Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor through which halogenated aromatic hydrocarbons such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) cause altered gene expression and toxicity. The AHR belongs to the basic helix–loop–helix/Per-ARNT-Sim (bHLH-PAS) family of transcriptional regulatory proteins, whose members play key roles in development, circadian rhythmicity, and environmental homeostasis; however, the normal cellular function of the AHR is not yet known. As part of a phylogenetic approach to understanding the function and evolutionary origin of the AHR, we sequenced the PAS homology domain of AHRs from several species of early vertebrates and performed phylogenetic analyses of these AHR amino acid sequences in relation to mammalian AHRs and 24 other members of the PAS family. AHR sequences were identified in a teleost (the killifish Fundulus heteroclitus), two elasmobranch species (the skate Raja erinacea and the dogfish Mustelus canis), and a jawless fish (the lamprey Petromyzon marinus). Two putative AHR genes, designated AHR1 and AHR2, were found both in Fundulus and Mustelus. Phylogenetic analyses indicate that the AHR2 genes in these two species are orthologous, suggesting that an AHR gene duplication occurred early in vertebrate evolution and that multiple AHR genes may be present in other vertebrates. Database searches and phylogenetic analyses identified four putative PAS proteins in the nematode Caenorhabditis elegans, including possible AHR and ARNT homologs. Phylogenetic analysis of the PAS gene family reveals distinct clades containing both invertebrate and vertebrate PAS family members; the latter include paralogous sequences that we propose have arisen by gene duplication early in vertebrate evolution. Overall, our analyses indicate that the AHR is a phylogenetically ancient protein present in all living vertebrate groups (with a possible invertebrate homolog), thus providing an evolutionary perspective to the study of dioxin toxicity and AHR function.
Halogenated aromatic hydrocarbons such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) are potent modulators of cellular growth and differentiation and thus are highly toxic to vertebrate animals (1). These effects are mediated by the aryl hydrocarbon receptor (Ah receptor, AHR, or “dioxin receptor”), a ligand-activated transcription factor that acts in concert with the Ah receptor nuclear translocator [ARNT (2)] to alter the expression of target genes, such as cytochrome P450 1A1 (1, 3). The AHR and ARNT belong to the Per-ARNT-Sim (PAS) family of transcriptional regulatory proteins (3, 4), whose members play key roles in development (5), adaptation to hypoxia (6, 7), control of circadian rhythmicity (8, 9), and phototransduction (8, 10, 11). The physiological function of the AHR is not yet known, but an important role in the developing liver and immune system has been suggested by the phenotypes of mice bearing a targeted disruption of the AHR locus (12, 13).
The AHR has been studied almost exclusively in mammals, in which a single gene has been identified (14, 15). The mammalian AHR contains basic helix–loop–helix (bHLH) and PAS homology domains that define the PAS family. The bHLH domain contains basic and HLH motifs involved in protein–DNA and protein–protein interactions, respectively. The PAS domain forms a secondary dimerization surface for heteromeric interactions between AHR and ARNT, as well as among other bHLH-PAS proteins (16, 17). It includes two imperfect repeats of 51 amino acids [PAS-A and PAS-B (18)] separated by an intervening sequence of approximately 110 amino acids. Importantly, the distal portion of this region (PAS-B) is part of the ligand-binding domain of the AHR (15, 19–22).
In contrast to the extensive literature on the mammalian AHR, knowledge of the AHR in other vertebrate and invertebrate animals is limited (23–25). The objective of the present work was to investigate the evolutionary history of the AHR and its relationship to other members of the PAS family. Our approach was to sequence the AHR PAS domains from early chordates and to assess their relationships by phylogenetic inference, a powerful tool for understanding the evolution and interrelationships of multigene families (26). We focused on early chordates because previous results had suggested the first appearance of an AHR protein in cartilaginous fish (24). The PAS domain was chosen because it is a well-conserved and functionally important region of the mammalian AHR and other members of the PAS family (8, 15, 16, 27); except for the bHLH domain (28), other regions of PAS proteins are not highly conserved (29) and, therefore, are less suitable for phylogenetic analysis.
The results of these studies show that the AHR is a phylogenetically ancient protein that exists in bony and cartilaginous fish, as well as lamprey, the most “primitive” (i.e., early diverging) living vertebrate. We also report a second AHR in two species of gnathostome (jawed) fish and provide evidence that an AHR gene duplication occurred early in vertebrate evolution. Possible invertebrate AHR and ARNT homologs are also described. We discuss these results in relation to the diversification of the PAS family.
METHODS
Animals and RNA Isolation.
