Abstract
The increased interest in metabolite profiling is driving the need for improved analytical techniques with greater performance for a variety of important applications. Despite their limited sensitivity NMR methods are attractive because of their simplicity, reproducibility, quantitative nature, and wide applicability. The use of chemoselective isotopic tags has the potential to advance the application of NMR for analyzing metabolites in complex biofluids by allowing detection of metabolites down to the low μM level with high resolution and specificity. Here, we report a new 13C-tagging method using 13C-formic acid that delivers high sensitivity, good quantitation and excellent resolution for 1H-13C 2D NMR profiling of amino metabolites. High reproducibility (CV = 2%) was observed for metabolites in urine with concentrations down to 10 μM. As amino compounds comprise an important class of metabolites and small molecules of biological roles, this new method therefore should be amenable to a variety of applications.
Keywords: Metabolic Profiling, Metabolomics, NMR, Heteronuclear Correlation, 13C Isotope Tagging, Amine, Amino Acid
The study of the metabolite profile or metabolome can reveal the status of biological systems from a variety of perspectives, including insights into the normal and abnormal (or disease) states of an organism. For example, concentrations and fluxes of metabolites are constrained to a certain extent by their metabolic pathways and related enzymes under homeostasis, but these metabolite quantities are also subject to stresses such as environmental factors and disease. A quantitative measurement of the metabolome therefore provides an important snapshot of the ongoing normal and abnormal processes in complex biological systems.1-7 A more accurate, precise and rapid profile of the metabolome in an organism will lead to a better understanding of the systems biology and potentially a series of biomarkers that can be used for a variety of practical purposes, including diagnostics, drug development, nutrition, and environmental studies.8-17 As recognition of the importance of metabolic profiling has grown, the need for advanced analytical methods that deliver higher sensitivity, resolution and throughput has become recognized.
It is believed that most eukaryotic organisms possess at least 3000-5000 metabolites with very diversified molecular structures and physical/chemical properties.5 The quantitative determination of so many compounds in a single analysis remains out of reach for current technologies. Only a small fraction, normally the most abundant species, of the metabolites can currently be accurately and precisely detected. The information derived from this small sampling of metabolites is very often too superficial and nonspecific to reveal enough biochemical detail. Alternatively, one can measure a larger number of metabolites semi-quantitatively; however, this approach is often limiting as well.
The primary analytical methods used for metabolic profiling are mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR) due to their ability to provide rich information of complex mixtures at high throughput.18 MS is very attractive because of its high sensitivity, experimental flexibility and ability to determine unknown molecules. While a high number of metabolites can be detected and identified by MS (typically a few hundred), far fewer can be quantified to better than 10% coefficient of variation (CV) on a routine basis because of ion suppression and other matrix effects. Compared to MS, NMR is much less sensitive, but produces data that is more easily reproduced and quantified using a single internal standard. The same nuclei (usually 1H) are detected with the same sensitivity in one NMR experiment, irrespective of molecular origin.
Nevertheless, serious impediments to more widespread use of NMR for quantitative metabolite profiling, especially for lower concentration species, are the serious spectral overlap and complexity. The signals of metabolites with low concentrations are often buried under nearby strong signals due to insufficient resolution. These species become non-detectable unless selective methods19, 20 or multidimensional experiments21, 22 are used. Methodological innovations that significantly improve the resolving power of NMR would reduce a current bottleneck and greatly advance the application of NMR in metabolic profiling.
