafect one transfection reagents (Dharmacon, http://www.dharmacon.com) were used according to the manufacturer’s instruction. MiR-204 or anti-miR-204 was transfected at the concentration of 50 nM, and Runx2 siRNA was transfected at the concentration of 50 nM. Methods of preparation of retrovirus and infection of C3H10T1/2, ST2, and C2C12 cells were reported by other labs and described in our previous studies. Briefly, Plat-E packaging cells were cultured in DMEM with 10% FBS and transfected with pMX retroviral vectors. After 48 hours, retroviral supernatant was collected to infect cells. Selection was performed using puromycin (5 μg/ml) for 2–3 weeks [16, 17].
Plasmids, Small RNA Oligos, and Antibodies
All retroviral vectors used in this study were constructed based on pMX vector [18]. Mir-204 (miRBase accession number: MI0000247) and miR-211 (miRBase accession number: MI0000708) overexpression vectors were constructed by ligating EcoRI/XhoI polymerase chain reaction (PCR) fragments with pMX-puro. Primers used for miR-204 and miR-211 amplification were 204F (TAT AGA ATT CGC TTC ATT CAG CAC CTA GTT, the restriction enzyme recognition site is marked by underline), 204R (TAT ACT CGA GTG GGT AAG GTT CTT TGA TGT), 211F (TAT AGA ATT CAG AGT GTG GTC AAC CTA TCA), and 211R (TAT ACT CGA GGA AAA TGG ATC AGG GTG GCA). Retroviral “sponge” vectors were constructed using ligation of two copies of miR-204/211 complementary oligos (AGG CAW AGG AAC TAA AGG GAA, W = A or T) and pMX-puro (supplemental online Fig. S1). MiR-204 and anti-miR-204 oligos were purchased from Dharmacon. Runx2 siRNA were purchased from Qiagen (Hilden, Germany, http://www1.qiagen.-com). Monoclonal antibodies specific for Runx2 and β-actin were purchased from MBL (http://www.mblintl.com) and Sigma (Saint Louis, MO, http://www.sigmaaldrich.com), respectively.
Quantitative reverse transcription-polymerase chain reaction
Total RNA was extracted from cell cultures using mirVana miRNA Isolation Kit (Ambion). Reverse transcription (RT) and quantitative PCR were performed as described [19]. Specific miR-204/211 RT primer sequence is GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA GGC AW (W = A or T), and PCR primer sequences for miR-204/211 are GGG CTT CCC TTT GTC ATC CT and GTG CAG GGT CCG AGG T. U6 RNA was used as an internal control for miRNA detection, and the primer sequences are CGC TTC GGC AGC ACA TAT AC and CAG GGG CCA TGC TAA TCT T. Expression levels of alkaline phosphatase (Alp), osteocalcin (OC), osteopontin (OPN), aP2, adipsin, and α-actin mRNA were examined using quantitative RT-PCR amplification.
Western Blot Analysis
Whole cell lysates for western blotting were extracted with lysis buffer containing 50 mM Tris (pH 7.6), 150 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.5% NP-40 and protease inhibitor cocktail (Sigma). Protein samples were resolved by 10% SDS-PAGE gel and transferred to the nitrocellulose membranes, which were incubated with individual antibodies according to manufacturer’s instruction and then incubated with a horseradish peroxi-dase-conjugated secondary antibody (Bio-Rad, Hercules, CA, http://www.bio-rad.com). The blots were visualized by an enhanced chemiluminesence system (Amersham Biosciences, Piscataway, NJ, http://www.amersham.com) or Femto (Pierce, Rockford, IL, http://www.piercenet.com) according to the manufacturer’s instructions.
Luciferase Reporter Assay
The empty 3′-UTR reporter vector (CMV-Luc2) was constructed by ligating Luc2 gene and pcDNA3 using HindIII/BamHI. Runx2 3′-UTR reporter was constructed by ligating 4-kb Runx2 3′-UTR fragment, which was amplified using primers (CAC CGG ATC CAA TTC GTC AAC CAT GGC CCA and TAT ACT CGA GGA AAA AAA AAG CCT TTT TAT TG) with CMV-Luc2 using BamHI/XhoI. Mutagenesis of the individual five binding sites was achieved according to the instructions for the Quik-Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, http://www.stratagene.com). Specifically, the bases of GGG that are complementary to CCC in the seed sequence (UUUCCCUU) of miR-204/211 were changed to CCC, abolishing the interaction between a specific binding site in the 3′-UTR and miRNA. Cells were transfected with combinations of CMV-Luc2 plasmids and control siRNA or miR-204 RNA oligos. After 48 hours, reporter assay were performed according to the manufacturer’s instruction.