Killifish (Fundulus heteroclitus), smooth dogfish (Mustelus canis), little skate (Raja erinacea), and Atlantic hagfish (Myxine glutinosa) were obtained as described (24, 25). Larval sea lamprey (Petromyzon marinus) were a gift from Gary Swain (University of Pennsylvania). Adult amphioxus (Branchiostoma floridae) were purchased from Gulf Specimens (Panacea, FL). Poly(A)+ RNA was prepared from frozen liver powders (killifish, skate, dogfish, and hagfish), from the anterior region of the lamprey, or from the total visceral organs of amphioxus, either directly using a FastTrack kit (InVitrogen) or by sequential isolation of total RNA (RNA STAT-60; Tel-Test, Friendswood, TX) and poly(A)+ mRNA [mini-oligo(dT)-cellulose spin column kit; 5 Prime → 3 Prime].
RT–PCR, Cloning, and Sequencing.
Degenerate inosine-containing oligonucleotides AHR-A1 and AHR-B1 were designed as described (25, 30). Reverse transcription coupled-PCR (RT–PCR) was performed by using the Gene-Amp RNA-PCR kit (Perkin–Elmer) and a GeneAmp 2400 thermocycler. Reverse transcription was primed with random hexamers; for the PCR, AHR-A1 and AHR-B1 were used at 1 μM. PCR conditions were optimized for each species. For hagfish, lamprey, and amphioxus, MgCl2 concentration was 3.0 mM rather than 2.0 mM. PCR cycles were as follows: 105 sec at 95°C; 35 cycles of 95°C for 15 sec and 50°C for 30 sec, followed by 7 min at 72°C. PCR products were analyzed by Southern blotting using oligonucleotide J2u (5′-GGCTAYCAGTTYATYCATGC-3′), targeted to the conserved sequence GYQFIHA (corresponding to amino acids 315–321 of the mouse AHR). Hybridizing bands were cloned into pCNTR (5 Prime → 3 Prime) or pT7BlueR (Novagen) and sequenced in both directions by using SequiTherm and SequiTherm Excel long-read cycle sequencing kits (Epicentre Technologies, Madison, WI) and an automated DNA sequencer (LI-COR). Three to seven clones were sequenced for each PCR fragment. The sequence of AHR1 from Fundulus was obtained from genomic DNA clones and confirmed by RT–PCR (S.I.K. and M.E.H., unpublished results).
Sequence Analysis.
The sequences of RT–PCR products were assembled and translated. Multiple alignment of the deduced amino acid sequences was performed by using clustalw version 1.6 (31). The aligned amino acid sequences (PAS domain only) were used to construct phylogenetic trees by using the Neighbor-Joining (NJ) algorithm (32) and maximum parsimony [paup 3.1 (33)]. Alignment positions with gaps were excluded. Bootstrap analysis (34) was performed to assess relative confidence in the topologies obtained.
RESULTS
AHRs in Early Vertebrates.
To examine the evolutionary history of the AHR, degenerate PCR primers (25, 30) were used to amplify cDNA sequences from the cephalochordate amphioxus and representative species of early chordates, including the teleost Fundulus, two cartilaginous fish (smooth dogfish and little skate), and two jawless fish (sea lamprey and Atlantic hagfish). [Lamprey are the most ancient living vertebrates, and hagfish are considered invertebrate chordates (35, 36).] Products of the predicted size (∼700 bp) that hybridized to an AHR-specific probe were obtained from Fundulus, dogfish, skate, and lamprey (data not shown). The nucleotide and deduced amino acid sequences of these RT–PCR products were most closely related to PAS domains of mammalian AHRs (60–76% amino acid identity; Table 1).
Table 1.