Towards this goal, we recently introduced several methods for in vitro chemical tagging of biofluids.23-25 A molecular tag containing an enriched or high abundance isotope such as 13C/15N or 31P can be introduced to the biofluid; it selectively reacts with metabolites with a certain functional group and therefore tags these metabolites with that isotope. The subsequent heteronuclear 2D NMR experiment selectively detects the tags and provides a simple spectrum free of the background signals from the rest of the tagged molecule as well as the untagged molecules. Instead of the crowded and coupled signals from all the protons into one dimension of ~10 ppm, one can easily resolve a smaller number of signals (each metabolite has only one cross-peak unless multiply tagged) in two dimensions. Since every metabolite molecule has at least one functional group in order to function in its involved biological processes, one can in principle use different isotopic tags to profile all metabolites in selective classes. The improved resolving power comes from the high-resolution heteronuclear 2D NMR, the chemoselective tagging and the reduced complexity and number of signals. It should be noted that chemoselective tags including the isotopic variant tags have also been successfully adopted for metabolic or proteomic profiling with MS for separation or quantification purposes.26-30 Previously we have reported the 13C-tagging of amino metabolites with 13C-acetic anhydride for 1H-13C HSQC 2D NMR profiling,23 15N-tagging of carboxyl-containing metabolites with 15N-ethanolamine for 1H-15N HSQC 2D NMR profiling,24 and 31P-tagging of active hydrogen moieties in lipid extracts with 2-chloro-4,4,5,5,-tetramethyldioxaphospholane for 31P NMR analysis.25 Because of the closeness of the tag to metabolite molecule and the relatively strong 1H-15N J-coupling, the use of 15N-ethanolamine has been especially successful for profiling over 100 carboxyl-containing metabolites in biofluids with 2D NMR. Here, 13C-formic acid is applied as the isotopic tag to further improve the performance of 2D NMR in profiling amino metabolites. Amino acids are the most important and common amino metabolites; they are not only the building blocks for proteins, but also the precursors for nucleotides,31 and they provide an energy source through transamination, the urea cycle, the citric acid cycle and gluconeogenesis.32-34 Other common amino metabolites include derivatives of amino acids, taurine, dimethylamine, methylamine, and many neurotransmitters such as dopamine, serotonin and histamine. Drugs such as amphetamine, procaine, rimantadine and their metabolites also belong to this group of compounds.
EXPERIMENTAL SECTION
Chemicals and Biological Samples
All metabolite standards, N, N-dicyclohexylcarbodiimide (DCC), N-hydroxysuccinimide (HOSu) (Sigma-Aldrich) and 13C-formic acid (Cambridge Isotope Laboratories) were purchased and used without further purification. Human serum and urine samples were obtained either from commercial sources (Innovative Research, Novi, MI) or from healthy volunteers in accordance with the Institutional Review Board at Purdue University.
General Procedure for 13C-Formylation
13C-formic acid (2 μL, 0.05 mmol) and N-hydroxysuccinimide (5 mg, 0.04 mmol) were dissolved in tetrahydrofuran (100 μL). N, N-dicyclohexylcarbodiimide (9 mg, 0.04 mmol) in tetrahydrofuran (50 μL) was added to the mixture and stirred at room temperature.35 After 15 min, the reaction mixture was centrifuged to remove the insoluble urea; the supernatant containing 13C-N-formyloxysuccinimide was then added to the biofluid sample (500 μL) along with 2 M NaHCO3 (50 μL, 0.1 mmol) aqueous solution. The reaction mixture was stirred at room temperature for 4 h and dried under vacuum. After redispersing the dried product mixture in D2O (500 μL), the pH of the solution was adjusted to 7.0 by adding 1 M HCl and then transferred to a standard 5mm NMR tube for analysis. 13C-N-formyloxysuccinimide can also be purified by recrystallization in ethanol31 and used for the tagging reaction instead of in situ generation.
NMR Spectroscopy and Data Processing
NMR experiments were carried out at 298 K on a Bruker Avance-III-800 spectrometer equipped with a room temperature 1H inverse detection Z-gradient probe. 1H NMR spectra were obtained using the water Pre-SAT180 sequence.36 The sensitivity-enhanced 1H-13C 2D HSQC experiments employed an INEPT transfer delay of 2.5 ms corresponding to a 1JC-H of 200 Hz. Spectral widths were approximately 10 kHz for the 1H dimension and 3 kHz for 13C. 128 or 256 free induction decays of 2,048 data points each were collected in the indirect (t1) dimension using 4 or 8 transients per increment. 13C decoupling during the direct detection dimension (t2) was achieved with the Globally Optimized Alternating-Phase Rectangular Pulses (GARP) sequence. The resulting 2D data were zero-filled to 1,024 points in the t1 dimension after forward linear prediction to 512 points. A 45°-shifted sinebell window function was then applied to both dimensions before Fourier transformation. NMR data were processed using Bruker Topspin 3.0 spectrometer software on a Redhat Linux platform. The reproducibility of the method was evaluated using the auto peak picking and integration functions of Topspin; the 2D signals in three spectra were then compared individually after normalization with respect to the most intense signal (L-alanine) in order to reduce errors from volumetric measurements and probe tuning.
