Abstract
Background
Anatid herpesvirus 1 (AHV-1) is known for the difficulty of monitoring and controlling, because it has a long period of asymptomatic carrier state in waterfowls. Furthermore, as a significant essential agent for viral attachment, release, stability and virulence, gC (UL44) gene and its protein product (glycoprotein C) may play a key role in the epidemiological screening. The objectives of this study were to rapidly, sensitively, quantitatively detect gC gene of AHV-1 and provide the underlying basis for further investigating pcDNA3.1-gC DNA vaccine in infected ducks by TaqMan™ fluorescent quantitative real-time PCR assay (FQ-PCR) with pcDNA3.1-gC plasmid.
Results
The repeatable and reproducible quantitative assay was established by the standard curve with a wide dynamic range (eight logarithmic units of concentration) and very good correlation values (1.000). This protocol was able to detect as little as 1.0 × 101 DNA copies per reaction and it was highly specific to AHV-1. The TaqMan™ FQ-PCR assay successfully detected the gC gene in tissue samples from pcDNA3.1-gC and AHV-1 attenuated vaccine (AHV-1 Cha) strain inoculated ducks respectively.
Conclusions
The assay offers an attractive method for the detection of AHV-1, the investigation of distribution pattern of AHV-1 in vivo and molecular epidemiological screening. Meanwhile, this method could expedite related AHV-1 and gC DNA vaccine research.
Background
Anatid herpesvirus 1 (AHV-1) infection alternatively known as duck virus enteritis (DVE), or duck plague (DP), is one of the most widespread and devastating diseases of waterfowls in the family Anatidae[1]. As an acute and contagious herpesvirus, AHV-1 can infect ducks, geese, and swans of all ages and species[2]. Since the first outbreak in the Netherlands in 1923, AHV-1 had a dramatic impact on international trade of waterfowls and waterfowl products throughout the world [3-5]. Like other herpesviruses, AHV-1 can be carried and periodically shed by recovered birds from the disease. Moreover, the reactivation of latent AHV-1 may threaten domestic and migrating waterfowls populations[6]. AHV-1 has already become an important potential risk factor for waterfowls health.
As a significant agent of AHV-1, gC (UL44) gene has seldom been reported about the research of its molecular biology, and its research level fall behind relatively in other herpesviruses[7]. Although gC is nonessential component for the viral replication, its protein product (glycoprotein C) has several important biological functions. As a multifunctional glycoprotein in Alphaherpesvirinae, glycoprotein C involves in viral attachment, release, stability, virulence and other functions [8-14]. Being situated on the envelope surface of mature virus particles, glycoprotein C contains many antigen determinants, and can adequately induce immune response [15-18]. Some DNA vaccines based on gC gene from other kinds of herpesviruses immunized in mice or other relative animals could receive good immune responses and protective efficacy [19-23], while the biological functions of AHV-1 glycoprotein C and DNA vaccine based on AHV-1 gC have not been reported. In this study, pcDNA3.1-gC plasmid is not only used as standard DNA to develop a standard curve for TaqMan™ FQ-PCR but also as a DNA vaccine to inoculate ducks.
Many diagnosis and detection methods about AHV-1 have been reported in a long time, such as epidemiological information, viral isolation and immunological methods [24-28]. These tests are laborious and time-consuming resulting from requiring strict operation. Thus, these methods can not be used to direct detection. In addition, the reliable diagnosis is difficult to obtain from mixed or secondary infected waterfowls. AHV-1 is difficult to be monitored and controlled because it has a long period of asymptomatic carrier state in waterfowls[29]. It is usually detected only during the intermittent shedding period of the virus. Thus, how to sensitively detect AHV-1 has become a significant factor from infected waterfowls. PCR is a useful tool with high sensitivity for detecting nucleic acids of virus from the ducks [30-33]. However, the traditional PCR assays still had some flaws, such as poor performance in quantitation and a relative waste of time. It is not suitable for large-scale applications.
In recent times, a more sensitive, time-saving and advanced method has emerged in the field, which is fluorescent quantitative real-time PCR (FQ-PCR). This technology accurately quantifies target DNA in a given sample and then could accurately detect viral loads in clinical samples[34]. Yang and Guo have reported the detection of AHV-1 with FQ-PCR method [35,36]. FQ-PCR based on TaqMan™ technology provides certain advantages including high sensitivity, high specificity, and reproducibility, and has been widely used to quantify the copies of viral genomic after optimization [37-44]. In this study, the developed FQ-PCR method was extremely valuable for AHV-1 detection. Moreover, the results provide some interesting basic data that may be beneficial to further investigate pcDNA3.1-gC DNA vaccine in vivo in ducks.
Results
Development and optimization of a TaqMan™ FQ-PCR
Final concentrations of primers each of 0.5 μmol/L and probe of 0.25 μmol/L were selected, and the optimized annealing temperature was 53°C. The combination of primers, probe and annealing temperature was used for subsequent experiments.
Standard curve establishment
The amplification curves (Figure 1.a.) and standard curve (Figure 1.b.) of the TaqMan™ FQ-PCR were generated by using the 10-fold dilutions of pcDNA3.1-gC, which has already known its copies to undertake FQ-PCR reaction under optimum conditions with the iCycler IQ Detection System. The curve covered a dynamic range of eight log units of concentration and displayed a clear linear relationship with a correlation coefficient of 1.000 and high amplification efficiency (100%). By using the following formula, we were able to quantify the amount of unknown samples: Y = -3.321X + 45.822 (Y = threshold cycle, X = log starting quantity).
Figure 1.
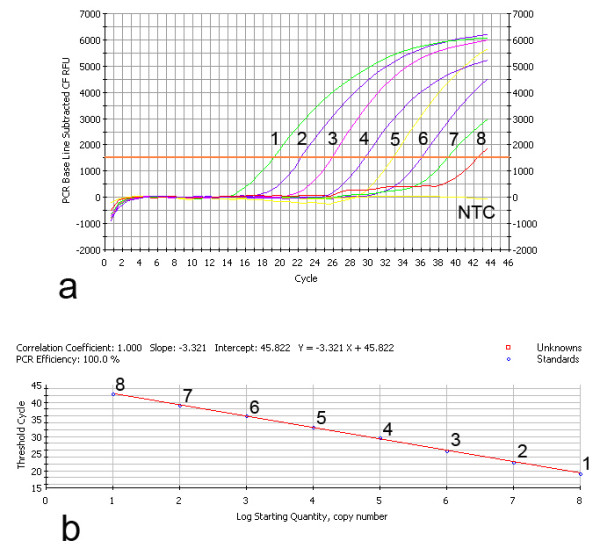
The amplification curves (Figure 1.a.) and standard curve (Figure 1.b.) of the TaqMan™ FQ-PCR detection. Ten-fold dilutions of standard DNA ranging from 1.0 × 108 to 1.0 × 101 copies/reaction were used (1-8), as indicated in the x-axis, whereas the corresponding Ct values are presented on the y-axis. The correlation coefficient and the slope value of the regression curve were calculated and indicated.