AHR PAS domains: Comparison of deduced amino acid sequences, % identity
| Fundulus AHR1 | Fundulus AHR2 | Dogfish AHR1 | Dogfish AHR2 | Skate AHR | Lamprey AHR | C. elegans C41G7.5 | Human AHR | Mouse AHRb-1 | Rat AHR | |
|---|---|---|---|---|---|---|---|---|---|---|
| Fundulus AHR1 | 100 | 63 | 71 | 69 | 61 | 70 | 36 | 70 | 70 | 70 |
| Fundulus AHR2 | 100 | 66 | 68 | 57 | 59 | 33 | 62 | 65 | 64 | |
| Dogfish AHR1 | 100 | 70 | 62 | 70 | 35 | 76 | 75 | 75 | ||
| Dogfish AHR2 | 100 | 63 | 67 | 35 | 68 | 67 | 67 | |||
| Skate AHR | 100 | 66 | 30 | 61 | 61 | 60 | ||||
| Lamprey AHR | 100 | 35 | 68 | 68 | 67 | |||||
| C. elegans C41G7.5 | 100 | 35 | 34 | 34 | ||||||
| Human AHR | 100 | 87 | 88 | |||||||
| Mouse AHRb-1 | 100 | 95 | ||||||||
| Rat AHR | 100 |
Initially, a single AHR sequence was obtained from Fundulus, as reported (25). Subsequently, a second Fundulus AHR sequence was obtained by screening a genomic DNA library; its expression was confirmed by RT–PCR. Similarly, RT–PCR using dogfish RNA revealed two different AHR-like sequences, which were confirmed by sequencing multiple clones from two independent RT–PCRs. We have designated the two putative AHRs in each of these species as AHR1 and AHR2; in each case AHR1 shares greater sequence identity with mammalian AHR sequences than does AHR2 (Table 1). Both Fundulus AHR1 and AHR2 possess bHLH motifs that are closely related to those of mammalian AHRs [AHR1, 83% amino acid identity; AHR2, 73% amino acid identity; both exhibit 100% identity of amino acids critical for DNA binding (37)]. The designation of these fish sequences as AHRs is based on the high bHLH and PAS sequence identities in comparisons with mammalian AHRs and on the phylogenetic analyses described below. The full-length sequence and other properties of both Fundulus AHRs will be described in detail elsewhere (S.I.K. and M.E.H., unpublished results). Herein, it is important to note that the sequence difference between the two apparently paralogous AHRs within each species is as great or greater than the interspecies differences (Table 1), suggesting an ancient duplication.
Alignment of the PAS domain sequences of all vertebrate AHRs reveals several conserved regions within and between the PAS-A and PAS-B boxes (Fig. 1). Overall, 82 residues (41%) are conserved in the PAS domains of all of these vertebrate AHRs. These include 20 amino acids in PAS-A, 23 in PAS-B, and 35 in the region between the two PAS boxes. At several positions, characteristic amino acids distinguish the fish and mammalian AHRs. Of the fish sequences, dogfish AHR1 appears to be most closely related to the mammalian AHRs (Fig. 1 and Table 1). The skate AHR is the most divergent, both overall and at certain residues conserved in all of the other sequences. For example, the sequence RCLLDNSSGFL is identical in all AHRs except skate, where five differences occur. At five positions the two AHR2 sequences share unique residues that are not present in any of the other AHRs.
Figure 1.

Alignment of PAS domain amino acid sequences of vertebrate AHRs and a possible invertebrate AHR homolog. Deduced amino acid sequences in the PAS domains of vertebrate AHRs and C. elegans C41G7.5 were aligned by using clustalw(1.6). GenBank accession numbers are listed in Table 2. Amino acids that are identical in five or more of the sequences are boxed and shaded. Similar amino acids are in boldface type. The PAS “A” and “B” imperfect repeats (as defined originally in refs. 15 and 18) are underlined. The consensus sequence (≥50%) is shown below the aligned sequences.
Molecular Phylogeny of AHR Genes.
Phylogenetic analyses were used to assess the relationship of the two putative AHR2 forms to each other and to the other AHR sequences. In both NJ (Fig. 2A) and maximum parsimony analyses (Fig. 2B), the AHR2 forms from Fundulus and dogfish form a monophyletic group. Bootstrap analysis provides strong (96%) support for the AHR2 cluster and, thus, for an orthologous relationship between the Fundulus and dogfish AHR2 forms. The relationship of the skate and lamprey AHRs to the other fish AHR sequences is not resolved in these unrooted trees. In a further attempt to assess the position of the skate and lamprey sequences, the Caenorhabditis elegans sequence C41G7.5 (for rationale, see below) was used as an outgroup to provide a root for the vertebrate AHR analysis (Fig. 2 C and D). The NJ tree is consistent with two AHR clades, one containing the two AHR2 sequences, and the second containing all other fish and mammal AHRs (Fig. 2C). The most parsimonious tree (Fig. 2D) is also consistent with orthology of the two AHR2 proteins, because the AHR2 group is monophyletic in each of the six shortest trees (194–196 steps). However, the other AHR sequences appear paraphyletic in this analysis; the shortest tree in which the AHR1 (including skate and lamprey AHR) and AHR2 clusters are each monophyletic requires 202 steps.
Figure 2.