Results
Post-Transcriptional Regulation of Runx2 During BMSC Differentiation
Mesenchymal stem cells (MSCs) have the potential to differentiate into multiple cell types upon encountering different environmental signals. C3H10T1/2 and ST2 cells are mesenchymal progenitor cell lines that have the capacity to differentiate into adipocytes when they are cultured with adipocyte differentiation medium. Runx2 protein level was significantly decreased when cells were cultured with adipocyte differentiation medium for 3 days compared with cells cultured without adipocyte differentiation medium (Fig. 1A and supplemental online Fig. S1A). In contrast, Runx2 mRNA level was only slightly reduced (Fig. 1B and supplemental online Fig. S1B). This observation suggests that Runx2 expression may be mainly regulated at post-transcriptional level, such as miRNA suppression of protein translation or ubiquitin-proteasome degradation.
Figure 1.
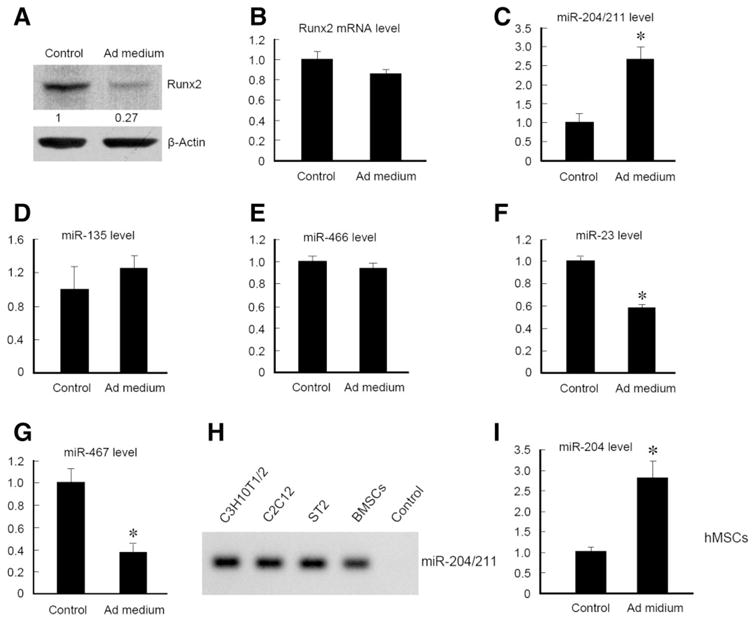
Runx2 regulation and miRNA expression during adipocyte differentiation. (A–G): C3H10T1/2 cells were induced with adipocyte differentiation medium for 3 days and then harvested for examination of Runx2 expression by western blotting (A) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays of Runx2 (B) or miRNA expression as indicated. (H) RT-PCR results showed that miR-204/211 were expressed in C3H10T1/2, C2C12, ST2, and BMSCs. (I) hMSCs were treated with adipocyte differentiation medium for 8 days and harvested for miR-204/211 qRT-PCR assays. *, p < .05, unpaired Student’s t-test, n = 3. Abbreviations: hMSCs, human mesenchymal stem cells.
Both human and mouse Runx2 mRNAs have a considerably long 3′-untranslated region (about 4-kb), based on RNA blot [20, 21] or the sequence of cDNA clones [22]. We hypothesized that Runx2 may be regulated by miRNAs through their binding to the 3′-UTR of Runx2 mRNA, decreasing Runx2 protein level and preventing cells to differentiate into osteoblasts. Thus, we performed bioinformatic analyses using PicTar [23] and miRanda [24] programs. We identified several miRNAs, such as miR-204/211, miR-135, miR-466, miR-23, and miR-467, that could potentially bind 3′-UTR of Runx2 mRNA through multiple binding sites conserved in multiple species with the optimal free energy. To determine if these miRNAs are regulated during adipocyte differentiation, we treated C3H10T1/2 cells with adipocyte differentiation medium for 4 days and extracted miRNA for RT-PCR analysis. The results showed that expression of miR-204/211 was significantly increased during adipocyte differentiation (Fig. 1C and supplemental online Fig. S1C). In contrast, expression of miR-135 and miR-466 was not changed (Fig. 1D and 1E). Expression of miR-23 and miR-467 was significantly reduced (Fig. 1F and 1G). MiR-23 and miR-466 were abundantly expressed; whereas miR-135 and miR-467 were expressed at a very low level in C3H10T1/2 cells. To further confirm miR-204/211 expression, we performed RT-PCR in multiple sources of mesenchymal progenitor cell lines such as C3H10T1/2, C2C12, ST2, and primary BMSCs. The results showed that miR-204/211 expression was detected in all these cells (Fig. 1H). We further examined miR-204 expression in hMSCs and found that miR-204 expression was significantly increased in hMSCs when the cells were cultured with adipocyte differentiation medium (Fig. 1I). Collectively, our results demonstrate that Runx2 protein level is negatively regulated, whereas expression of miR-204/211 are induced during the differentiation of mesenchymal progenitor cells and hMSCs into adipocytes, suggesting that miR-204/211 could be the suppressors of Runx2 in these cells.
