Abstract
Nrf2 (NF-E2-related factor 2) is a master transcription factor containing a powerful acidic transcriptional activation domain. Nrf2-dependent gene expression impacts cancer chemoprevention strategies, inflammatory responses, and progression of neurodegenerative diseases. Under basal conditions, association of Nrf2 with the CUL3 adaptor protein Keap1 results in the rapid Nrf2 ubiquitylation and proteasome-dependent degradation. Inhibition of Keap1 function blocks ubiquitylation of Nrf2, allowing newly synthesized Nrf2 to translocate into the nucleus, bind to ARE sites and direct target gene expression. Site-directed mutagenesis experiments coupled with proteomic analysis support a model in which Keap1 contains at least 2 distinct cysteine motifs. The first is located at Cys 151 in the BTB domain. The second is located in the intervening domain and centers around Cys 273 & 288. Adduction or oxidation at Cys151 has been shown to produce a conformational change in Keap1 that results in dissociation of Keap1 from CUL3, thereby inhibiting Nrf2 ubiquitylation. Thus, adduction captures specific chemical information and translates it into biochemical information via changes in structural conformation.
Introduction
Proteins that compose the Phase II superfamily and antioxidant enzymes provide an enzymatic line of defense against electrophiles and reactive oxygen species, two important contributors to the etiology of many human diseases. Proteins such as NAD(P)H: quinone oxidoreductase (DT-diaphorase), glutathione S-transferases, UDP-glucoronosyltransferses, MnSOD, catalase, thioredoxin, and glutamate cysteine ligase (gamma glutamylcysteine synthetase) are canonical members of the Phase II and antioxidant enzyme family and are regulated by a common cis-acting regulatory element located in the proximal promoter region. This cis-acting element was identified in mouse models and named the Electrophile-Response Element (EpRE; (Friling et al., 1990), as well as identified in rat models as the Antioxidant Response Element ARE; (Favreau and Pickett, 1991). Heterodimeric binding of Nrf2 (NF-E2-related factor 2) to AREs induces target gene expression (Venugopal and Jaiswal, 1996; Venugopal and Jaiswal, 1998). Initially, Nrf2 was discovered in a screen that utilized a tandem repeat of the consensus sequences for AP1 and NF-E2 present in the locus control region in the 5’ region of β-globin (Moi et al., 1994). Nrf2 belongs to the basic leucine zipper (bZIP) transcription factor family and contains a powerful acidic transcriptional activation domain. Nrf2 activity impacts cancer chemoprevention strategies, inflammatory responses, and progression of neurodegenerative diseases.
Sulfhydryl chemistry represents the basis for strategies designed to induce Nrf2-mediated Phase II gene expression (Prestera et al., 1993). McMahon et al., (McMahon et al., 2001) studied Phase II gene expression in the intestine of wild type and Nrf2 null animals. They found that expression of Nrf2 was required for induction of Phase II genes by synthetic sulfhydryl cancer chemopreventive agents. Sulforaphane[(−)-1-isothiocyanato-(4R0-(methylsulfinyl)butante] is a representative example. Conjugation to thiols represents a major metabolic pathway for isothiocyanates such as sulforaphane (Jiao et al., 1996). This putative cancer chemoprotective agent induces the transcription of many Phase II enzymes in an Nrf2-mediated manner thereby inhibiting benzo[a]pyrene-induced tumors (Fahey et al., 2002). Studies such as these demonstrate the importance of sulfhydryl chemistry and Nrf2 activation.
Nrf2 undergoes rapid ubiquitination and proteasome degradation
Under homeostatic cellular conditions Nrf2 is maintained at low levels due to CUL3-dependent E3-ubiquitin ligase mediated ubiquitination of Nrf2's amino terminus Neh2 domain at lysines 44, 50, 52, 53, 56, 64, and 68 (Itoh et al., 2003; Kobayashi et al., 2004; Zhang et al., 2004). Ubiquitination directs proteasome-dependent degradation of Nrf2 (Sekhar et al., 2002; Itoh et al., 2003). Proteins targeted for Cullin-dependent ubiquitination are captured by a substrate receptor module that provides a protein recognition site and appropriate positioning within a Cullin-E3 complex. Current models of ubiquitin ligase complexes indicate that an assembled Cullin/substrate receptor module serves as a rigid scaffold to position the charged ubiquitin-conjugating E2 enzyme and its substrate. As discussed by Duda et al. (Duda et al., 2008) and Saha and Deshaies (Saha and Deshaies, 2008) the distance between the E2 active site and the substrate is on the order of 50 Å, which decreases during the course of polyubiquitination. Nedd8 conjugation to the Cullin allows the E3/Ub-E2 complex to exhibit significant structural flexibility that accommodates dramatic changes in substrate geometry, thereby promoting polyubiquitination (Saifee and Zheng, 2008).