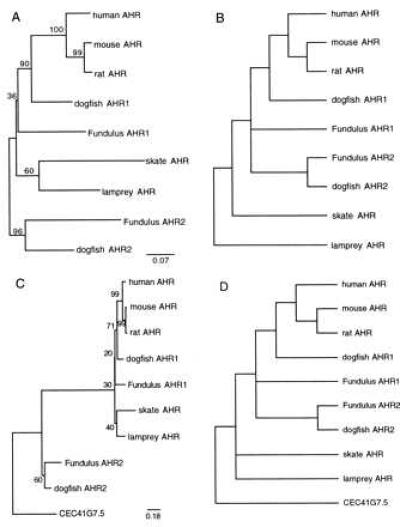
Phylogenetic analysis of vertebrate AHR proteins. Gene trees were inferred from the amino acid alignment in Fig. 1 by using distance (NJ) (A and C) or maximum parsimony methods (B and D). The trees in A and B are unrooted, whereas in C and D the C. elegans C41G7.5 sequence was used as an outgroup (see Fig. 3 and text for explanation). (A and C) Distance (NJ) trees. Positions with gaps were excluded and corrections were made for multiple substitutions. Numbers in boldface type next to branch points are bootstrap values based on 1,000 samplings. The distance between sequences is the sum of the horizontal distances separating them. (B and D) Maximum parsimony trees. Exhaustive searches were performed by using paup 3.1.1 (33). The tree shown in B is the strict consensus of the three shortest trees (177 steps), based on 61 informative characters. The tree shown in D is the strict consensus of the six shortest trees (194–196 steps), based on 72 informative characters. In D, the shortest tree that could be constructed with the AHR2s (fish only) and all other AHRs (mammals and fish) as two monophyletic groups was 202 steps. In B and D, the length of the branches does not correspond to distance between the sequences.
The PAS domain examined herein is expected to be highly conserved based on its important roles in ligand binding and protein–protein interactions. Because rates of evolutionary change are known to differ between functionally distinct regions of the same gene [mosaic evolution (38)], phylogenetic analysis of the complete sequences of these and additional AHR1 and AHR2 genes will be necessary to establish their relationships with greater certainty. Nevertheless, the present results provide support for orthology of the AHR2 genes in Fundulus and dogfish.
Phylogenetic Analysis of the PAS Superfamily.
To examine the relationship of these new AHR sequences to sequences of other PAS proteins, we conducted phylogenetic analyses of all AHR PAS domain sequences and representative sequences of the 24 other PAS proteins from vertebrate and invertebrate animals, plants, and bacteria, as identified from the literature and through searches of the nonredundant GenBank protein sequence database (Table 2).† In these analyses, the vertebrate AHRs, including all AHR1 and AHR2 genes, form a distinct clade that is strongly supported by bootstrap analysis (NJ tree, 100%; MP tree, 85%; Fig. 3). This strong clustering is the basis for our designation of all the fish sequences reported herein as AHRs.
Table 2.
PAS family members, GenBank accession numbers, and synonymous genes
| Human AHR (hAHR; L19872), mouse AHRb-1 allele (mAHR; M94623), rat AHR (rAHR; U09000), Fundulus AHR1 (fAHR1; AF024591), Fundulus AHR2 (fAHR2; U29679), dogfish AHR1 (dfAHR1; AF024592), dogfish AHR2 (dfAHR2; AF024593), skate AHR (skAHR; AF024594), and lamprey AHR (lampAHR; AF024595) |
| Mouse single-minded 1 (mSIM1; D79209) |
| Mouse single-minded 2 (mSIM2, D63383) |
| Human hypoxia-inducible factor 1α (hHIF-1α; U22431) |
| Human member of PAS family 2 (hMOP2; U51626), human endothelial PAS protein (hEPAS1; U81984), and mouse HIF-1 α-like factor (mHLF; D89787) |
| Human MOP-5 (hMOP5; U51628) and human neuronal PAS protein 1 (hNPAS1; U77968) |
| Human MOP-4 (hMOP4; U51625) and human neuronal PAS protein 2 (hNPAS2; U77970) |
| Mouse CLOCK (mCLOCK; AF000998) |
| Mouse AHR nuclear translocator (mARNT1; A56241) |
| Mouse ARNT2 (mARNT2; D63644) |
| Rainbow trout ARNTb (rtARNTb; U73841) |
| Human MOP-3 (hMOP3; U51627); human JAP3 (hJAP3; U60415), and human brain and muscle ARNT-like protein (hBMAL1a; D89722) |
| Human transcriptional intermediary factor (hTIF2; X97674) |
| Human steroid receptor coactivator (hSRC-1; U59302) |
| Drosophila Similar (dSIMA; U43090) |
| Drosophila trachealess (dTRH; U33427) |
| Drosophila Sim (dSIM; A29945) |
| Drosophila Per (dPER; A26427) |
| Caenorhabditis elegans C41G7.5 (CEC41G7.5; Z81048) |
| C. elegans C15C8.2 (CEC15C8.2; Z75527) |
| C. elegans T01D3.2 (CET01D3.2; Z81110) |
| C. elegans C25A1.11 (CEC25A1.11; Z81038) |
| Arabidopsis thaliana phytochrome A (phyA; P14712) |
| Mesotaenium caldariorum phytochrome 1b (MESPHY 1b; U31284) |
| Bacillus subtilis kinase A (kinA; M31067) |
Note: Where orthologous genes have been cloned from more than one species (e.g., mouse SIM1 and human SIM1), only one sequence is referenced, except in the case of the AHRs and where different names have been given to the same gene. A comparison of amino acid sequences in the PAS domain shows that hMOP3 = hJAP3 = hBMAL1, hMOP2 = hEPAS1, mEPAS1 = mHLF, hMOP4 and hNPAS2 differ by two amino acids, and hMOP5 and hNPAS1 differ by two amino acids.