Amplification sensitivity, specificity, repeatability and reproducibility
Ten-fold dilution series of pcDNA3.1-gC standard DNA (from 1.0 × 105 to 1.0 × 100 copies/reaction) were tested by the established FQ-PCR assay to evaluate the sensitivity of the system, the mean threshold cycle (Ct) values were 29.60, 33.10, 36.43, 39.30, 42.57 and N/A respectively. The results showed that the assay could detect down to 1.0 × 101 copies per reaction (Figure 2.). All liver samples were retested positive for AHV-1 from infected ducks with the established FQ-PCR assay, it indicated that this method was sensitive for clinical cases. Comparisons were made between the established FQ-PCR method and conventional PCR method by using 10-fold dilutions of viral DNA from infected allantoic fluid to calculate the end-point sensitivity of each assay. The results showed that the established FQ-PCR could detect viral DNA down to dilutions of 2.730 × 101, while the dilutions of only 2.730 × 104 for conventional PCR.
Figure 2.
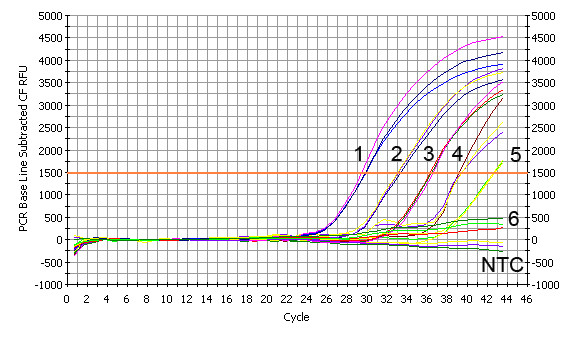
The sensitivity of TaqMan™ FQ-PCR detection. Ten-fold serial dilutions of AHV-1 standard template were used (1-6), 1.0 × 105-1.0 × 100 copies/reaction of AHV-1 standard template. As shown in the figure, the detection limit for the assay was 1.0 × 101 copies.
The specificity test showed that pcDNA3.1-gC, AHV-1 attenuated vaccine (AHV-1 Cha) strain virus and AHV-1 virulent (AHV-1 Chv) strain virus were found positive for AHV-1 by the established FQ-PCR assay, while the bacteria, remaining viruses including negative control (liver sample of the healthy duck) were negative (Figure 3.). The results were confirmed by gel electrophoresis, there was a band of the expected size (78 bp) observed exclusively from samples of pcDNA3.1-gC, AHV-1 Cha and AHV-1 Chv. It indicated that the established FQ-PCR assay was highly specific.
Figure 3.
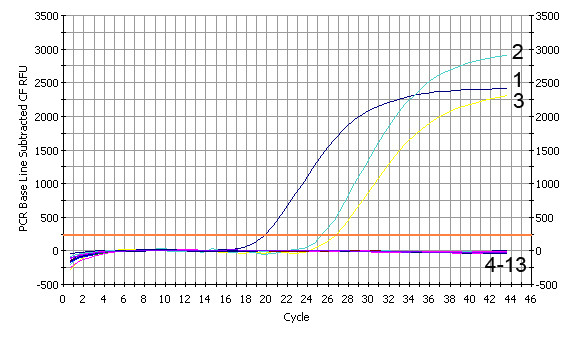
The specificity of TaqMan™ FQ-PCR detection. The pcDNA3.1-gC (1), AHV-1 Cha (2), AHV-1 Chv (3), gosling new type viral enteritis virus (4), duck hepatitis virus type1 (5), duck adenovirus (6), goose parvovirus (7), Marek's disease virus (8), Pasteurella multocida (5:A) (9), Escherichia coli (O78) (10), Salmonella enteritidis (No. 50338) (11), the liver DNA of the healthy duck (12) and NTC (13) were tested to evaluate the specificity of the assay by FQ-PCR.
In the intra-assay and inter-assay, the mean Ct values and standard deviations (SD) values were calculated. As shown in Table 1, the coefficient of variation (CV) values ranged from 0.44% to 2.03%, indicating that this assay was highly repeatable and reproducible.
Table 1.
Intra-assay and inter-assay of the TaqMan™ FQ-PCR assay
| Variation | Copies of standard | Crossing point | ||
|---|---|---|---|---|
| Mean | SDa | CV (%)b | ||
| Intra-assay | 1.00E+08 | 19.73 | 0.15 | 0.77 |
| 1.00E+07 | 23.10 | 0.20 | 0.87 | |
| 1.00E+06 | 26.37 | 0.29 | 1.09 | |
| 1.00E+05 | 29.63 | 0.21 | 0.70 | |
| 1.00E+04 | 33.07 | 0.31 | 0.92 | |
| 1.00E+03 | 36.33 | 0.31 | 0.84 | |
| 1.00E+02 | 39.50 | 0.17 | 0.44 | |
| 1.00E+01 | 42.67 | 0.32 | 0.75 | |
| Inter-assay | 1.00E+08 | 19.70 | 0.28 | 1.41 |
| 1.00E+07 | 23.09 | 0.47 | 2.03 | |
| 1.00E+06 | 26.31 | 0.32 | 1.23 | |
| 1.00E+05 | 29.59 | 0.42 | 1.42 | |
| 1.00E+04 | 33.04 | 0.34 | 1.03 | |
| 1.00E+03 | 36.35 | 0.41 | 1.14 | |
| 1.00E+02 | 39.52 | 0.43 | 1.10 | |
| 1.00E+01 | 42.70 | 0.41 | 0.97 | |
Repeatability and reproducibility (R & R) of the TaqMan™ FQ-PCR assay.
a Standard deviation.
b Coefficient of variation.
Detection of AHV-1 gC gene in samples for practical applications
AHV-1 gC gene and viral load quantification demonstrated that the AHV-1 gC copies of each sample could be calculated by using the Ct value determined from the standard curve. As shown in Table 2, AHV-1 gC can be detected in all analyzed tissues at 1 hour postinoculation. gC copies of all tissues reached a peak at 1 hour postinoculation in gC DNA vaccine-inoculated ducks, while the copies of most tissues (other than kidney) reached a peak at 4 hours postinoculation in AHV-1 Cha strain-infected ducks. The concentration of nucleic acid in DNA vaccine-inoculated ducks maintained 107 copies/g level at 4 weeks postinoculation. The copies of the liver, spleen and thymus were more than other tissues in gC DNA vaccine-inoculated ducks, while the copies of the duodenum and rectum were relatively low in AHV-1 Cha strain-infected ducks.
Table 2.