Unlike other Cullin-targeted substrates, Nrf2 ubiquitination is a constitutive process that does not require post-translational modification. Nrf2's Neh2 domain is captured by the carboxyl Kelch domain of the CUL 3 ligase substrate adaptor protein, Keap1 (Kelch-like ECH-associated protein 1) (Itoh et al., 1999; Kobayashi et al., 2004). Keap1 functions as a homodimer, a consequence of amino-terminal BTB (Broad complex Tramtrack, Bric-a-brac) domain interactions (Zipper and Mulcahy, 2002) that allow homodimeric binding to the amino-terminus of CUL3 (Furukawa and Xiong, 2005). Cell-based studies suggest that Nrf2 binds to Keap1 with a 2:2 stoichiometry (Lo et al., 2006) whereas biophysical and biochemical studies support a 1:2 stoichiometry (Tong et al., 2006a).
Induction of Nrf2-Directed Gene Expression: Identification of A Common Chemistry
Inhibition of Nrf2 ubiquitination is followed by induction of Nrf2-directed gene expression (Kobayashi et al., 2004). The pioneering work of Talalay and colleagues (Talalay et al., 1988; Dinkova-Kostova et al., 2002; Holtzclaw et al., 2004) demonstrated that induction of Nrf2-directed gene expression can be accomplished by 10 chemically diverse classes of compounds. These compounds exhibit a common chemical feature: they react with sulfhydryl groups. Thus, it was hypothesized that protein thiol modification induced Nrf2-dependent transcription (Prestera et al., 1993). The knowledge that Keap1 directs the ubiquitination of Nrf2 and contains either 25 (murine) or 27 (human) cysteine residues made it the prime candidate for the hypothesized thiol sensor.
Thiol-Based Redox Sensors
In the last 10 years a significant effort has been undertaken to provide a biophysical understanding of how thiol-based redox sensors distinguish between different types of chemistries and translate chemical information into biochemical signals (Paget and Buttner, 2003; Salmeen et al., 2003; Codreanu et al., 2008). Yap1 represents a well characterized model (Georgiou, 2002). Distinct cysteine residues within the transcription factor can be used to capture specific chemical information and translate it into biochemical information via changes in structural conformation, allowing redox specific modifications to induce gene transcription.
Based on the knowledge that Keap1 directs the ubiquitination of Nrf2 and that Keap1 contains a number of reactive cysteine residues, significant effort has been made to identify reactive residues and biochemically link adduction/oxidation of those residues to changes in Keap1 function. Mutation studies involving Keap1 cysteine residues have been very informative. Mutation of Cys to either Ser or Ala would not be expected to affect protein conformation significantly and consistent with this concept, the majority of the mutations did not affect Keap1 function (Zhang and Hannink, 2003). However, the absence of an effect when specific Cys residues are mutated does not demonstrate that adduction at those residues would not affect Keap1 function (Holland et al., 2008). Furthermore, the observation that mutation of a specific Cys residue affects function does not demonstrate that that particular residue is reactive. But, when a mutation does affect Keap1 function, one can hypothesize that if that residue is adducted, then adduction will affect function.
The mutation studies identified 3 important Cys residues in Keap1: Cys 151, Cys 273, and Cys 288 (Zhang and Hannink, 2003). Whereas electrophiles and oxidants induce disassociation of Keap1 from CUL3, with subsequent inhibition of Nrf2 ubiquitination, mutation of Cys 151 to Ser abolished electrophile and oxidant-mediated disassociation and Keap1-directed ubiquitination (Zhang and Hannink, 2003). The role of Cys 151 in electrophile/oxidant repression of Nrf2 ubiquitination has been verified in vivo (Yamamoto et al., 2008). Mutation of either Cys 273 or Cys 288 to Ala inhibits Keap1's ability to direct the constitutive ubiquitination of Nrf2. Inhibition of Nrf2 ubiquitination is followed by Nrf2 nuclear localization, with subsequent Nrf2-directed transcription (Zhang and Hannink, 2003). Ectopic expression of Keap1 containing a C273A mutation has been shown to complement (50%) a Keap1 molecule containing a C288A mutation (Wakabayashi et al., 2004). This has been recapitulated in vivo (Yamamoto et al., 2008).