MiR-204 and MiR-211 Act As Attenuators of Runx2 Expression
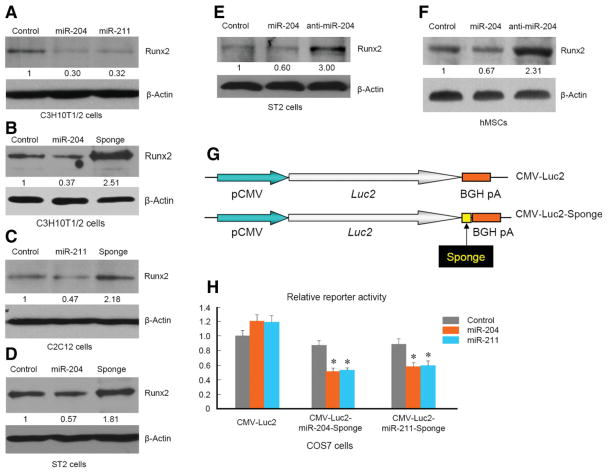
MiR-204 and miR-211 are close homologues and have 1 nt (in mouse) or 2 nt (in human) difference. To determine if miR-204/211 gain-of-function results in decreased Runx2 protein levels, we retrovirally over-expressed miR-204 or miR-211 gene. Quantitative RT-PCR results demonstrated that both miR-204 and miR-211 were overexpressed in C3H10T1/2 cells. In these miRNA overexpressing cells, Runx2 protein levels were significantly reduced (Fig. 2A), suggesting that miR-204/211 target Runx2 gene to suppress its protein translation. To investigate if endogenous miR-204/211 act as attenuators of Runx2 expression, we designed antagomir sequence against miR-204 or miR-211 (supplemental online Fig. S2) and retrovirally overexpressed them to achieve loss-of-function of miRNA, a technique called “sponge” [25]. Introducing sponges against miR-204 or miR-211 resulted in higher protein levels of Runx2 (Fig. 2B and data not shown). Because the sponge sequence against miR-204 or miR-211 is almost identical due to the high similarity of these two miR-NAs and both of them worked to inhibit endogenous miR-204/211 (supplemental online Fig. S2), we mainly used miR-204-sponge in the following experiments to achieve loss-of-function of miR-204/211. To determine if miR-204/211 have a negative effect on Runx2 protein levels in other mesenchymal progenitor cells, we also retrovirally expressed miR-204 or miR-204-sponge in ST2 and C2C12 cells. Western blot analysis showed that miR-204 overexpression decreased Runx2 protein levels, and inhibition of miR-204/211 by miR-204-sponge led to elevated Runx2 protein levels in these cells (Fig. 2C and 2D). These results suggest that miR-204/211 are expressed in mesenchymal progenitor cells from different origins and suppress Runx2 expression.
Figure 2.
MiR-204 downregulates Runx2 protein levels. (A): C3H10T1/2 cells were infected with control (empty vector), miR-204, or miR-211 expressing retrovirus individually. (B): C3H10T1/2 cells were infected with control (empty vector), miR-204, or miR-204 sponge expressing retrovirus individually. (C, D): C2C12 or ST2 cells were infected with control (empty vector), miR-204, or miR-204-sponge expressing retrovirus. For (A–D), cells were harvested for western blot analysis to determine changes in Runx2 protein levels after 15-day selection with puromycin. (E): ST2 cells were transfected with control siRNA, miR-204 mimic, or anti-miR-204 oligos for 2 days and then harvested for western blotting. (F): Human mesenchymal stem cells were transfected with control oligo, miR-204, or anti-miR-204 oligos for 2 days and then harvested for western blot analysis. (G): A diagram of CMV-luc2-sponge. (H): COS7 cells were transfected with combinations of CMV-luc2-sponge and miR-204 or -211 expressing vectors.