Mean AHV-1 gC copies and viral loads in different samples for practical applications
| Samples (lg copies/g) |
Groups | 1 h | 4 h | 8 h | 12 h | 1 d | 3 d | 5 d | 7 d | 2 wk | 4 wk |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | 1 | 9.38 ± 0.23 | 9.15 ± 0.15 | 8.93 ± 0.18 | 8.62 ± 0.09 | 8.41 ± 0.19 | 8.18 ± 0.08 | 8.06 ± 0.20 | 8.03 ± 0.12 | 8.03 ± 0.25 | 7.98 ± 0.14 |
| 2 | 8.45 ± 0.01 | 8.62 ± 0.13 | 8.58 ± 0.08 | 8.37 ± 0.07 | 8.23 ± 0.10 | 7.95 ± 0.19 | 7.72 ± 0.16 | 7.57 ± 0.16 | 7.28 ± 0.14 | 6.87 ± 0.19 | |
| Pancreas | 1 | 8.32 ± 0.03 | 8.21 ± 0.07 | 8.13 ± 0.04 | 8.01 ± 0.12 | 7.95 ± 0.09 | 7.87 ± 0.05 | 7.73 ± 0.17 | 7.56 ± 0.07 | 7.42 ± 0.15 | 7.35 ± 0.05 |
| 2 | 8.16 ± 0.16 | 8.38 ± 0.10 | 8.22 ± 0.14 | 8.08 ± 0.08 | 8.00 ± 0.11 | 7.98 ± 0.06 | 7.86 ± 0.14 | 7.61 ± 0.18 | 7.2 ± 0.03 | 6.69 ± 0.05 | |
| Spleen | 1 | 9.23 ± 0.11 | 9.06 ± 0.06 | 8.75 ± 0.10 | 8.52 ± 0.04 | 8.32 ± 0.16 | 8.26 ± 0.14 | 8.15 ± 0.18 | 8.12 ± 0.24 | 7.94 ± 0.08 | 7.87 ± 0.17 |
| 2 | 9.26 ± 0.08 | 9.49 ± 0.06 | 9.21 ± 0.13 | 8.92 ± 0.16 | 8.75 ± 0.20 | 8.51 ± 0.13 | 8.33 ± 0.07 | 8.07 ± 0.81 | 7.68 ± 0.13 | 7.53 ± 0.12 | |
| Kidney | 1 | 8.47 ± 0.02 | 8.36 ± 0.05 | 8.24 ± 0.14 | 8.13 ± 0.13 | 7.94 ± 0.11 | 7.62 ± 0.05 | 7.48 ± 0.18 | 7.31 ± 0.27 | 7.33 ± 0.13 | 7.29 ± 0.11 |
| 2 | 8.41 ± 0.13 | 8.33 ± 0.17 | 8.23 ± 0.08 | 8.05 ± 0.15 | 7.97 ± 0.04 | 7.82 ± 0.09 | 7.79 ± 0.11 | 7.58 ± 0.14 | 6.96 ± 0.12 | 6.84 ± 0.06 | |
| Lung | 1 | 8.89 ± 0.07 | 8.77 ± 0.10 | 8.44 ± 0.14 | 8.14 ± 0.11 | 8.02 ± 0.05 | 7.88 ± 0.18 | 7.51 ± 0.21 | 7.50 ± 0.16 | 7.42 ± 0.11 | 7.30 ± 0.16 |
| 2 | 8.34 ± 0.03 | 8.58 ± 0.03 | 8.42 ± 0.17 | 8.21 ± 0.04 | 8.14 ± 0.16 | 7.86 ± 0.13 | 7.82 ± 0.18 | 7.62 ± 0.21 | 7.35 ± 0.21 | 7.11 ± 0.15 | |
| Thymus | 1 | 9.2 ± 0.16 | 9.04 ± 0.11 | 8.86 ± 0.18 | 8.52 ± 0.05 | 8.4 ± 0.19 | 8.23 ± 0.15 | 8.11 ± 0.17 | 8.03 ± 0.01 | 7.95 ± 0.05 | 7.84 ± 0.08 |
| 2 | 9.27 ± 0.14 | 9.41 ± 0.18 | 9.22 ± 0.06 | 8.94 ± 0.20 | 8.72 ± 0.13 | 8.38 ± 0.07 | 8.04 ± 0.17 | 7.77 ± 0.05 | 7.56 ± 0.14 | 7.39 ± 0.15 | |
| Heart | 1 | 8.68 ± 0.07 | 8.59 ± 0.14 | 8.44 ± 0.17 | 8.20 ± 0.03 | 8.13 ± 0.02 | 8.07 ± 0.13 | 8.02 ± 0.25 | 7.94 ± 0.17 | 7.82 ± 0.22 | 7.76 ± 0.14 |
| 2 | 9.06 ± 0.05 | 9.14 ± 0.16 | 9.08 ± 0.16 | 8.95 ± 0.14 | 8.63 ± 0.11 | 8.34 ± 0.18 | 8.12 ± 0.06 | 7.71 ± 0.11 | 7.52 ± 0.21 | 7.23 ± 0.12 | |
| Brain | 1 | 8.25 ± 0.09 | 8.04 ± 0.07 | 7.83 ± 0.18 | 7.77 ± 0.16 | 7.71 ± 0.12 | 7.67 ± 0.04 | 7.64 ± 0.09 | 7.48 ± 0.13 | 7.39 ± 0.15 | 7.24 ± 0.07 |
| 2 | 9.05 ± 0.11 | 9.18 ± 0.13 | 9.02 ± 0.05 | 8.93 ± 0.08 | 8.64 ± 0.02 | 8.36 ± 0.05 | 7.84 ± 0.10 | 7.57 ± 0.16 | 7.33 ± 0.19 | 7.14 ± 0.18 | |
| Duodenum | 1 | 8.77 ± 0.02 | 8.59 ± 0.07 | 8.29 ± 0.23 | 8.05 ± 0.11 | 7.94 ± 0.13 | 7.85 ± 0.07 | 7.81 ± 0.21 | 7.78 ± 0.27 | 7.63 ± 0.20 | 7.61 ± 0.19 |
| 2 | 8.25 ± 0.16 | 8.33 ± 0.03 | 8.23 ± 0.07 | 8.16 ± 0.09 | 8.04 ± 0.02 | 7.69 ± 0.15 | 7.41 ± 0.19 | 7.26 ± 0.16 | 6.81 ± 0.09 | 6.64 ± 0.11 | |
| Rectum | 1 | 8.68 ± 0.07 | 8.47 ± 0.16 | 8.26 ± 0.10 | 7.98 ± 0.23 | 7.94 ± 0.23 | 7.88 ± 0.17 | 7.91 ± 0.27 | 7.83 ± 0.14 | 7.74 ± 0.03 | 7.70 ± 0.18 |
| 2 | 8.23 ± 0.05 | 8.27 ± 0.08 | 8.18 ± 0.11 | 8.12 ± 0.05 | 8.02 ± 0.10 | 7.78 ± 0.17 | 7.52 ± 0.15 | 7.21 ± 0.06 | 6.95 ± 0.09 | 6.78 ± 0.05 | |
| Harderian gland | 1 | 8.32 ± 0.08 | 8.11 ± 0.04 | 7.94 ± 0.21 | 7.72 ± 0.14 | 7.62 ± 0.09 | 7.54 ± 0.24 | 7.5 ± 0.08 | 7.38 ± 0.22 | 7.41 ± 0.29 | 7.33 ± 0.19 |
| 2 | 8.23 ± 0.15 | 8.35 ± 0.07 | 8.21 ± 0.11 | 8.13 ± 0.14 | 8.05 ± 0.19 | 7.77 ± 0.14 | 7.52 ± 0.07 | 7.18 ± 0.20 | 6.55 ± 0.01 | 6.39 ± 0.04 | |
| Bursa of Fabricius | 1 | 8.86 ± 0.07 | 8.80 ± 0.05 | 8.50 ± 0.10 | 8.17 ± 0.09 | 8.02 ± 0.17 | 7.85 ± 0.19 | 7.78 ± 0.11 | 7.72 ± 0.09 | 7.6 ± 0.22 | 7.63 ± 0.25 |
| 2 | 8.83 ± 0.11 | 8.92 ± 0.13 | 8.88 ± 0.07 | 8.79 ± 0.17 | 8.44 ± 0.19 | 8.25 ± 0.15 | 7.88 ± 0.19 | 7.47 ± 0.10 | 6.68 ± 0.08 | 6.53 ± 0.13 |
Group 1: pcDNA3.1-gC
Group 2: AHV-1 Cha strain vaccine
Discussion
The accurate and prompt diagnosis of AHV-1 infection in waterfowls is a vital part of surveillance and disease control strategy. Currently, the diagnosis of AHV-1 usually depends on epidemiological information, clinical symptoms, pathological changes and serological methods [45-47]. However, these methods are time-consuming, inconvenient, and requiring special collection and transport conditions to maintain the viability of the virus, and the whole process may take 1 to 2 weeks. Virus can not be promptly detected from infected waterfowls with these methods. The conventional qualitative PCR method is also developed for the diagnosis of AHV-1 infection, which may not provide the sensitivity that is needed to detect low-level of viral loads. FQ-PCR is based on the conventional principles of PCR and has being become an increasingly popular way for the diagnosis of bacteria and viruses infection. The diagnostic process requires only 4 hours for detection and quantitation of bacteria and viruses from nucleic acid extraction to FQ-PCR.