Keap1 has been shown to be a Zn2+ binding protein (Dinkova-Kostova et al., 2005). Mutation of Cys 273 or Cys 288 decreased the Ka for Zn2+ binding from 1.02 (+/− 0.19) × 1011 M−1 to approximately 1 × 1010 M−1, yet the physiological significance of these observations is not understood. Mutation of Cys 273 or Cys 283 to Ser does not affect the stability of Keap1 nor its subcellular localization (Zhang and Hannink, 2003). Mutation of Cys 226, 241, 257, 273, 288, and 197 to Ala does not affect the ability of Keap1 to bind CUL3 (Kobayashi et al., 2004). Mutation of Cys 273 and Cys 288 to Ala does not affect the ability of Keap1 to bind Nrf2. Using a BIAcore interaction assay, Kobayashi et al., (Kobayashi et al., 2006) found that the Kd for association of wildtype Keap1 with a GFP/Nrf2-Neh2 domain was not significantly different from the Kd that was obtained when Keap1 contained C273A and C288A mutations (Kobayashi et al., 2006). Currently, the mechanisms by which C273 and C288 mutations affect Keap1 function are not well understood.
A two-site recognition model has been proposed, in which oxidative modification or electophilic adduction of Keap1, particularly in the intervening domain, results in disassociation of Keap1's Kelch domain from a low affinity Nrf2/DLG binding site. The high affinity ETGE binding site in the Neh2 domain would not be affected by adduction/oxidiation of Cys residues. Loss of Kelch/DLG interaction is postulated to affect orientation of the Neh2 domain with respect to the Ub-E2 conjugating enzyme, thereby inhibiting ubiquitination (McMahon et al., 2006; Tong et al., 2006b). Using this model, one can hypothesize that mutation of either C273 or C288 in Keap1 results in a conformational change that impacts Keap1/DLG interactions. However, such a model needs to reconcile the question of Nrf2/Keap1 stoichiometry and the plasticity outlined in current models of CUL-based ubiquitination.
Adduction of Keap1
Table 1, adapted from (Holland et al., 2008) summarizes the current state of knowledge concerning cysteine adduction or oxidation of recombinant Keap1. The first column lists the domains within Keap1. The second column lists the amino acid residues in Keap1's Kelch domain shown by mutation analysis to regulate binding to Nrf2 and/or repression of Nrf2-directed gene expression (Lo et al., 2006). The third column lists Cys residues that have been shown to be adducted. Cys residues list in column 3 that are in bold font have been shown by mutational studies to impact Nrf2 ubiquitination. The remaining columns (4–12) list the various reagents shown to adduct human Keap1 Cys residues. Columns (13–16) list the various reagents shown to adduct murine Keap1. Data concerning the kinetics of reactivity are not presented in this Table. Adduction results are somewhat methodology-based. Further information concerning methodology used to measure adduction of Keap1 can be obtained from the following references (Hong et al., 2005a; Hong et al., 2005b; Eggler et al., 2007; Luo et al., 2007).
Table 1.