We also transfected mesenchymal progenitor cells with miRNA mimic or antagomir oligos to determine if introduction of chemically synthesized miRNA or antagomir could also regulate Runx2 protein level as efficient as retroviral infection. Western blot analysis showed that ST2 cells and hMSCs had lower Runx2 protein levels when transfected with miR-204 and had higher Runx2 levels when transfected with miR-204 antagomir (Fig. 2E and 2F), confirming the consistency of Runx2 downregulation by miR-204, regardless of different gene-delivery methods, and demonstrating that miR-204 functions as a negative regulator of Runx2 in hMSCs. These results suggest that endogenous miR-204/211 can reduce Runx2 protein levels in ST2 cells and hMSCs.
To determine if miR-204/211 sponges specifically interact with miR-204 and miR-211, we cloned miR-204 or miR-211 sponge into CMV-Luc2 reporter at the 3′-site of Luc2 cDNA (Fig. 2G). Transfection of miR-204 or miR-211 had no effect on wild-type (WT) CMV-Luc reporter (Fig. 2H). In contrast, CMV-Luc2-sponge activity was significantly downregulated by the transfection of miR-204 or miR-211 in COS cells (Fig. 2H). We also infected ST2 cells with retrovirus expressing green fluorescent protein (GFP) alone or GFP with sponge at the 3′-UTR and found that ST2 cells infected with GFP-sponge showed less GFP signals than cells with GFP alone (supplemental online Fig. S3). These results suggest that miR-204/211 sponges can specifically interact with miR-204 or miR-211 and miR-204 sponge can bind to both miR-204 and miR-211.
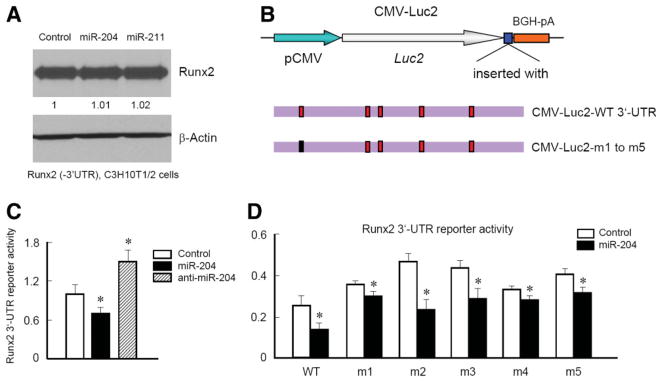
MiR-204 and MiR-211 Bind 3′-UTR of Runx2 mRNA to Downregulate Runx2 Expression
Runx2 has a long 3′-UTR which is ~4 kb and contains at least five putative binding sites with high probability for miR-204/211 interaction. We expressed Runx2 with the SV40 poly(A) sequences to replace the original 3′-UTR of Runx2 in C3H10T1/2 cells, infected cells with retrovirus expressing miR-204 or miR-211, and examined Runx2 protein levels by western blotting. The exclusion of the 3′-UTR of Runx2 abolished the inhibitory effect of miR-204 or miR-211 on Runx2 expression (Fig. 3A), suggesting that miR-204/211 suppress Runx2 through interaction with 3′-UTR of Runx2. To further determine if miR-204 binds 3′-UTR of Runx2 to downregulate Runx2 protein levels, we performed 3′-UTR reporter assay and mutagenesis analysis (Fig. 3B). Luciferase activities of 3′-UTR reporter were examined using C3H10T1/2 cells transfected with control siRNA (the oligo with the identical length as miR-204 but with nonspecific nucleotide sequence), miR-204, or anti-miR-204 oligos. Consistent with the results of Runx2 western blotting, the reporter assays showed that transfection of miR-204 inhibited reporter activity while transfection of anti-miR-204 oligos upregulated the reporter activity (Fig. 3C), suggesting that the 3′-UTR of Runx2 is required for miR-204-induced downregulation of Runx2 protein expression. To further determine which site may be the actual binding site of miR-204/211, we mutated five potential binding sites individually (Fig. 3D) and then co-transfected these 3′-UTR reporters with miR-204 oligo. The results of luciferase activity demonstrated that all five 3′-UTR mutant reporters (m1 to m5) had higher luciferase activity compared with the WT 3′-UTR reporter (Fig. 3D). Transfection of miR-204 retained its inhibitory effects on the reporter even when the putative binding sites were mutated, although the inhibitory potency was different (Fig. 3D). Since m1 and m4 mutated reporters had less responses to the miR-204 transfection, we further construct a reporter with m1 and m4 double mutations. Combination of m1 and m4 mutations abolished the effect of miR-204 on Runx2 3′-UTR reporter in a greater degree than m1 or m4 alone (data not shown), suggesting that the binding sites of m1 and m4 may play more important roles in miR-204 regulated Runx2 protein levels.