Figure 3.
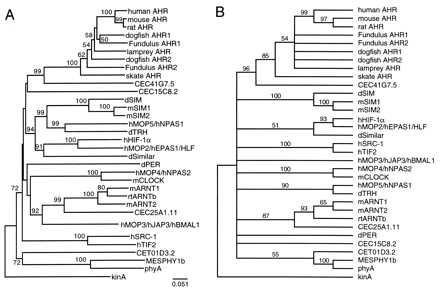
Phylogenetic analysis of PAS family proteins. (A) Distance (NJ) tree. The tree was inferred from an alignment of PAS domains of all PAS proteins; the alignment is available upon request to M.E.H. or can be viewed at http://www.whoi.edu/biology/hahnm.html. GenBank accession numbers are listed in Table 2. Positions with gaps were excluded and no corrections were made for multiple substitutions. The bacterial kinA sequence was treated as the outgroup. Numbers in boldface type next to branch points are bootstrap values based on 1,000 samplings (values <50% are not shown). (B) Maximum parsimony tree. An heuristic search was performed by using paup 3.1.1 (33). The tree shown is the 50% majority rule tree, based on 163 informative characters. Numbers in boldface type next to branch points are bootstrap values from 100 samplings. Where multiple names are shown for synonymous proteins, the first name is that of the sequence used in the alignment.
Interestingly, a search of the GenBank protein database for sequences related to AHR PAS domains revealed that the CEC41G7 locus in the C. elegans genome (39) encodes a predicted protein (C41G7.5) that, although clearly distinct from vertebrate AHRs, is more closely related to them than to any other known member of the PAS gene family (bootstrap values of ≥96% using either criterion; Fig. 3). Of the 82 amino acid residues that are conserved in all of the vertebrate AHR PAS domains, 40 are also conserved in this nematode protein (Fig. 1). Overall, this region of C41G7.5 shares 30–36% identity with the PAS domains of the AHR sequences (Table 1). C41G7.5 also possesses a bHLH domain that shares 57% amino acid identity with that of mammalian AHRs (data not shown). In addition to C41G7.5, blast searches revealed three other putative PAS family proteins in C. elegans, including a possible ARNT homolog (C25A1.11) that also possesses a bHLH domain (data not shown) and forms a monophyletic group with mammalian and fish ARNT proteins (Fig. 3).
The trees shown in Fig. 3 suggest that the PAS family is organized into several distinct clades. In addition to the AHR/C41G7.5 group, several other monophyletic groups occur in the distance tree (Fig. 3A). Most of these groups contain both invertebrate (Drosophila, C. elegans) and vertebrate representatives. One group consists of dSIM, mSIM1, mSIM2, MOP5, and Drosophila Trachealess (dTRH). A second lineage contains Drosophila Similar, MOP2, and HIF-1α. A third cluster includes ARNT1, ARNT2, MOP3, and C25A1.11. MOP4 and CLOCK cluster together, as do SRC-1 and TIF2. The sequences of phyA, MesPHY1b, and C. elegans T01D3.2 also form a monophyletic group. Drosophila Per and C. elegans C15C8.2 do not fall into any natural group.
Parsimony analysis also provides support for the AHR + C41G7.5 clade, the ARNT1 + ARNT2 + C25A1.11 clade (minus MOP3), the phyA + MesPHY1b + CET01D3.2 clade, and the pairs HIF-1α/MOP2, MOP4/CLOCK, and SRC-1/TIF2. Two distinct clades containing dSIM + mSIM1 + mSIM2 and MOP5 + dTRH were strongly supported, and in some analyses clustered together as in the NJ tree. Other relationships suggested by the distance tree are unresolved (bootstrap value <50%) in the parsimony tree.