Adduction/oxidation of recombinant Keap1, adapted from (Holland et al., 2008) with permission.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Domain | Cys | BMCC | DEX | GSSG | IAB | ISO | Liqustilide | SHO | SULF | XAN | DEM | DEX# | NAPQ1 | 15d-PGJ2 | |
| N-terminus | 12 | X | X | ||||||||||||
| 13 | X* | X | X | ||||||||||||
| 23 | X | X | |||||||||||||
| 38 | X | X | X | X | |||||||||||
| BTB | 77 | X | X | X | X | X | X | ||||||||
| 151 | X | X* | X | X | X | X | X | ||||||||
| 171 | X | ||||||||||||||
| IVR | 196 | X | X | X | X | X | |||||||||
| 226 | X | X | X | X | X | X | X | ||||||||
| 241 | X | X | |||||||||||||
| 249 | X | X | X | ||||||||||||
| 257 | X | X* | X | X | X | X | |||||||||
| 273 | X | X | X | X | X | X | X | ||||||||
| 288 | X | X* | X | X | X | X | X | ||||||||
| 297 | X | X | X* | X | X | ||||||||||
| Kelch | 319 | X | X | X | X | X | X | X | X | ||||||
| Tyr 334 | |||||||||||||||
| 368 | X | X | X | X | X | X | |||||||||
| Arg 380 | |||||||||||||||
| Asn 382 | |||||||||||||||
| 395 | X | ||||||||||||||
| 406 | |||||||||||||||
| Arg 415 | |||||||||||||||
| 434 | X | X | X | X | X | X | |||||||||
| His 436 | |||||||||||||||
| Phe 478 | |||||||||||||||
| Arg 483 | |||||||||||||||
| 489 | X | X | X | X | X | X | X | X | X | X | |||||
| 513 | X | X | X | ||||||||||||
| 518 | X | X | X | ||||||||||||
| Tyr 525 | |||||||||||||||
| Tyr 572 | |||||||||||||||
| C-terminus | 583 | X | X | X | X | ||||||||||
| 613 | X | X* | X | X | X | X | X | X | |||||||
| 622 | X* | X | |||||||||||||
| 624 | X | X |
Human Keap1 denoted in columns identified using red numbers
BMCC: 1-biotinamido-4-(4′-[maleimidiethylcyclohexane]-carboxamido)butane: (Hong et al., 2005b; Luo et al., 2007)
DEX: dexamethasonemesylate adduction to human Keap1: (Liebler, 2006)
GSSG: glutathione disulfide: (Holland et al., 2008)
IAB: N-iodoacetyl-N-biotinylhexylenediamine: (Hong et al., 2005b; Eggler et al., 2007)
ISO: Isoliquiritigenin: (Luo et al., 2007)
Liqustilide: (Dietz et al., 2008)
SHO: 10-Shogaol:(Luo et al., 2007)
SUL: Sulforaphane: (Hong et al., 2005a)
XAN: Xanthohumol: (Luo et al., 2007)
X* IAB addition to Cys residue in vivo
Murine Keap1 denoted in columns identified using blue numbers
DEM:Diethylmaleate: (Kobayashi et al., 2009)
DEX#: dexamethasonemesylate adduction to murine Keap1: (Dinkova-Kostova et al., 2002; Copple et al., 2008)
NAPQ1: N-acetyl-p-benzoquinoneimine: (Copple et al., 2008)
15d-PGJ2: 15-deoxy-Δ12,14-prostaglandin J2:(Copple et al., 2008; Kobayashi et al., 2009)
For the sake of completeness it should be pointed out that 8-nitroguanosine 3',5' – cyclic monophosphate (Sawa et al., 2007), carnosic acid (Satoh et al., 2008), and S-nitrosocysteine (Buckley et al., 2008) have all been shown to adduct Keap1 but specific residues have not been identified.
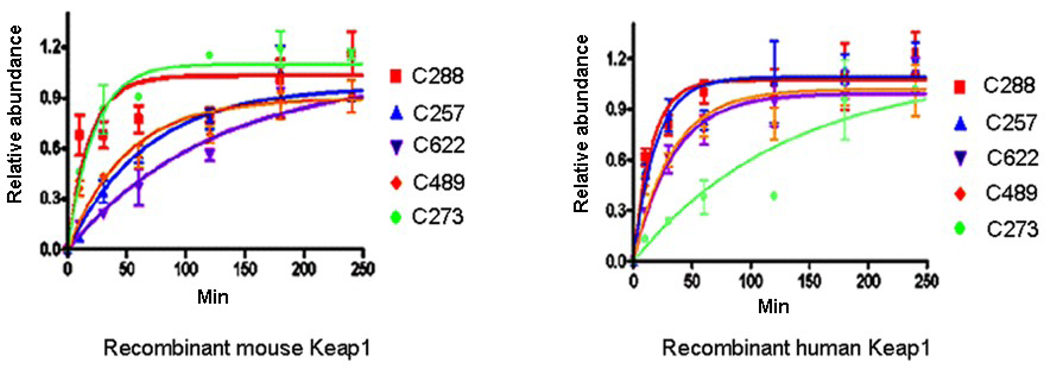
Complicating interpretation is the knowledge that some studies have used human recombinant Keap1 while others have used recombinant murine Keap1. Liebler and colleagues (Xiong, Y., Liebler, D.C., and Freeman, M., unpublished results) performed a head-to-head comparison of adduction kinetics in recombinant human and mouse Keap1 and found significant species-specific differences in reactivity. The most glaring difference relates to adduction at Cys273. As shown in Figure 1, this residue is highly reactive to IAB in murine Keap1. However, in human Keap1 reactivity of Cys273 is significantly diminished.