Calibration
Stock solutions of 9 standard compounds were made individually at 50 mM with their precise concentrations determined using 1D 1H NMR against a calibrated sodium d6-2, 2-dimethyl-2-silapentane-5-sulfonate (DSS) solution. The individual stock solutions were then mixed at equal volumes and diluted with water to make a mixed stock solution at 4 mM, which was further diluted to make a calibration series ranging from 5 μM to 2 mM. A fixed amount of ethanolamine (0.5 μmol, 1 mM internal reference) was added to 500 μL of each calibration solution to make the final calibration samples; which were then subjected to 13C-formylation and 1H-13C HSQC analysis. The 2D peaks were integrated and referenced to the integration volume of 13C-formylated ethanolamine and plotted against the concentrations previously determined by 1D 1H NMR.
RESULTS
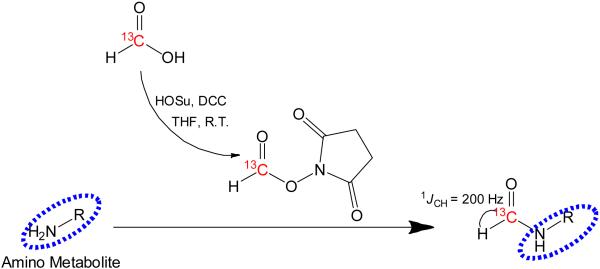
After conversion to the active NHS (N-hydroxysuccinimidyl) ester, 13C-formic acid readily reacted with amino metabolites and introduced a 1H-13C pair with a one-bond J-coupling of 200 Hz into the metabolite molecule as shown in Figure 1. The newly introduced HCO- moeity was highly polar and therefore retained good solubility in water. An HSQC experiment was then applied to provide a highly resolved spectrum of the 1H-13C pairs in the tagged metabolite molecules. The stuctural variations of the metabolites led to a good dispersion of the 2D signals in both 1H and 13C dimensions. At the same time, the J-couplings and relaxation rates do not vary significantly across different metabolites, which is essential for unbiased detection of all tagged amino metabolites.
Figure 1.
Amino metabolites in biofluids are isotopically tagged by 13C-formylation. The tagged metabolites are subsequently detected using sensitivity-enhanced 2D 1H-13C correlation NMR.
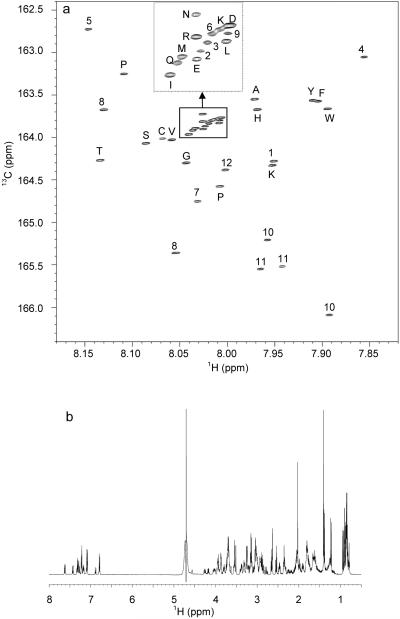
The performance of the method was first evaluated with standard compounds that are commonly seen in biofluids. A mixture of 32 standard compounds with amino groups was isotopically tagged by 13C-formylation and analyzed using a 1H-13C HSQC experiment at 800 MHz. The resulting spectrum is shown in Figure 2 with peak assignments, and the chemical shifts for signals of the tagged standard compounds are listed in Table 1. Each compound has one signal in the 1H-13C 2D spectrum except for metabolites such as lysine and spermidine that have more than one unique amino group. Proline, 4-hydroxyproline and sarcosine also have two signals due to the cis- and trans- isomers formed by 13C-formylation. The signals of the 13C-fomylated compounds are dispersed over 4 ppm in the 13C dimension and over 0.3 ppm in the 1H dimension with good spectral separation.23 By comparison, the 1D 1H NMR detection of the same mixture before 13C-tagging produces a complex spectrum with significant serious signal overlap (Figure 2b).