Figure 3.
Mir-204 and miR-211 bind to the 3′-UTR of the Runx2 gene. (A): Clonal C3H10T1/2 cells, in which Runx2 is overexpressed under the control of the CMV promoter and SV40 3′-UTR (Runx2 3′-UTR deletion), were infected with control (empty vector), miR-204, or miR-211 expressing retrovirus. After 15-day selection with puromycin, cells were harvested for Runx2 protein expression using western blot analysis. Overexpression of miR-204 or miR-211 did not change Runx2 protein levels, suggesting that Runx2 3′-UTR may be the target of miR-204/211. (B): A diagram of Runx2 3′-UTR reporters. m1, m2, m3, m4, and m5 denote five different Runx2 3′-UTR reporters, which contain one mutated site that cannot be bound to miR-204. (C): The wild-type (WT) Runx2 3′-UTR reporter was cotransfected with miR-204 or anti-miR-204 oligos into C3H10T1/2 cells. Forty-eight hours after transfection, luciferase activities were measured (n = 6). (D): The wild-type Runx2 3′-UTR reporter or mutated 3′-UTR reporter was cotransfected with miR-204 or control oligos into C3H10T1/2 cells. Forty-eight hours after transfection, luciferase activities were measured. *, p < .05, compared with WT for mutated reporters, ANOVA followed by Dunnett’s test.
MiR-204 and MiR-211 Inhibit Osteoblast Differentation
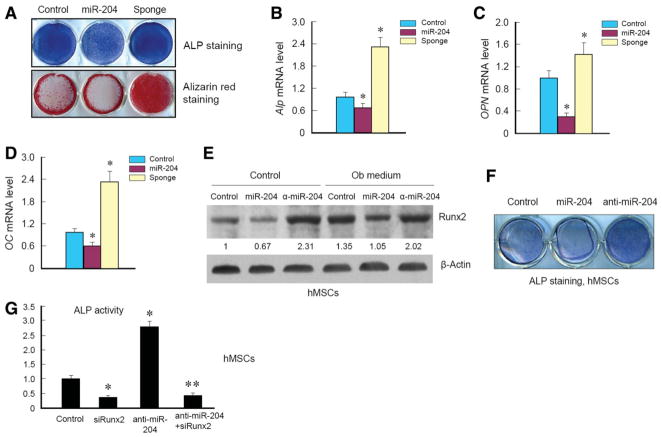
Because miR-204 has an inhibitory effect on Runx2 expression in C3H10T1/2 and ST2 cells and in hMSCs, we next performed a series of experiments to determine if miR-204 has an effect on osteoblast differentiation. We cultured ST2 cells, which stably expressed empty vector, miR-204, or miR-204-sponge by retroviral infection, and treated cells with osteoblast differentiation medium containing BMP-2, ascorbic acid, and β-glycerophosphate. After 3 days of treatment, cells were subjected to alkaline phosphatase (ALP) staining. Next, we treated cells for 10 days and stained cells with Alizarin red to evaluate changes in bone matrix mineralization. Expression of miR-204 significantly decreased ALP activity and mineralized bone matrix formation, and expression of miR-204-sponge resulted in an enhanced ALP activity and mineralized bone matrix formation, compared with cells infected with retroviral empty vector (Fig. 4A). These results suggest that miR-204 has a negative effect on osteoblast differentiation and subsequent mineralization, possibly through negative regulation of Runx2 protein levels. To further confirm these findings, we performed quantitative RT-PCR assay to determine if osteoblast marker genes, such as Alp, OPN, and OC, were altered in a manner correlated with miR-204 perturbation. With BMP-2 treatment, Alp, OPN, and OC were significantly induced in all three groups of cells (control, miR-204, and miR-204-sponge). Expression of miR-204 significantly inhibited Alp, OPN, and OC expression. In contrast, expression of miR-204-sponge significantly upregulated expression of Alp, OPN, and OC in ST2 cells (Fig. 4B–4D).
Figure 4.