The FQ-PCR assay has more advantages than conventional qualitative PCR assays, including rapidity, higher sensitivity, higher specificity, quantitive measurement, decreased risk of cross-contamination through absence of post-PCR handling and automated product detection[48]. An oligonucleotide probe of the TaqMan™ FQ-PCR assay is not included in conventional qualitative PCR, and is labelled at 5' with FAM dye as reporter and labelled at 3' with TAMRA as quencher. It facilitates highly specific binding to the targeted sequence, and results in greater accuracy in the measurement.
Previous studies have detected AHV-1 by FQ-PCR in infected ducks [35,36]. However, Yang et al. developed a relatively narrowed dynamic range for FQ-PCR, it may not be beneficial to large-scale detection in various infected cases. Guo et al. established a similar dynamic range (from 1.0 × 109 to 1.0 × 102 copies), but the end-point sensitivity (1.0 × 101 copies) was not included in the standard curve, the method may not be reliable to quantitate a low viral load (<1.0 × 102 copies). In this study, the comparisons were carried out between the established FQ-PCR method and conventional PCR method for AHV-1 detection from infected allantoic fluid, the results indicated that the established FQ-PCR method is approximately 103 times more sensitive and reliable than the conventional PCR method for clinical cases. A FQ-PCR assay was established to be highly specific for AHV-1, and had a sensitive detection limit of 1.0 × 101 DNA copies per reaction in this study, which produced excellent linear with the DNA concentration from 1.0 × 108 to 1.0 × 101 copies, with correlation coefficient of 1.000 and a reaction efficiency of 100%. The linear amplification of this assay covered a wide dynamic range suitable for quantitative applications.
The potential contamination of AHV-1 DNA that could lead to false-positive results and it was a major concern in this study. This problem was successfully avoided through the findings of high Ct values (low copies) in this assay. Furthermore, no template controls (NTCs) always be included on every plate in every experiment, which can identify the extent of pollution during the test [49]. NTCs and template controls from healthy ducks had no amplification signal in this assay, it is reasonable to think that the sample amplification is real.
The distribution and concentration of AHV-1 has been investigated in AHV-1 Cha strain-infected ducks by Qi [50]. This assay was similar with Qi's report about the distribution of the different kinds of tissues. AHV-1 attenuated vaccine can be distributed in various tissues and organs of ducks within 1 hour by subcutaneous route in this study, furthermore, the concentration of nucleic acid maintained at least 106 copies/g level at 4 weeks postinoculation. They revealed that AHV-1 attenuated vaccine can play an important role against the virulent AHV-1 in the immune ducks, but the copies of the duodenum and rectum were relatively low in infected ducks, it implyed that the various inoculate routes have large impact on the replication of vaccine virus in digestive tracts, and consistents with the gradual circulation of lymphocytes [6].
Plasmid DNA has been confirmed to widely distributed in the thymus, heart, lung, kidney, liver, mesenteric lymph nodes and other organs in a short time by intramuscular injection of DNA vaccine [51,52]. In this study, AHV-1 gC can be detected in all analyzed tissues at 1 hour postinoculation, and the concentration of gC maintained 107 copies/g level at 4 weeks postinoculation. The copies of gC in the liver, spleen and thymus were more than other tissues, and it may be due to plasmid was widely distributed in all tissues through the lymphatic flow and blood circulation in a short time [53]. These basic data can set the stage for further research about gC DNA vaccine.
Currently, the surveillance of AHV-1 becomes difficult because of the inability to differentiate the infected from vaccinated animals (DIVA). The DIVA strategy has only been recently put into practice for avian influenza virus (AIV) [54,55]. In this study, the virus loads and gC gene copy number can be accurately detected by the established FQ-PCR from inoculated ducks, because the animals were certificated as AHV-1-free by qualitative PCR assay before being infected with AHV-1 Cha and pcDNA3.1-gC. Among the different DIVA strategies, one approach is to use a DNA vaccine based on an incomplete gC gene against AHV-1. If this vaccine will be successfully constructed in the future, the developed TaqMan™ FQ-PCR assay will become perfect for the surveillance of AHV-1.
Conclusions
In summary, the established TaqMan™ FQ-PCR was a rapid, highly specific, sensitive, repeatable and reproducible assay than conventional PCR method, and it was extremely valuable for AHV-1 detection and quantitation on the purpose of the disease transmission studies, diagnostic assays and efficacy evaluation of drugs. Also it provided some significant basic data that may be beneficial to further investigate pcDNA3.1-gC DNA vaccine. We are currently studying the dynamic distribution of gC in AHV-1-infected and DNA vaccine-inoculated ducks by using this method. We believe that this approach could expedite related AHV-1 and gC DNA vaccine research.