Figure 2.
(a) The 1H-13C HSQC spectrum of a mixture of 32 standard amino compounds after 13C-formylation (pH 7.0, 298 K). Inset shows the expanded view of the boxed region (1H: 8.00-8.05 ppm, 13C: 163.65-164.00 ppm). (b) 1D 1H NMR spectrum of the same mixture in water before 13C-formylation (pH 7.0, 298 K).
Table 1.
Chemical shifts for 32 13C-formylated amino metabolites standard compounds (D2O, pH 7.0, 298K)
| Label | Name | 1H (ppm) | 13C (ppm) |
|---|---|---|---|
| A | L-alanine | 7.972 | 163.521 |
| 1 | β-alanine | 7.951 | 164.252 |
| 2 | L-2-aminoadipic Acid | 8.023 | 163.841 |
| 3 | L-2-aminobutyric Acid | 8.020 | 163.807 |
| 4 | L-2-aminoisobutyric Acid | 7.856 | 163.025 |
| 5 | 4-aminophenol | 8.147 | 162.699 |
| R | L-arginine | 8.026 | 163.783 |
| N | L-asparagine | 8.026 | 163.695 |
| D | L-aspartic Acid | 8.007 | 163.738 |
| 6 | L-citrulline | 8.017 | 163.772 |
| C | L-cysteine | 8.069 | 163.986 |
| 7 | ethanolamine | 8.032 | 164.723 |
| E | L-glutamic Acid | 8.026 | 163.872 |
| Q | L-glutamine | 8.037 | 163.887 |
| G | glycine | 8.044 | 164.272 |
| H | L-histidine | 7.964 | 163.644 |
| 8 | 4-hydroxy-L-proline | 8.130 | 163.645 |
| 8.054 | 165.331 | ||
| I | L-isoleucine | 8.041 | 163.935 |
| L | L-leucine | 8.009 | 163.803 |
| K | L-lysine | 8.013 | 163.754 |
| 7.953 | 164.299 | ||
| M | L-methionine | 8.034 | 163.862 |
| 9 | L-norleucine | 8.008 | 163.771 |
| F | L-phenylalanine | 7.905 | 163.544 |
| P | L-proline | 8.109 | 163.224 |
| 8.008 | 164.545 | ||
| 10 | sarcosine | 7.958 | 165.178 |
| 7.893 | 166.057 | ||
| S | L-serine | 8.086 | 164.042 |
| 11 | spermidine | 7.965 | 165.521 |
| 7.942 | 165.494 | ||
| 12 | taurine | 8.002 | 164.353 |
| T | L-threonine | 8.134 | 164.24 |
| W | L-tryptophan | 7.895 | 163.635 |
| Y | L-tyrosine | 7.910 | 163.534 |
| V | L-valine | 8.059 | 164.000 |
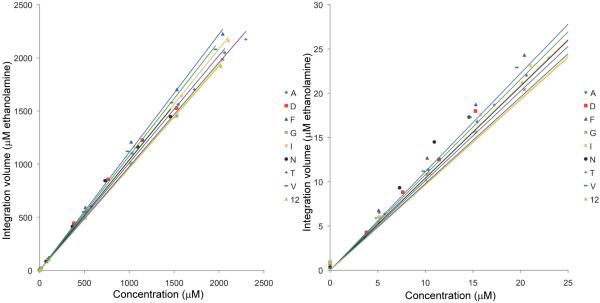
Because of the reproducibility and good sensitivity due to the strong J-coupling between 1H and 13C, HSQC detection of the 13C-formylated amines is quantitative as shown in Figure 3. Good linearity (R2 > 0.995) and quantitation (average deviation from trend line for the 9 standard compounds was 6% for concentrations above 10 μM and 12% for concentrations below 10 μM) was observed using the integrated 2D signal intensities for the standard compounds over a large range of concentrations. The strong J-coupling also ensures sensitive detection of trace amounts of metabolites. The 1H-13C HSQC experiment run at 800 MHz can detect 13C-formylated alanine with a concentration down to 4 μM (SNR ~4) within 8 min (1 scan, 128 increments). Though conventional 1D 1H NMR can detect substances at low μM levels, its performance is often reduced by 1H-1H couplings and spectral overlap in complex samples, as well as residual solvent signals. As a result, 1H NMR is normally utilized for profiling metabolites above 100 μM. The improved sensitivity and excellent detection linearity for the 13C-formylated amines result in part from the relatively large 200 Hz JC-H coupling between the 1H and 13C of the formyl tag.