DISCUSSION
The AHR Is an Ancient Protein.
Identification of AHR cDNA sequences in living representatives of early vertebrates (jawless, cartilaginous, and bony fish) provides evidence that the AHR is an ancient protein that existed early in vertebrate evolution, at least 450–510 million years ago. Its conservation in all vertebrate groups suggests that it serves an important function, as suggested also by recent findings of liver and immune system dysfunction after targeted disruption of an AHR gene in mice (12, 13). Although originally of interest because of its role in dioxin toxicity, the AHR likely has a more fundamental significance with regard to gene regulation, development, or other aspects of cellular homeostasis.
The identification of AHR cDNA sequences in the dogfish Mustelus confirms our previous report of an AHR protein in this species (24). In pairwise comparisons (Table 1) and phylogenetic analyses (Fig. 2), dogfish AHR1 consistently appears as the fish sequence most closely related to the mammalian AHRs. The AHR from skate, another cartilaginous fish, is the most divergent of the vertebrate AHR sequences. The AHR phylogeny does not match accepted phylogenetic relationships of these species, suggesting unequal rates of change in some lineages. A similar lack of concordance of gene and species phylogenies has been seen with the LDH-A genes of mammals, Fundulus, and another species of dogfish (40, 41).
In previous studies, we failed to detect AHR proteins by photoaffinity labeling of hepatic cytosol from adult lamprey (24). Similarly, induction of CYP1A in response to planar aromatic hydrocarbons—the “classical” AHR-dependent response—is not apparent in adult lamprey (60). The lamprey AHR sequence reported herein was obtained from the anterior section of larvae (ammocoetes), suggesting that expression of the AHR may be regulated developmentally or in a cell- or tissue-specific manner in this species. Our inability to identify an AHR in adult hagfish liver in the present study is consistent with our earlier ligand-binding results (24) and with the lack of CYP1A inducibility in adult animals (42, 60). However, in light of the lamprey results, a similar AHR-related gene may yet be found in hagfish or in other invertebrate chordates.
The presence of a gene in the nematode C. elegans that bears strong similarity to vertebrate AHRs is intriguing. Because this sequence (C41G7.5) contains both bHLH and PAS domains, it appears to represent a structural homolog of the vertebrate AHR—the first such invertebrate sequence identified. Interestingly, closer examination of this sequence reveals that the PAS-B box, which is part of the ligand-binding domain of the mammalian AHR (15, 19–22), is poorly conserved in the C. elegans C41G7.5 sequence. Thus, although the PAS-A box of C41G7.5 shares 43–50% amino acid identity with the homologous region of the vertebrate AHRs, the PAS-B box is only 25–29% identical to those of vertebrate AHRs (Fig. 1). The bHLH domain of C41G7.5 is more highly conserved with respect to the bHLH regions of vertebrate AHRs, including conservation of amino acids that have been shown (37) to be critical for DNA binding of the murine AHR (data not shown). These observations suggest that the C. elegans protein may participate in protein–protein and protein–DNA interactions that are qualitatively like those of the vertebrate AHRs but that its ligand-binding properties could be substantially different. Thus, this apparent AHR homolog in C. elegans and the possible ARNT homolog C25A1.11 may provide a system with which to examine possible ancestral functions of the AHR, especially those that may be ligand-independent.
PAS Family Gene Duplications.
The presence of a duplicated AHR gene in cartilaginous and bony fish and the degree of difference between the paralogous forms are consistent with a duplication event occurring early in vertebrate evolution. Phylogenetic analysis using two different methods support the orthology of Fundulus AHR2 and dogfish AHR2, suggesting that this duplication occurred prior to the divergence of bony and cartilaginous fish. The two AHR genes may have arisen as an isolated gene duplication or, alternatively, as a result of the genome duplications that are thought to have occurred early in chordate evolution (43). Such duplications have contributed to the diversification of Hox gene clusters (44, 45) and other gene families (46–49). Because the complexity of such gene families is similar in fish and mammals (50), multiple AHRs may also occur in other vertebrates. A recent report of a second AHR in mice (51) is consistent with this hypothesis.