Methods
Viruses and bacteria
AHV-1 Cha strain and Escherichia coli JM109 were obtained from Key Laboratory of Animal Diseases and Human Health of Sichuan Province. According to the gene libraries of AHV-1 constructed by the Avian Disease Research Center of Sichuan Agricultural University[56], the pMD18-gC plasmid was obtained through a 1296 bp fragment (gC gene) of PCR amplification was cloned into the pMD18-T vector (Takara, Japan), and then the result of sequencing compared with the sequences of AHV-1 in GenBank. Sequence was submitted to GenBank [GenBank: EU076811] by the Avian Disease Research Center of Sichuan Agricultural University [57].
Gosling new type viral enteritis virus, duck hepatitis virus type1, duck adenovirus, goose parvovirus, Marek's disease virus, AHV-1 Chv strain virus, Pasteurella multocida (5: A), Escherichia coli (O78) and Salmonella enteritidis (No. 50338) were provided by Key Laboratory of Animal Diseases and Human Health of Sichuan Province. They were propagated and the nucleic acid was extracted [58-60].
Standard templates preparation
The purified gC gene was obtained from pMD18-gC by using restriction enzymes (EcoR I and Xho I) (Takara, Japan), and was inserted into the eukaryotic expression vector pcDNA3.1(+) (Invitrogen, USA) according to the manufacturer's protocol. The constructed pcDNA3.1-gC plasmid was transformed into Escherichia coli JM109 cells. pcDNA3.1-gC plasmid was extracted by TIANprep plasmid extraction kit (Tiangen, China) according to manufacturer's protocol. The presence of target DNA was confirmed by PCR amplification with primers P1 and P2 (generated by Takara, Japan) targeting the gC gene on a Mycycler™ thermo cycler system (Bio-Rad, USA), their sequences were listed in Table 3. The product size was 1296 bp. DNA sequencing showed that pcDNA3.1-gC is real.
Table 3.
Oligonucleotide sequences of primers and probe used in AHV-1 FQ-PCR detection
| Name | Type | Sequences (5' to 3') | Length (nt) |
Amplicon size (bp) |
|
|---|---|---|---|---|---|
| P1 | Forward | CGGAATTCCAAAACGCCGCACAGATGAC | 28 | 1296 | |
| P2 | Reverse | CCCTCGAGGTATTCAAATAATATTGTCTGC | 30 | ||
| P3 | Forward | GAAGGACGGAATGGTGGAAG | 20 | 78 | |
| P4 | Reverse | AGCGGGTAACGAGATCTAATATTGA | 25 | ||
| P | Probe | FAM-CCAATGCATCGATCATCCCGGAA-TAMRA | 23 | ||
The underlined sequences of P1 and P2 are EcoR I and Xho I restriction sites respectively
PCR primers and probe design
The FQ-PCR assay primers and TaqMan™ probe (named P3, P4 and P respectively, generated by Genecore Corporation, China) design was carried out by using the Primer Express™ software supplied by Applied Biosystems according to the sequence of gC gene [GenBank: EU076811] and their sequences were listed in Table 3. The forward and reverse primers amplified a 78 bp fragment of AHV-1 gC gene. The fluorogenic probe was labelled at 5' with FAM (6-carboxyfluorescein) dye as reporter and labelled at 3' with TAMRA (tetra-methylcarboxyrhodamine) as quencher.
Protocol optimization
FQ-PCR was performed in an iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad, USA) with a reaction mixture (20 μL) containing 10 μL 2 × Premix Ex Taq™ (Takara, Japan) and 2 μL standard template according to the manufacturer's protocol. Autoclaved double-filtered nanopure water was added to get the final volume to 20 μL. The reactions were optimized in triplicate based on primers (P3 and P4) and TaqMan™ probe (P) concentration selection criteria, which was performed according to 5 × 5 matrix of primers concentrations (0.2, 0.3, 0.4, 0.5 and 0.6 μmol/L) and probe concentrations (0.1, 0.2, 0.25, 0.3 and 0.35 μmol/L). The two-step PCR cycling condition as follows: initial denaturation and hot-start Taq DNA polymerase activation at 95°C for 5 min, 45 cycles of denaturation at 94°C for 5 s, primer annealing and extension at 53°C for 30 s with fluorescence acquisition during each annealing and extension stage. The tests were carried out by using the 0.2 mL PCR tubes (Axygen, USA).
standard curve establishment
The recombinant plasmid pcDNA3.1-gC was used to establish standard curve as standard DNA of FQ-PCR. pcDNA3.1-gC concentration was determined by taking the absorbance at 260 nm by using a Smartspec 3000 spectrophotometer (Bio-Rad, USA) and purity was confirmed by using the 260/280 nm ratio. The pcDNA3.1-gC copies/μL was calculated and the purified plasmid DNA was serially diluted 10-fold in TE buffer, pH 8.0, from 5.0 × 107 to 5.0 × 100 plasmid copies/μL. The Primers (P3 and P4) were used for this amplification, These dilutions were used as amplification standards to construct the standard curve by plotting the plasmid copy number logarithm against the Ct values under optimum conditions. The standard curve and its correlation coefficient were generated through the software of iCycler IQ Detection System (Bio-Rad, USA) according to the manufacturer's protocol.
Amplification sensitivity, specificity, repeatability and reproducibility
The sensitivity of the assay was used as the limit extent of detection when testing 10-fold diluted DNA standards in triplicate. The dilution of plasmid pcDNA3.1-gC was ranging from 1.0 × 105 to 1.0 × 100 copies/reaction. This test was performed under optimum conditions.
The different 40 liver samples had been confirmed positive for AHV-1 by using the conventional PCR from infected ducks, these samples were retested with the established FQ-PCR method to evaluate the sensitivity of this method for clinical cases.
AHV-1 Cha strains was propagated in the allantoic cavity of 10-day-old SPF duck embryo. The allantoic fluid was harvested from dead embryo. Viral DNA from allantoic fluid was extracted by using TIANamp viral Genomic (DNA/RNA) extracting kit (Tiangen, China) according to the manufacture's instructions, then examined by the established FQ-PCR method and conventional PCR under same circumstance in triplicate after it was 10-fold diluted with sterile ultrapure water. The detection limit of the FQ-PCR was determined based on the highest dilution that resulted in the presence of Ct value in real-time PCR detection. The detection limit of the conventional PCR was determined through the highest dilution that resulted in the presence of clear amplified fragments (78 bp) on the agarose gel. The end-point sensitivity of both assays were calculated.
The specificity of the assay was evaluated by testing the different kinds of templates including pcDNA3.1-gC, AHV-1 Cha, AHV-1 Chv, gosling new type viral enteritis virus, duck hepatitis virus type1, duck adenovirus, goose parvovirus, Marek's disease virus, Pasteurella multocida (5: A), Escherichia coli (O78) and Salmonella enteritidis (No. 50338), then the liver DNA of the healthy duck should be added in this experiment as a negative control.