Figure 3.
Calibration curves for 9 standard amino compounds detected by 1H-13C HSQC after 13C-formylation, with an expanded view of 0-25 μM concentration range shown in the right panel. The 2D peaks were integrated and referenced to the integration volume of 13C-formylated ethanolamine (internal standard) and plotted against the concentrations previously determined by 1D 1H NMR. The integration volume of 13C-formylated 1 μM ethanolamine serves as the unit for the vertical axis. The trend lines were generated by linear regression; R2 values were greater than 0.995 for all 9 compounds; Good quantitation was observed: the average deviation from the calibration trend line is 6% for concentrations above 10 μM and 12% for concentrations below 10 μM.
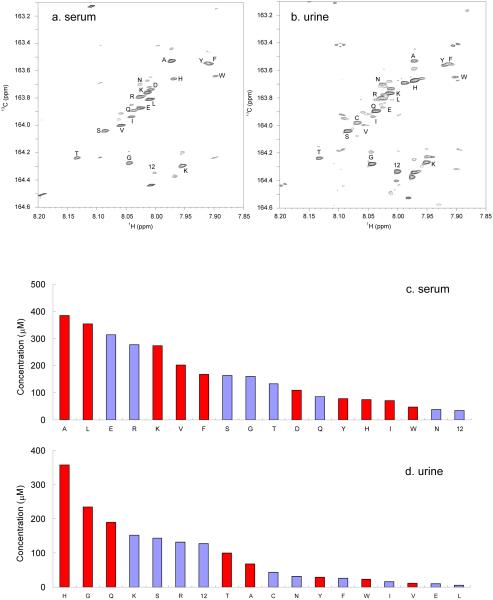
13C-formylation was then applied to profile amino metabolites in human biofluid samples. More than 40 signals, mostly of amino acids, were clearly seen and well resolved from a human serum sample as shown in Figure 4a. The urine spectrum (Figure 4b.) showed not only the presence of most amino acids that were found in serum at different levels but also additional signals representing unidentified amino metabolites. However, as a collection of bio-wastes and by-products rather than a circulating body fluid, urine has a much greater variation in chemical content compared to blood.37 Very often the urinary metabolites are reflections of what has been introduced into the human body. Therefore, the extra signals in the studied urine spectrum could arise from amino compounds generated via the metabolism of nitrogen-containing dietary intake.
Figure 4.
1H-13C HSQC spectra of a healthy human serum sample (a) and a healthy human urine sample (b) after 13C-isotopic tagging with 13C-formic acid. Eighteen metabolites were indentified and quantified in each spectrum. Concentrations for amino metabolites identified and quantified using the 2D 1H-13C HSQC spectra for the serum sample (c) and the urine sample (d) after 13C-formylation. Metabolites that have resolved 1D 1H signals are marked in red and others are marked in light blue.