MiR-204 inhibits osteoblast differentiation. (A–D): ST2 cells were infected with control virus or retrovirus expressing miR-204 or miR-204-sponge and treated with BMP-2 (300 ng/ml), ascorbic acid, and β-glycerophosphate. ALP staining was performed 3 days after BMP-2 treatment and Alizarin red staining was performed 10 days after BMP-2 treatment. The expression of Alp (B), OPN (C), and OC (D) was measured by quantitative reverse transcription polymerase chain reaction. *, p < .05, unpaired Student’s t-test, n = 3. (E, F): Human mesenchymal stem cells were transfected with control siRNA, miR-204, or anti-miR-204 oligos for 2 days and then treated with osteoblast differentiation medium for additional 12 days and subjected to western blotting or stained for ALP activity measurement. (G): hMSCs were transfected with control siRNA, Runx2 siRNA, anti-miR-204, or a combination of Runx2 siRNA and anti-miR-204 for 2 days and then treated with osteoblast differentiation medium for additional 12 days and ALP activity was measured. *, p < .05, unpaired Student’s t-test, n = 3. Abbreviations: ALP, alkaline phosphatase; hMSCs, human mesenchymal stem cells.
To determine the effects of miR-204 on hMSC differentiation, we transfected control oligo, miR-204, and anti-miR-204 oligo into hMSCs and found that expression of miR-204 reduced Runx2 protein levels and transfection of anti-miR-204 oligo increased Runx2 protein levels under normal conditions or under culture conditions with osteoblast differentiation medium (Fig. 4E). Expression of miR-204 inhibited ALP activity and transfection of anti-miR-204 oligo enhanced ALP activity in hMSCs (Fig. 4F).
To determine if miR-204-regulated osteoblast differentiation is Runx2-dependent, we examined the effect of Runx2 siRNA on miR-204-regulated osteoblast differentiation. We found that transfection of Runx2 siRNA significantly reduced ALP activity and transfection of anti-miR-204 oligo significantly upregulated ALP activity in hMSCs (Fig. 4G). Cotransfection of Runx2 siRNA with anti-miR-204 oligo completely blocked anti-miR-204-induced ALP activity (Fig. 4G), suggesting that the stimulatory effect of anti-miR-204 on ALP activity is Runx2-dependent. Taken together, our results suggest that miR-204/211 play a negative role in osteoblast differentiation through downregulation of Runx2.
MiR-204 Promotes Adipocyte Differentiation
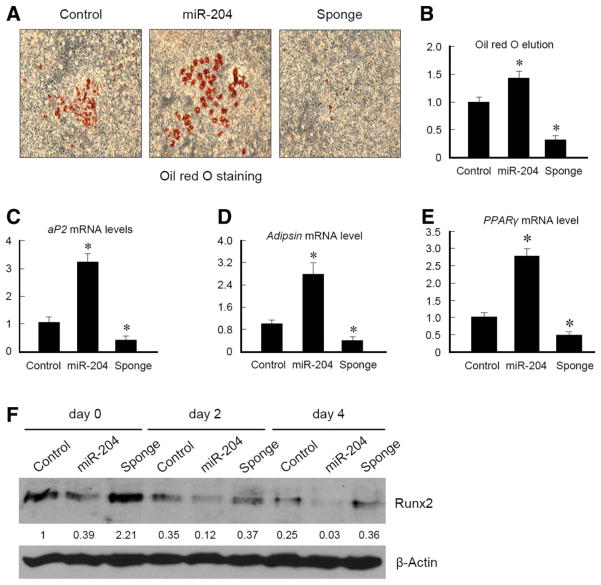
The commitment and differentiation of MSCs are determined by regulation of the key factors such as Runx2 and PPARγ in various differentiation processes [12–14]. It has been reported that Runx2 deficiency causes enhanced adipogenesis [12]. Thus, we were interested if Runx2 regulation by miR-204/211 has an impact on adipocyte differentiation. We used ST2 cells, in which miR-204 or miR-204-sponge was overexpressed, for adipocyte differentiation assay. Cells were treated with adipocyte differentiation medium for 10 days. Results of Oil red O staining showed that miR-204 expression induced adipocyte formation, demonstrated by more oil droplet staining, and expression of miR-204-sponge almost completely inhibited adipocyte formation (Fig. 5A). Quantification of Oil red O eluted from cells demonstrated that miR-204 overexpressing cells globally formed more adipocytes (Fig. 5B). This finding was further confirmed by analysis of expression of adipocyte marker genes such as aP2, adipsin, and PPARγ (Fig. 5C–5E). In miR-204 overexpressing cells, expression of aP2, adipsin, and PPARγ was upregulated. In contrast, expression of aP2, adipsin, and PPARγ was suppressed in miR-204-sponge overexpressing cells (Fig. 5C–5E). Consistent with the above findings, Runx2 protein levels were significantly regulated by miR-204 and miR-204-sponge when ST2 cells were cultured with adipocyte differentiation medium (Fig. 5F). Collectively, our results suggest that miR-204 is a positive regulator of adipocyte differentiation, possibly by downregulation of Runx2.