The existence of a second AHR is reminiscent of the two forms of other PAS proteins (ARNT, Sim) recently described in mammals (52, 53). We suggest that these and several other pairs of PAS proteins are paralogs, i.e., homologous by gene duplication (54). Thus, the following pairs of proteins share extensive amino acid identity (64–90%) in the PAS domain and cluster together in both NJ and MP trees: AHR1 + AHR2, ARNT1 + ARNT2, SIM1 + SIM2, HIF-1α + MOP2, SRC-1 + TIF2, and CLOCK + MOP4. Duplication of these PAS genes may have occurred at about the same time as the proposed AHR gene duplication (i.e., near the origin of the gnathostomes), consistent with the genome duplication scenario. Thus, the PAS gene family—like other gene families (47, 49)—appears to contain sets of related genes (paralog groups), which might exhibit some degree of functional redundancy (28, 55). Such redundancy has been suggested to occur within the ARNT1 + ARNT2 pair (56), possibly in conjunction with the hypoxia-responsive paralogs HIF-1α and MOP2/EPAS1/HLF (6, 7, 57).
Molecular Evolution of the PAS Gene Family.
Recent findings suggest that the PAS domain had its origin in early photoreceptor proteins, the descendants of which exist in modern bacteria, fungi, and plants (8, 10, 11, 27). Some of these proteins may have subsequently become involved in regulation of circadian rhythms (8, 9, 27). In animals, PAS domain-containing proteins and their functions have diversified further, evolving roles in development and the response to environmental variables, including oxygen tension (hypoxia) and small ligands (dioxin). In the phylogenetic analysis reported herein, we identify several clusters of metazoan PAS proteins, including invertebrate orthologs of vertebrate PAS proteins, that suggest evolutionary and possibly functional relationships. Bradfield and coworkers (29) recently presented a phylogenetic analysis of 16 PAS members. Our analysis of 26 PAS proteins confirms some, but not all, of their groupings and reveals additional relationships. Our trees are consistent with an initial diversification of the PAS family in invertebrates, followed by extensive gene duplication and further diversification in early vertebrates. Because of the rapid pace at which new PAS family members are being discovered (e.g., refs. 8, 9, 29, 57, and 58), a definitive description of evolutionary relationships within this family must await a more complete cataloguing of its members and will require continuing phylogenetic analyses.
Conclusions.
The vertebrate AHR plays a critical role in susceptibility to dioxin toxicity (59), but its conservation in all vertebrate groups suggests that it has a more fundamental role in cellular physiology. The existence of a second AHR-like gene in fish and mammals raises questions concerning the functions and possible interactions of these two genes. Understanding the phylogenetic relationships among these AHR genes and other members of the PAS family may provide an evolutionary context within which to interpret the functions of these proteins in gene regulation, development, environmental homeostasis, and toxicity.
Acknowledgments
We thank Drs. J. Stegeman, W. Powell, and H. Morrison for comments on the manuscript. This work was supported in part by the National Institute of Environmental Health Sciences (Grants R29 ES06272, F32 ES05644, and P42 ES07381), the Donaldson Charitable Trust, and a Christopher Haebler Frantz Fellowship (to M.A.S.). The automated sequencer was obtained through National Science Foundation Grant BIR-9419673. This is contribution 9,560 from the Woods Hole Oceanographic Institution.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AHR, aryl hydrocarbon (Ah) receptor; ARNT, AHR nuclear translocator; bHLH, basic helix–loop–helix; NJ, neighbor-joining; PAS, PER-ARNT-SIM; RT–PCR, reverse transcription-coupled PCR.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF024591–AF024595).
The sequences of wc-1, wc-2, and PYP proteins from Neurospora crassa and Ectothiorhodospira halophila, recently shown to contain partial PAS domains (8), were excluded from these analyses.