In order to assess intra-assay variability, eight dilutions of pcDNA3.1-gC (1.0 × 108-1.0 × 101 copies/reaction) were prepared separately. These samples were assayed simultaneously in triplicate in a same experiment. Five experiments were performed on different days in order to assess inter-assay variability, using eight pcDNA3.1-gC dilutions (1.0 × 108-1.0 × 101 copies/reaction). All tests were performed under optimum conditions. The mean Ct values, SD values and CV values were calculated independently for each DNA dilution.
Detection of AHV-1 gC gene in samples for practical applications
This study was conducted with 90 AHV-1-free Peking ducks (28 days old) from a AHV-1-free farm which were certificated with qualitative PCR as described by Song[61]. 60 ducks were randomly divided into two equal groups in this study (30 in each group). Thirty non-immunized ducks in Groups 3 used as controls. Ducks in Groups 1-2 were inoculated with 200 μg pcDNA3.1-gC (2.714 × 1013 copies) as DNA vaccine by intramuscular route and with 0.2 mL AHV-1 Cha strain vaccine (6.692 × 1011 copies) by subcutaneous route respectively. At each of ten sampling times, three vaccinated ducks of each immune group were chosen randomly for sampling. The liver, pancreas, spleen, kidney, lung, thymus, heart, brain, duodenum, rectum, Harderian gland and bursa of Fabricius were collected at 1 h, 4 h, 8 h, 12 h, 1 d, 3 d, 5 d, 7 d, 2 wk and 4 wk postinoculation respectively. DNA of all these samples were extracted by Animal cell/tissue DNA magnetic bead extraction kit (Bioeasy Technology, China) in Thermo Scientific KingFisher (mL) (Thermo, USA) from 15 mg tissues according to the manufacturer's protocol, followed by being dissolved in 50 μL sterile ultrapure water. Then, 2 μL DNA of each sample was prepared to detect AHV-1 gC accumulation in triplicate by TaqMan™ FQ-PCR.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
QZ and KS carried out most of the experiments. QZ drafted the manuscript. AC and MW strictly revised the manuscript and the experiment design. CX, DZ, RJ, QL, YZ, ZC and XC assisted with the experiments. All of the authors read and approved the final manuscript.
Contributor Information
Qing Zou, Email: sirius_zou@163.com.
Kunfeng Sun, Email: sunkunfeng1981@163.com.
Anchun Cheng, Email: chenganchun@vip.163.com.
Mingshu Wang, Email: mshwang@163.com.
Chao Xu, Email: xuc06@yahoo.com.cn.
Dekang Zhu, Email: zdk24@163.com.
Renyong Jia, Email: cqrc_jry@163.com.
Qihui Luo, Email: luoqihui80@163.com.
Yi Zhou, Email: zhouyi1963@163.com.
Zhengli Chen, Email: chzhli75@163.com.
Xiaoyue Chen, Email: chenganchun@hotmail.com.
Acknowledgements
This work was supported by the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT0848); the earmarked fund for Modern Agro-industry Technology Research System (nycytx-45-12).
References
- Hess JC, Pare' JA. Viruses of waterfowl. Seminars in Avian and Exotic Pet Medicine. 2004;13:176–183. doi: 10.1053/j.saep.2004.04.002. [DOI] [Google Scholar]
- Sandhu TS, Shawky SA. In: Diseases of Poultry. 11. Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editor. Iowa State Press, Ames; 2003. Duck virus enteritis (Duck plague) pp. 354–363. [Google Scholar]
- Baudet A. Mortality in ducks in the Netherlands caused by a filtrable virus; fowl plague. Tijdschr Diergeneeskd. 1923;50:455–459. [Google Scholar]
- Montgomery RD, Stein G Jr, Novilla MN, Hurley SS, Fink RJ. An outbreak of duck virus enteritis (duck plague) in a captive flock of mixed waterfowl. Avian Dis. 1981;25:207–213. doi: 10.2307/1589844. [DOI] [PubMed] [Google Scholar]
- Campagnolo ER, Banerjee M, Panigrahy B, Jones RL. An outbreak of duck viral enteritis (duck plague) in domestic Muscovy ducks (Cairina moschata domesticus) in Illinois. Avian Dis. 2001;45:522–528. doi: 10.2307/1592999. [DOI] [PubMed] [Google Scholar]
- Shawky S, Schat KA. Latency sites and reactivation of duck enteritis virus. Avian Dis. 2002;46:308–313. doi: 10.1637/0005-2086(2002)046[0308:LSAROD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Liu F, Ma B, Zhao Y, Zhang Y, Wu Y, Liu X, Wang J. Characterization of the gene encoding glycoprotein C of duck enteritis virus. Virus Genes. 2008;37:328–332. doi: 10.1007/s11262-008-0266-5. [DOI] [PubMed] [Google Scholar]
- Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whealy M, Robbins A, Enquist L. Pseudorabies virus glycoprotein gIII is required for efficient virus growth in tissue culture. J Virol. 1988;62:2512. doi: 10.1128/jvi.62.7.2512-2515.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger A, Schmidt J, Mettenleiter TC. Infectivity of a pseudorabies virus mutant lacking attachment glycoproteins C and D. J Virol. 1998;72:7341–7348. doi: 10.1128/jvi.72.9.7341-7348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rux AH, Lou H, Lambris JD, Friedman HM, Eisenberg RJ, Cohen GH. Kinetic analysis of glycoprotein C of herpes simplex virus types 1 and 2 binding to heparin, heparan sulfate, and complement component C3b. Virology. 2002;294:324–332. doi: 10.1006/viro.2001.1326. [DOI] [PubMed] [Google Scholar]
- Rue CA, Ryan P. A role for glycoprotein C in pseudorabies virus entry that is independent of virus attachment to heparan sulfate and which involves the actin cytoskeleton. Virology. 2003;307:12–21. doi: 10.1016/S0042-6822(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Kirisawa R, Hosoi Y, Yamaya R, Taniyama H, Okamoto M, Tsunoda N, Hagiwara K, Iwai H. Isolation of equine herpesvirus-1 lacking glycoprotein C from a dead neonatal foal in Japan. Arch Virol. 2005;150:2549–2565. doi: 10.1007/s00705-005-0587-9. [DOI] [PubMed] [Google Scholar]
- Robbins A, Ryan J, Whealy M, Enquist L. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J Virol. 1989;63:250. doi: 10.1128/jvi.63.1.250-258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiuk LA, L'Italien J, van Drunen Littel-van den Hurk S, Zamb T, Lawman JP, Hughes G, Gifford GA. Protection of cattle from bovine herpesvirus type I (BHV-1) infection by immunization with individual viral glycoproteins. Virology. 1987;159:57–66. doi: 10.1016/0042-6822(87)90347-3. [DOI] [PubMed] [Google Scholar]