Figure 5.
MiR-204 promotes adipocyte differentiation. (A): ST2 cells were infected with control virus or retrovirus expressing miR-204 or miR-204-sponge, cultured in adipocyte differentiation medium for 9 days, and then subjected to Oil red O staining. (B): Oil red O was eluted with isopropanol from stained cells and quantified to measure which sample has more Oil red O-positive staining. (C–E): mRNA was extracted from ST2 cells cultured with adipocyte differentiation medium for 9 days and quantitative RT-PCR was performed using aP2, adipsin, and PPARγ primers. *, p < .05, unpaired Student’s t-test, n = 3. (F): ST2 cells infected with control virus or retrovirus expressing miR-204 or miR-204-sponge were cultured in adipocyte differentiation medium for indicated days and harvested for western blotting to examine Runx2 protein levels.
Discussion
In this work, we identified miR-204 and miR-211 as negative regulators of osteoblast differentiation in mesenchymal progenitor cells and BMSCs. MiR-204/211 are expressed in these cells and have multiple conserved binding sites within 4-kb 3′-UTR of the Runx2 gene. Inhibition of miR-204/211 significantly elevated the Runx2 protein levels in these cells, demonstrating that miR-204 and miR-211 are endogenous attenuators of Runx2 expression to restrain cells from differentiating into osteoblasts. The regulation of Runx2 protein also affects the adipogenic potential of progenitor cells, suggesting an inhibitory effect of Runx2 on preventing progenitor cells from being committed to adipocyte lineage. Taken together with findings from other groups about inverse relationship between osteoblast and adipocyte differentiation, our results confirmed that cross-talk among key lineage-specific regulators may contribute to the maintenance of stem cell status.
MicroRNAs, as small molecular regulators of gene expression, play critical roles in stem cell function [15, 26]. Their ability of fine-tuning protein levels is often exploited against key proteins involved in self-renewal or differentiation, such as nanog targeted by miR-21 in ES cells [27] and serum response factor (SRF) targeted by miR-133 in myogenesis [28]. As a positive regulator of myogenesis, miR-133 is specifically expressed in adult cardiac and skeletal muscle tissues and robustly upregulated during the myoblast differentiation of C2C12 cells [28, 29]. Interestingly, miR-133 was recently shown to be downregulated in BMP-2 treated C2C12 cells [30], suggesting that miR-133 may have an additional role in suppressing BMP-2 induced osteoblast differentiation of C2C12 cells besides its well documented function in myogenesis. In the study described above, a reporter containing ~400 bp Runx2 3′-UTR fragment and Runx2 protein levels were regulated by miR-133. In addition, miR-133 also regulates expression of osteoblast marker genes, such as Alp and osteocalcin [30]. Thus, miR-133 may also have a regulatory effect on Runx2 expression, along with miR-204/211, in mesenchymal progenitor cells and BMSCs.
As a key regulator in osteogenesis, Runx2 is likely subjected to multiple levels of regulation in BMSCs [31, 32]. Intriguingly, Runx2 has a considerably long 3′-UTR (~4 kb), presumably containing multiple regulatory elements. This prompted us to study whether there are miRNAs binding to the 3′-UTR of Runx2 and suppressing its translation, since it was recognized that miRNAs have emerged as a new regulatory mechanism of gene expression in recent years. As shown in our results, miR-204/211 are recruited to regulate the expression of Runx2, an important factor in osteogenesis, to regulate differentiation of BMSCs. However, it should be noted that Runx2 protein expression is not exclusively regulated by miRNAs in BMSC differentiation, as our data (Fig. 5F) demonstrated that decrease of Runx2 protein levels during adipocyte differentiation cannot be reversed by miR-204 sponge expression, although miR-204 perturbation did have significant effects on Runx2 protein levels. Thus, our results may suggest that Runx2 expression in BMSC differentiation is regulated by multiple mechanisms, which could be translational suppression by miRNAs and other post-transcriptional mechanism like ubiquitin-proteasome dependent degradation.