Figure 1.
Inspection of the Table indicates that Cys 489 in human recombinant Keap1 is adducted by all reagents except GSSG and liqustilide (Dietz et al., 2008; Holland et al., 2008). Cys 489 is located in Strand C of Blade IV, between a conserved Glu (G488) and a conserved Tyr (Y490) (Li et al., 2004). Less work has been done using murine Keap1 and different results have been obtained with regard to adduction at Cys 489. Whereas dexamethasone (DEX) adducts human Keap1 at Cys 489 (Liebler, 2006), it appears not to adduct Cys489 in murine Keap1 (Dinkova-Kostova et al., 2002). Fifteen-deoxy-PGJ2 is a potent Nrf2 inducting agent in human and murine systems (Levonen et al., 2004; Hosoya et al., 2005). Yet, this reagent does not appear to adduct at mKeap1 Cys 489. Nor is there evidence that NAPQ1, another potent Nrf2 inducing agent, adducts at this residue in murine models (Copple et al., 2008). Therefore, the significance of adduction at Cys 489 is not currently understood.
It is clear from Table 1 that the pattern of adduction of Keap1 Cys residues is electrophile dependent. In fact, one can classify adduction patterns into classes, analogous to the classification system developed by Prestera et al., (Prestera et al., 1993). Many of the compounds shown in Table 1 adduct at Cys 151, 273, and or Cys 288. Based on the mutational analysis, it may be expected that adduction at Cys 273 or Cys 288 would inhibit Keap1-directed ubiquitination of Nrf2. Yet the molecular mechanisms that underlie inhibition are not well understood.
Cys151 is a non-conserved amino acid (Furukawa et al., 2003) that resides in the BTB domain of Keap1, the domain that binds CUL3. The core BTB structure consists of 95 amino acid residues that fold into five alpha-helices that end with a three-stranded beta sheet (Perez-Torrado et al., 2006). BTB domains can have N-terminal or C-terminal extensions to the core domain that contribute to dimerization or tetramerization. The observation that a C151S mutation inhibits electrophile and oxidant-mediated disassociation of Keap1 from CUL3 suggests that the BTB fold in Keap1 is intolerant to adduction or oxidation at this residue. Use of recombinant protein systems, as well as ectopically expressed protein in cell culture models have shown that adduction at Cys 151 induces a conformational shift in Keap1's structure that is accompanied by disassociation of Keap1 from CUL3 (Zhang and Hannink, 2003; Rachakonda et al., 2008). The C151S mutation in Keap1 was shown to inhibit adduction-mediated conformational changes, as well as adduction-mediated disassociation of Keap1 from CUL3.
Relevant to this discussion is the question whether recombinant models of adduction mirror the in vivo situation. Rachakonda et al., (Rachakonda et al., 2008) addressed this question using N-iodoacetyl-N-biotinylhexylenediamine (IAB) as a surrogate electrophile. Their results are presented in Table 1. IAB adducted Cys residues in recombinant human Keap1 are denoted by Xs whereas adduction in vivo is denoted by an (*). Cys residues C77, C151, C257, C288, C297, C613, and C622 were found to be adducted in vitro and in vivo. These data show that two key residues, Cys 151 and Cys 288 represent two critical targets in vivo and that their adduction can be modeled using in vitro systems. Furthermore, Rachakonda et al., (Rachakonda et al., 2008) found that both Cys151 and Cys288 were rapidly adducted in vivo.