- Denis M, Slaoui M, Keil G, Babiuk LA, Ernst E, Pastoret PP, Thiry E. Identification of different target glycoproteins for bovine herpes virus type 1-specific cytotoxic T lymphocytes depending on the method of in vitro stimulation. Immunology. 1993;78:7–13. [PMC free article] [PubMed] [Google Scholar]
- Denis M, Hanon E, Rijsewijk FA, Kaashoek MJ, van Oirschot JT, Thiry E, Pastoret PP. The role of glycoproteins gC, gE, gI, and gG in the induction of cell-mediated immune responses to bovine herpesvirus 1. Vet Microbiol. 1996;53:121–132. doi: 10.1016/S0378-1135(96)01240-0. [DOI] [PubMed] [Google Scholar]
- Ober BT, Summerfield A, Mattlinger C, Wiesmuller KH, Jung G, Pfaff E, Saalmuller A, Rziha HJ. Vaccine-induced, pseudorabies virus-specific, extrathymic CD4+CD8+ memory T-helper cells in swine. J Virol. 1998;72:4866–4873. doi: 10.1128/jvi.72.6.4866-4873.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes A, Alber DG, Cameron RS, Marshall RN, Allen GP, Killington RA. The production of a truncated form of baculovirus expressed EHV-1 glycoprotein C and its role in protection of C3H (H-2Kk) mice against virus challenge. Virus Res. 1996;44:97–109. doi: 10.1016/0168-1702(96)01339-1. [DOI] [PubMed] [Google Scholar]
- Packiarajah P, Walker C, Gilkerson J, Whalley JM, Love DN. Immune responses and protective efficacy of recombinant baculovirus-expressed glycoproteins of equine herpesvirus 1 (EHV-1) gB, gC and gD alone or in combinations in BALB/c mice. Vet Microbiol. 1998;61:261–278. doi: 10.1016/S0378-1135(98)00189-8. [DOI] [PubMed] [Google Scholar]
- Monteil M, Le Pottier MF, Ristov AA, Cariolet R, L'Hospitalier R, Klonjkowski B, Eloit M. Single inoculation of replication-defective adenovirus-vectored vaccines at birth in piglets with maternal antibodies induces high level of antibodies and protection against pseudorabies. Vaccine. 2000;18:1738–1742. doi: 10.1016/S0264-410X(99)00545-9. [DOI] [PubMed] [Google Scholar]
- Fischer T, Planz O, Stitz L, Rziha HJ. Novel recombinant parapoxvirus vectors induce protective humoral and cellular immunity against lethal herpesvirus challenge infection in mice. J Virol. 2003;77:9312–9323. doi: 10.1128/JVI.77.17.9312-9323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Saini M, Gupta LK, Rao VDP, Bandyopadhyay SK, Butchaiah G, Garg GK, Garg SK. Induction of immune responses in cattle with a DNA vaccine encoding glycoprotein C of bovine herpesvirus-1. Vet Microbiol. 2001;78:293–305. doi: 10.1016/S0378-1135(00)00304-7. [DOI] [PubMed] [Google Scholar]
- Brand CJ, Docherty DE. Post-epizootic surveys of waterfowl for duck plague (duck virus enteritis) Avian Dis. 1988;32:722–730. doi: 10.2307/1590991. [DOI] [PubMed] [Google Scholar]
- Deng MY, Burgess EC, Yuill TM. Detection of duck plague virus by reverse passive hemagglutination test. Avian Dis. 1984;28:616–628. doi: 10.2307/1590230. [DOI] [PubMed] [Google Scholar]
- Islam MR, Nessa J, Halder KM. Detection of duck plague virus antigen in tissues by immunoperoxidase staining. Avian Pathol. 1993;22:389–393. doi: 10.1080/03079459308418929. [DOI] [PubMed] [Google Scholar]
- Pearson GL, Cassidy DR. Perspectives on the diagnosis, epizootiology, and control of the 1973 duck plague epizootic in wild waterfowl at Lake Andes, South Dakota. J Wildl Dis. 1997;33:681–705. doi: 10.7589/0090-3558-33.4.681. [DOI] [PubMed] [Google Scholar]
- Wolf K, Burke CN, Quimby MC. Duck viral enteritis: microtiter plate isolation and neutralization test using the duck embryo fibroblast cell line. Avian Dis. 1974;18:427–434. doi: 10.2307/1589110. [DOI] [PubMed] [Google Scholar]
- Burgess EC, Ossa J, Yuill TM. Duck plague: a carrier state in waterfowl. Avian Dis. 1979;23:940–949. doi: 10.2307/1589610. [DOI] [PubMed] [Google Scholar]
- Plummer PJ, Alefantis T, Kaplan S, O'Connell P, Shawky S, Schat KA. Detection of duck enteritis virus by polymerase chain reaction. Avian Dis. 1998;42:554–564. doi: 10.2307/1592682. [DOI] [PubMed] [Google Scholar]
- Hansen WR, Brown SE, Nashold SW, Knudson DL. Identification of duck plague virus by polymerase chain reaction. Avian Dis. 1999;43:106–115. doi: 10.2307/1592768. [DOI] [PubMed] [Google Scholar]
- Pritchard LI, Morrissy C, Van Phuc K, Daniels PW, Westbury HA. Development of a polymerase chain reaction to detect Vietnamese isolates of duck virus enteritis. Vet Microbiol. 1999;68:149–156. doi: 10.1016/S0378-1135(99)00071-1. [DOI] [PubMed] [Google Scholar]
- Hansen WR, Nashold SW, Docherty DE, Brown SE, Knudson DL. Diagnosis of duck plague in waterfowl by polymerase chain reaction. Avian Dis. 2000;44:266–274. doi: 10.2307/1592539. [DOI] [PubMed] [Google Scholar]
- Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Jia W, Yue H, Luo W, Chen X, Xie Y, Zen W, Yang W. Development of quantitative real-time polymerase chain reaction for duck enteritis virus DNA. Avian Dis. 2005;49:397–400. doi: 10.1637/7338-020305R.1. [DOI] [PubMed] [Google Scholar]
- Guo Y, Cheng A, Wang M, Shen C, Jia R, Chen S, Zhang N. Development of TaqMan MGB fluorescent real-time PCR assay for the detection of anatid herpesvirus 1. Virol J. 2009;6:71. doi: 10.1186/1743-422X-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehée A, Césaire R, Désiré N, Lézin A, Bourdonné O, Béra O, Plumelle Y, Smadja D, Nicolas JC. Quantitation of HTLV-I proviral load by a TaqMan real-time PCR assay. J Virol Methods. 2002;102:37–51. doi: 10.1016/S0166-0934(01)00445-1. [DOI] [PubMed] [Google Scholar]
- Yun Z, Lewensohn-Fuchs I, Ljungman P, Ringholm L, Jonsson J, Albert J. A real-time TaqMan PCR for routine quantitation of cytomegalovirus DNA in crude leukocyte lysates from stem cell transplant patients. J Virol Methods. 2003;110:73–79. doi: 10.1016/S0166-0934(03)00103-4. [DOI] [PubMed] [Google Scholar]