Besides other post-transcriptional/translational regulations, miRNAs provide a tool for fine-tuning protein levels, and this attenuation would be precisely controlled by miRNA expression, processing, or accessibility [33]. Nevertheless, these micro-rheostats play significant roles in diverse signaling pathway and physiological/pathological processes, demonstrated by genetic studies of knockout mice of miRNA processor Dicer and miRNA genes [15, 34]. This raises a possibility that administration of miRNA mimic or miRNA inhibitors may be used as therapies for diseases as consequences of BMSC malfunction.
Conclusion
In mesenchymal progenitor cells and in BMSCs, transcription factor Runx2 is downregulated by miR-204. This attenuation of Runx2 expression has a negative effect on osteoblast differentiation but induces adipocyte differentiation, suggesting that Runx2 regulation by miR-204 represents a novel mechanism regulating progenitor cell and BMSC differentiation.
Supplementary Material
Acknowledgments
This work was supported in part by Grant N08G-070 to D.C. from New York State Department of Health and the Empire State Stem Cell Board and by Grants R01 AR051189, R01 AR054465, and K02 AR052411 to D.C. from the National Institute of Health.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author contributions: J.H.: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; L.Z.: Provision of study material, collection and/or assembly of data; L.X.: Provision of study material, discussion of results; D.C.: Conception and design, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
References
- 1.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 2.Tuli R, Tuli S, Nandi S, et al. Characterization of multipotential mesenchymal progenitor cells derived from human trabecular bone. Stem Cells. 2003;21:681–693. doi: 10.1634/stemcells.21-6-681. [DOI] [PubMed] [Google Scholar]
- 3.Duque G. Bone and fat connection in aging bone. Curr Opin Rheumatol. 2008;20:429–434. doi: 10.1097/BOR.0b013e3283025e9c. [DOI] [PubMed] [Google Scholar]
- 4.Meunier P, Aaron J, Edouard C, et al. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A Quantitative Study Of 84 Iliac Bone Biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- 5.Verma S, Rajaratnam JH, Denton J, et al. Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J Clin Pathol. 2002;55:693–698. doi: 10.1136/jcp.55.9.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu W, Andersen TE, Bollerslev J, et al. Patients with high bone mass phenotype exhibit enhanced osteoblast differentiation and inhibition of adipogenesis of human mesenchymal stem cells. J Bone Miner Res. 2007;22:1720–1731. doi: 10.1359/jbmr.070721. [DOI] [PubMed] [Google Scholar]
- 7.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida CA, Yamamoto H, Fujita T, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen ED, Walkey CJ, Puigserver P, et al. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 10.Bennett CN, Longo KA, Wright WS, et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto H, Furuichi T, Zanma A, et al. Runx2 deficiency in chondrocytes causes adipogenic changes in vitro. J Cell Sci. 2004;117:417–425. doi: 10.1242/jcs.00866. [DOI] [PubMed] [Google Scholar]
- 13.Akune T, Ohba S, Kamekura S, et al. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J Clin Invest. 2004;113:846–855. doi: 10.1172/JCI19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong JH, Hwang ES, McManus MT, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 15.Gangaraju VK, Lin H. MicroRNAs: Key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Guo R, Lu Y, et al. VEGF-C, a lymphatic growth factor, is a RANKL target gene in osteoclasts that enhances osteoclastic bone resorption through an autocrine mechanism. J Biol Chem. 2008;283:13491–13499. doi: 10.1074/jbc.M708055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura T, Koshino Y, Shibata F, et al. Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 19.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi H, Shimizu K, Kozu T, et al. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takazawa Y, Tsuji K, Nifuji A, et al. An osteogenesis-related transcription factor, core-binding factor A1, is constitutively expressed in the chondrocytic cell line TC6, and its expression is upregulated by bone morphogenetic protein-2. J Endocrinol. 2000;165:579–586. doi: 10.1677/joe.0.1650579. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa E, Maruyama M, Kagoshima H, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 24.John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. Plos Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakshmipathy U, Hart RP. Concise review: MicroRNA expression in multipotent mesenchymal stromal cells. Stem Cells. 2008;26:356–363. doi: 10.1634/stemcells.2007-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 28.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bialek P, Kern B, Yang X, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 32.Shen R, Wang X, Drissi H, et al. Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J Biol Chem. 2006;281:16347–16353. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.