References
- 1.Poland A, Knutson J C. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 3.Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt J V, Bradfield C A. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 5.Nambu J R, Lewis J O, Wharton K A, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosthwaite S K, Dunlap J C, Loros J J. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 9.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M P, Steeves T D L, Vitaterna M H, Kornhauser J M, Lowrey P L, Turek F W, Takahashi J S. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linden H, Macino G. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagarias D M, Wu S-H, Lagarias J C. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Salguerro P, Pineau T, Hilbert D M, McPhail T, Lee S S T, Kimura S, Nebert D W, Rudikoff S, Ward J M, Gonzalez F J. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt J V, Su G H-T, Reddy J K, Simon M C, Bradfield C A. Proc Natl Acad Sci USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fuji-Kuriyama Y. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 15.Burbach K M, Poland A, Bradfield C A. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 17.Dolwick K M, Swanson H I, Bradfield C A. Proc Natl Acad Sci USA. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crews S T, Thomas J B, Goodman C S. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- 19.Whitelaw M, Gottlicher M, Gustafsson J A, Poellinger L. EMBO J. 1993;12:4169–4179. doi: 10.1002/j.1460-2075.1993.tb06101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poland A, Palen D, Glover E. Mol Pharmacol. 1994;46:915–921. [PubMed] [Google Scholar]
- 21.Coumailleau P, Poellinger L, Gustafsson J-A, Whitelaw M L. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga B N, Probst M R, Reiszporszasz S, Hankinson O. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzen A, Okey A B. Toxicol Appl Pharmacol. 1990;106:53–62. doi: 10.1016/0041-008x(90)90105-4. [DOI] [PubMed] [Google Scholar]
- 24.Hahn M E, Poland A, Glover E, Stegeman J J. Arch Biochem Biophys. 1994;310:218–228. doi: 10.1006/abbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 25.Hahn M E, Karchner S I. Biochem J. 1995;310:383–387. doi: 10.1042/bj3100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillis D M. Science. 1997;276:218–219. doi: 10.1126/science.276.5310.218. [DOI] [PubMed] [Google Scholar]
- 27.Kay S A. Science. 1997;276:753–754. doi: 10.1126/science.276.5313.753. [DOI] [PubMed] [Google Scholar]
- 28.Atchley W R, Fitch W M. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 30.Karchner S I, Hahn M E. Mar Environ Res. 1996;42:13–17. [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Swofford D L. paup 3.1.1; Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. [Google Scholar]
- 34.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 35.Gee H. Before the Backbone. Views on the Origin of the Vertebrates. London: Chapman & Hall; 1996. [Google Scholar]
- 36.Maisey J G. Cladistics. 1986;2:201–256. doi: 10.1111/j.1096-0031.1986.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 37.Swanson H I, Yang J-H. J Biol Chem. 1996;271:31657–31665. doi: 10.1074/jbc.271.49.31657. [DOI] [PubMed] [Google Scholar]
- 38.Atchley W R, Fitch W M. Proc Natl Acad Sci USA. 1995;92:10217–10221. doi: 10.1073/pnas.92.22.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 40.Stock D W, Powers D A. Mol Mar Biol Biotechnol. 1995;4:284–294. [PubMed] [Google Scholar]
- 41.Quattro J M, Pollock D D, Powell M, Woods H A, Powers D A. Mol Mar Biol Biotechnol. 1995;4:224–231. [PubMed] [Google Scholar]
- 42.Goksøyr A, Andersson T, Buhler D R, Stegeman J J, Williams D E, Forlin L. Fish Physiol Biochem. 1991;9:1–13. doi: 10.1007/BF01987606. [DOI] [PubMed] [Google Scholar]
- 43.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 44.Ruddle F H, Bartels J L, Bentley K L, Kappen C, Murtha M T, Pendleton J W. Annu Rev Genet. 1994;28:423–442. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 45.Holland P W H, Garcia-Fernandez J. Dev Biol. 1996;173:382–395. doi: 10.1006/dbio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 46.Ruddle, F. H., Bentley, K. L., Murtha, M. T. & Risch, N. (1994) Development Supplement, 155–161. [PubMed]
- 47.Iwabe N, Kuma K, Miyata T. Mol Biol Evol. 1996;13:483–493. doi: 10.1093/oxfordjournals.molbev.a025609. [DOI] [PubMed] [Google Scholar]
- 48.Sharman A C, Holland P W H. Netherlands J Zool. 1996;46:47–67. [Google Scholar]
- 49.Spring J. FEBS Lett. 1997;400:2–8. doi: 10.1016/s0014-5793(96)01351-8. [DOI] [PubMed] [Google Scholar]
- 50.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 51.Fujii-Kuriyama Y, Kobayashi A, Ema M, Mimura J, Morita M, Sogawa K. FASEB J. 1997;11:A780. (Abstract P56). [Google Scholar]
- 52.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. Mol Cell Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan C-M, Kuwana E, Bulfone A, Fletcher C F, Copeland N G, Jenkins N A, Crews S, Martinez S, Puelles L, Rubenstein J J R, Tessier-Lavigne M. Mol Cell Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- 54.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 55.Atchley W R, Fitch W M, Bronner-Fraser M. Proc Natl Acad Sci USA. 1994;91:11522–11526. doi: 10.1073/pnas.91.24.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maltepe E, Schmidt J V, Baunoch D, Bradfield C A, Simon M C. Nature (London) 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 57.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y D, Barnard M, Tian H, Li X, Ring H Z, Francke U, Shelton J, Richardson J, Russell D W, McKnight S L. Proc Natl Acad Sci USA. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Salguerro P, Hilbert D M, Rudikoff S, Ward J M, Gonzalez F J. Toxicol Appl Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- 60.Hahn, M. E., Stegeman, J. J. & Tillitt, D. E. (1998) Comp. Biochem. Physiol., in press. [DOI] [PubMed]