- Gravitt P, Peyton C, Wheeler C, Apple R, Higuchi R, Shah K. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J Virol Methods. 2003;112:23–33. doi: 10.1016/S0166-0934(03)00186-1. [DOI] [PubMed] [Google Scholar]
- Brunborg I, Moldal T, Jonassen C. Quantitation of porcine circovirus type 2 isolated from serum/plasma and tissue samples of healthy pigs and pigs with postweaning multisystemic wasting syndrome using a TaqMan-based real-time PCR. J Virol Methods. 2004;122:171–178. doi: 10.1016/j.jviromet.2004.08.014. [DOI] [PubMed] [Google Scholar]
- La Fauce KA, Layton R, Owens L. TaqMan real-time PCR for detection of hepatopancreatic parvovirus from Australia. J Virol Methods. 2007;140:10–16. doi: 10.1016/j.jviromet.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Hussein IT, Field HJ. Development of a quantitative real-time TaqMan PCR assay for testing the susceptibility of feline herpesvirus-1 to antiviral compounds. J Virol Methods. 2008;152:85–90. doi: 10.1016/j.jviromet.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Yang J, Cheng A, Wang M, Pan K, Li M, Guo Y, Li C, Zhu D, Chen X. Development of a fluorescent quantitative real-time polymerase chain reaction assay for the detection of Goose parvovirus in vivo. Virol J. 2009;6:142. doi: 10.1186/1743-422X-6-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Li XK, Cui BA, Wei ZY, Li XS, Wang YB, Zhao L, Wang ZY. A TaqMan-based real-time polymerase chain reaction for the detection of porcine parvovirus. J Virol Methods. 2009;156:84–88. doi: 10.1016/j.jviromet.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Converse KA, Kidd GA. Duck plague epizootics in the United States, 1967-1995. J Wildl Dis. 2001;37:347–357. doi: 10.7589/0090-3558-37.2.347. [DOI] [PubMed] [Google Scholar]
- Shawky S, Sandhu T, Shivaprasad HL. Pathogenicity of a low-virulence duck virus enteritis isolate with apparent immunosuppressive ability. Avian Dis. 2000;44:590–599. doi: 10.2307/1593098. [DOI] [PubMed] [Google Scholar]
- Cheng A, Han X, Zhu D, Wang M, Yuan G, Liao Y, Xu C. Application of indirect immuno-fluorescent staining method for detection and antigen location of duck enteritis virus in paraff in sections. Chin J Vet Sci. 2008;28:871–875. [Google Scholar]
- Apfalter P, Barousch W, Nehr M, Makristathis A, Willinger B, Rotter M, Hirschl AM. Comparison of a new quantitative ompA-based real-Time PCR TaqMan assay for detection of Chlamydia pneumoniae DNA in respiratory specimens with four conventional PCR assays. J Clin Microbiol. 2003;41:592–600. doi: 10.1128/JCM.41.2.592-600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- Qi X, Yang X, Cheng A, Wang M, Guo Y, Jia R. Replication kinetics of duck virus enteritis vaccine virus in ducklings immunized by the mucosal or systemic route using real-time quantitative PCR. Res Vet Sci. 2009;86:63–67. doi: 10.1016/j.rvsc.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Manam S, Ledwith BJ, Barnum AB, Troilo PJ, Pauley CJ, Harper LB, Griffiths TG, Niu Z, Denisova L, Follmer TT, Pacchione SJ, Wang Z, Beare CM, Bagdon WJ, Nichols WW. Plasmid DNA vaccines: tissue distribution and effects of DNA sequence, adjuvants and delivery method on integration into host DNA. Intervirology. 2000;43:273–281. doi: 10.1159/000053994. [DOI] [PubMed] [Google Scholar]
- Tuomela M, Malm M, Wallen M, Stanescu I, Krohn K, Peterson P. Biodistribution and general safety of a naked DNA plasmid, GTU-MultiHIV, in a rat, using a quantitative PCR method. Vaccine. 2005;23:890–896. doi: 10.1016/j.vaccine.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Coelho-Castelo AA, Trombone AP, Rosada RS, Santos RR Jr, Bonato VL, Sartori A, Silva CL. Tissue distribution of a plasmid DNA encoding Hsp65 gene is dependent on the dose administered through intramuscular delivery. Genet Vaccines Ther. 2006;4:1. doi: 10.1186/1479-0556-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I, Terregino C, Cattoli G, Mutinelli F, Rodriguez J. Development of a DIVA (Differentiating Infected from Vaccinated Animals) strategy using a vaccine containing a heterologous neuraminidase for the control of avian influenza. Avian Pathol. 2003;32:47–55. doi: 10.1080/0307945021000070714. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Alvarez R, Swayne DE, Suarez DL. Diagnostic approach for differentiating infected from vaccinated poultry on the basis of antibodies to NS1, the nonstructural protein of influenza A virus. J Clin Microbiol. 2005;43:676–683. doi: 10.1128/JCM.43.2.676-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wang M, Wen M, Zhou W, Guo Y, Jia R, Xu C, Yuan G, Liu Y. Construction of duck enteritis virus gene libraries and discovery, cloning and identification of viral nucleocapsid protein gene. High Technol Lett. 2006;16:948–953. [Google Scholar]
- Xu C, Li X, Xin H, Lian B, Cheng A, Wang M, Zhu D, Jia R, Luo Q, Chen X. Cloning and molecular characterization of gC gene of duck plague virus. Vet Sci Chin. 2008;38:1038–1044. [Google Scholar]
- Chen S, Cheng A, Wang M. Morphologic observations of new type gosling viral enteritis virus (NGVEV) virulent isolate in infected duck embryo fibroblasts. Avian Dis. 2008;52:173–178. doi: 10.1637/8080-072107-ResNote. [DOI] [PubMed] [Google Scholar]
- Yang M, Cheng A, Wang M, Xing H. Development and application of a one-step real-time Taqman RT-PCR assay for detection of Duck hepatitis virus type1. J Virol Methods. 2008;153:55–60. doi: 10.1016/j.jviromet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Deng S, Cheng A, Wang M, Cao P. Serovar-specific real-time quantitative detection of Salmonella Enteritidis in the gastrointestinal tract of ducks after oral challenge. Avian Dis. 2008;52:88–93. doi: 10.1637/8102-090107-Reg. [DOI] [PubMed] [Google Scholar]
- Song Y, Cheng A, Wang M, Liu F, Liao Y, Yuan G, Han X, Xu C, Chen X. Development and application of PCR to detect duck plague virus. Chin J Vet Med. 2005;41:17–20. [Google Scholar]