The assignment of 2D 1H-13C signals could be accomplished by simply comparing the 1H and 13C chemical shifts of a particular peak with the library of 32 compounds (Table 1). This is in contrast to the sometimes challenging and often time-consuming assignment of low level 1D 1H NMR signals in complex samples that involves checking chemical shifts, coupling constants, integrations or even spiking standard compounds. Quantification of these amino metabolites is straightforward as the integration volume of a 2D NMR signal is proportional to the number of protons. Eighteen amino metabolites were identified and quantified in both serum and urine samples with their concentrations shown in Figure 4c and 4d, respectively. Taurine and sixteen amino acids were identified from both samples; in addition, aspartic acid was quantified in the serum and cysteine was quantified in the urine. In general, the observed metabolite concentrations in serum were within the healthy concentration ranges appearing in the Human Metabolome Database.38 Profiling of amino metabolites using 1D 1H spectra was challenging especially for metabolites of low concentrations that appeared in crowded spectral regions. As a result, more than half of the amino-metabolites profiled using the 13C tagging and 2D detection approach could not be identified and quantified using 1D 1H detection (Figure 4c, 4d, S1 and S2). Most of the signals that were clearly resolved and quantifiable in the 1D 1H spectrum had relatively high intensities and appeared either in the methyl region (~1-2 ppm) where signals had narrow line widths and simple splitting patterns or the aromatic region (~6-8 ppm) where fewer signals were present.
The reproducibility of the method was tested in triplicate experiments on a split sample of the same urine. As shown in Figure S3 and S4, the three spectra are very similar. Peak integrations indicated that the average coefficient of variation (CV) was 2% for 12 metabolites above 10 μM and 11% for 6 metabolites below 10 μM (SNR are less than 8 for 3 metabolites below 4 μM). Again, only 7 of the 18 amino metabolites had recognizable 1D 1H NMR signals (Figure S5). This high reproducibility results from the quantitative nature of both the NMR detection and the 13C-tagging reaction, which is essential for metabolic profiling.
DISCUSSION
Successful metabolic profiling methods rely on the sensitive and unbiased detection of numerous metabolites. The combination of 13C-formyl tagging and 2D 1H-13C HSQC detection provide a sensitive, unbiased method with excellent quantitation and resolution. In the 13C-formylated metabolite molecule, the short, one-bond distance between the labeled 13C and its closest 1H produces a large (200 Hz) J-coupling constant that improves polarization transfer that is essential for the HSQC experiments. The 1JC-H of 200 Hz allows for a short 2.5 ms INEPT transfer delay compared to the 83 ms delay (corresponding to the 2JC-H of 6 Hz) that was used in our previous work involving acetic anhydride tagging.23 The faster polarization transfer significantly reduces polarization losses due to relaxation or competing transfer pathways involving 2JC-H or 3JC-H of 2-4 Hz and therefore leads to better detection sensitivity, reproducibility and linearity.
To further improve the resolving ability of 1H-13C HSQC 2D NMR for metabolic profiling, one can improve the performance of the spectrometer (i.e. using higher magnetic fields, more increments in the indirect 13C dimension), or make the 2D signals disperse in a wider region of chemical shifts across different metabolites. Chemical shifts originate from the impact of varying local electronic environments on the resonance frequency of the nuclear spin. For the 13C-tagged molecules, the chemical shifts of the 2D 1H-13C signals vary because of the structural differences between metabolites. The close distance between the isotope labeled 1H-13C nuclear pair and the rest of the metabolite leads to an increase in the chemical shift sensitivity of the tag, a better dispersion of signals in two dimensions, and less chance for spectral overlap. In addition to the improvements in NMR detection, the tagging reaction is now carried out in a simple mix-and-stir procedure that is easy to repeat using the NHS ester. The reaction time for this mild tagging reaction is longer than the previous acetylation using acetic anhydride, however no acetic acid is generated during the reaction and there is no need to control the pH by addition of NaOH throughout the reaction. Therefore both the time and labor for setting up one reaction are reduced. Moreover 13C-enriched formic acid is relatively inexpensive and has a long shelf life, making the method economically attractive.
Though the assignments of 2D signals for known metabolites can be easily accomplished by comparing chemical shifts to the library, the identification of unknown signal remains challenging. Efforts are currently underway to expand this library and to identify unassigned peaks in the spectrum using HPLC-NMR and microcoil NMR approaches.39-41 While 1D 1H NMR metabolic profiling primarily emphasizes signals from highly concentrated (mM) species that are often too nonspecific to be used as biomarkers, the new method allows the quantitative NMR profiling of metabolites at lower levels (μM) that should lead to new findings in future applications. The limit of detection may be further improved using preconcentration methods and microcoil NMR,39-41 or potentially new technologies such as dynamic nuclear polarization42 and para-hydrogen induced polarization.43 Though 2D NMR experiments can require longer times than simple 1D NMR experiments, the experiments here could be conducted in only 8 min per sample. In addition, advanced data collection and processing algorithms such as ultrafast 2D NMR,44 covariance NMR,45 nonlinear sampling and forward maximum entropy reconstruction46 can greatly reduce the acquisition time for 2D NMR experiments and allow high-throughput operation.
In conclusion, a new method to incorporate 13C nuclei into metabolites in vitro has been introduced for conducting quantitative 2D NMR metabolic profiling with improved resolution and sensitivity stemming from the structure of 13C-formic acid. The performance of method has been demonstrated with a mixture of 32 standard compounds, human urine and serum. High reproducibility and detection linearity were observed for metabolites down to 10 μM, therefore we anticipate that this method will be a useful tool for quantitative profiling of low-concentration amino metabolites in complex mixtures. As amino-containing compounds constitute an important class of molecules in biological processes, this new method is expected to find a number of applications in the quantitative analysis of these molecules.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health Grant 1 R01GM085291-01. DR is a member of the Purdue Center for Cancer Research and Oncological Sciences Center.
REFERENCES
- (1).Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- (2).Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Nat. Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- (3).Fiehn O. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- (4).Kell DB. Curr. Opin. Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- (5).Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- (6).van der Greef J, Smilde AK. J. Chemom. 2005;19:376–386. [Google Scholar]
- (7).Van Dien S, Schilling CH. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100078. 2006.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Serkova NJ, Niemann CU. Expert Rev. Mol. Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- (9).Griffin JL. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2006;361:147–161. doi: 10.1098/rstb.2005.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Gowda GAN, Zhang SC, Gu HW, Asiago V, Shanaiah N, Raftery D. Expert Rev. of Mol. Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang S, Gowda GAN, Asiago V, Shanaiah N, Barbas C, Raftery D. Anal. Biochem. 2008;383:76–84. doi: 10.1016/j.ab.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bedair M, Sumner LW. Trends Anal. Chem. 2008;27:238–250. [Google Scholar]
- (13).Bundy JG, Davey MP, Viant MR. Metabolomics. 2009;5:3–21. [Google Scholar]
- (14).Lindon JC, Holmes E, Nicholson JK. FEBS J. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- (15).Robertson DG. Toxicol. Sci. 2005;85:809–822. doi: 10.1093/toxsci/kfi102. [DOI] [PubMed] [Google Scholar]
- (16).Rezzi S, Ramadan Z, Fay LB, Kochhar S. J. Proteome Res. 2007;6:513–525. doi: 10.1021/pr060522z. [DOI] [PubMed] [Google Scholar]
- (17).Viant MR. Biochem. Biophys. Res. Commun. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- (18).Pan Z, Raftery D. Anal. Bioanal. Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- (19).Sandusky P, Raftery D. Anal. Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- (20).Sandusky P, Raftery D. Anal. Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- (21).Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Sussman MR, Markley JL. Anal. Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chikayama E, Suto M, Nishihara T, Shinozaki K, Hirayama T, Kikuchi J. PLoS ONE. 2008;3:e3805. doi: 10.1371/journal.pone.0003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Shanaiah N, Desilva MA, Gowda GAN, Raftery MA, Hainline BE, Raftery D. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ye T, Mo H, Shanaiah N, Gowda GAN, Zhang S, Raftery D. Anal. Chem. 2009;81:4882–4888. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).DeSilva MA, Shanaiah N, Gowda GAN, Rosa-Pérez K, Hanson BA, Raftery D. Magn. Reson. Chem. 2009 doi: 10.1002/mrc.2480. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Carlson EE, Cravatt BF. Nat. Methods. 2007;4:429–435. doi: 10.1038/nmeth1038. [DOI] [PubMed] [Google Scholar]
- (27).Zhang H, Li XJ, Martin DB, Aebersold R. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- (28).Brittain SM, Ficarro SB, Brock A, Peters EC. Nat. Biotechnol. 2005;23:463–468. doi: 10.1038/nbt1076. [DOI] [PubMed] [Google Scholar]
- (29).Lamos SM, Shortreed MR, Frey BL, Belshaw PJ, Smith LM. Anal. Chem. 2007;79:5143–5149. doi: 10.1021/ac062416m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Huang X, Regnier FE. Anal. Chem. 2008;80:107–114. doi: 10.1021/ac071263f. [DOI] [PubMed] [Google Scholar]
- (31).Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5th ed. W. H. Freeman & Co.; New York: 2002. pp. 693–698. [Google Scholar]
- (32).Sakami W, Harrington H. Annu. Rev. Biochem. 1963;32:355–398. doi: 10.1146/annurev.bi.32.070163.002035. [DOI] [PubMed] [Google Scholar]
- (33).Brosnan JT. J. Nutri. 2000;130:988S–990S. doi: 10.1093/jn/130.4.988S. [DOI] [PubMed] [Google Scholar]
- (34).Young VR, Ajami AM. J. Nutri. 2001;131:2449S–2459S. doi: 10.1093/jn/131.9.2449S. [DOI] [PubMed] [Google Scholar]
- (35).Hecht SM, Werner D. J. Chem. Soc., Perkin Trans. 1. 1973:1903–1906. doi: 10.1039/p19730001903. [DOI] [PubMed] [Google Scholar]
- (36).Mo H, Raftery D. J. Magn. Reson. 2008;190:1–6. doi: 10.1016/j.jmr.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Saude EJ, Adamko D, Rowe BH, Marrie T, Sykes BD. Metabolomics. 2007;3:439–451. [Google Scholar]
- (38).Wishart D, Knox C, Guo A, et al. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Djukovic D, Liu S, Henry I, Tobias B, Raftery D. Anal. Chem. 2006;78:7154–7160. doi: 10.1021/ac0605748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Djukovic E, Appiah-Amponsah E, Shanaiah N, Gowda GAN, Henry I, Everly M, Tobias B, Raftery D. J. Pharm. Biomed. Anal. 2008;47:328–334. doi: 10.1016/j.jpba.2007.12.035. [DOI] [PubMed] [Google Scholar]
- (41).Kc R, Henry I, Park GHJ, Raftery D. J. Magn. Reson. 2009;197:186–192. doi: 10.1016/j.jmr.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wilson DM, Hurd RE, Keshari K, Van Criekinge M, Chen AP, Nelson SJ, Vigneron DB, Kurhanewicz J. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5503–5507. doi: 10.1073/pnas.0810190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Adams RW, Aguilar JA, Atkinson KD, Cowley MJ, Elliott PIP, Duckett SB, Green GGR, Khazal IG, López-Serrano J, Williamson DC. Science. 2009;323:1708–1711. doi: 10.1126/science.1168877. [DOI] [PubMed] [Google Scholar]
- (44).Frydman L, Blazina D. Nat. Phys. 2007;3:415–419. [Google Scholar]
- (45).Zhang FL, Brüschweiler R. J. Am. Chem. Soc. 2004;126:13180–13181. doi: 10.1021/ja047241h. [DOI] [PubMed] [Google Scholar]
- (46).Hyberts SG, Heffron GJ, Tarragona NG, Solanky K, Edmonds KA, Luithardt H, Fejzo J, Chorev M, Aktas H, Colson K, Falchuk KH, Halperin JA, Wagner G. J. Am. Chem. Soc. 2007;129:5108–5116. doi: 10.1021/ja068541x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.