Kobayashi et al., (Kobayashi et al., 2009) have investigated the role of individual Keap1 Cys residues using an elegant zebrafish model, as well as interrogating recombinant murine Keap1. Using mutation analysis and ectopically expressed protein, they compared the ability of diethyl maleate, 1,2-dithiole-3-thione, sulforaphane, tert-butylhydroquinone, 15-deoxy-Δ12,14 prostaglandin J2, ebselen, 1,2-naphthoquinone, prostaglandin A2, hydrogen peroxide, CdCl2, and auranofin to stabilize Nrf2 and induce Nrf2 target gene expression in the presence of Keap1. The mutation analysis was combined with a MALDI-TOF MS analysis of murine recombinant Keap1 exposed to 15-deoxy-Δ12,14 prostaglandin J2, prostaglandin A2, or DEM. These combined analyzes resulted in the inducers being grouped into 6 classes. Classes 1 and 2 react with Cys 151 (diethyl maleate, 1,2-dithiole-3-thione, sulforaphane, tert-butylhydroquinone, 1,2-naphthoquinone, ebselen). Classes 3 – 6 react with Cys 273 but not C151 (15-deoxy-Δ12,14 prostaglandin J2, ebselen, 1,2-naphthoquinone, prostaglandin A2, hydrogen peroxide, CdCl2, auranofin). Kobayashi et al., (Kobayashi et al., 2009) hypothesize that Keap1 contains at least 2 distinct cysteine motifs that capture specific chemical information and translate it into biochemical information via changes in structural conformation, similar to Yap1 (Georgiou, 2002).
Inspection of Table 1 indicates that neither sulforaphane nor GSSG adducts Cys 151, Cys 273, or Cys 288. GSSG, the oxidized form of glutathione, represents a physiological oxidant that could form either a glutathione-mixed disulfide with Keap1 or generate inter or intramolecular disulfides in Keap1. Although the Table does not present the disulfide data, Holland et al., (Holland et al., 2008) found that GSSG induces disulfide bond formation between Cys residues 23 and 38, as well as residues 257 and 249 in human Keap1. GSSG induces a C319 intermolecular disulfide. Cys 23 has been shown to be mutated in breast cancer (C23Y) and inhibit Keap1-directed ubiquitination of Nrf2 (Nioi and Nguyen, 2007). Thus, Holland et al. (Holland et al., 2008), postulate that disulfide formations that involve Cys 23 have the potential to affect Nrf2 stability. Further work by Holland et al., (Holland et al., 2008) using molecular docking analysis supports a model in which formation of a mixed glutathione disulfide with Cys 434 and 368, which is also adducted by sulphoraphane, would occlude Nrf2 binding to the Kelch domain. This model is consistent with the multiple cysteine sensing motif model of Kobayashi et al., (Kobayashi et al., 2009).
BMCC and IAB exhibit similar adduction profiles (Table 1). BMCC adducts at Cys151, Cys273 and Cys288. Yet, BMCC does not induce Nrf2-directed gene expression in cell culture models (Hong et al., 2005b). It is known that electrophile and oxidant-mediated induction of Nrf2-directed gene expression requires activation of kinase signaling (Yu et al., 2000), as well as, adduction of Keap1 cysteine residues. In addition, Li and Kong (Li and Kong, 2009) have hypothesized that Nrf2 itself may be redox sensor. C183 in Nrf2 is located in a functional nuclear export signal motif. The motif is redox sensitive: oxidants and electrophiles inhibited nuclear export. A C183A mutation attenuated the redox sensitivity of Nrf2 (Li and Kong, 2009). These observations have led to the hypothesis that electrophiles and oxidants modify Nrf2 subcellular localization, in addition to modifying Keap1 function. Thus, one can envision multiple levels of redox regulation: adduction of Keap1 to inhibit Nrf2 ubiquitination, activation of signaling kinases, and modification of Nrf2 that direct its subcellular location.
Summary
Nrf2 is a master transcription factor containing a powerful acidic transcriptional activation domain. Nrf2-dependent gene expression impacts cancer chemoprevention strategies, inflammatory responses, and progression of neurodegenerative diseases. Under basal conditions, association of Nrf2 with Keap1 results in the rapid Nrf2 ubiquitylation and proteasome-dependent degradation. Inhibition of Keap1 function blocks ubiquitylation of Nrf2, allowing newly synthesized Nrf2 to translocate into the nucleus, bind to ARE sites and direct target gene expression. Site-directed mutagenesis experiments coupled with proteomic analysis support a model in which Keap1 contains at least 2 distinct cysteine motifs that capture specific chemical information and translate it into biochemical information via changes in structural conformation.
Acknowledgements
NIH/NCI grant CA104590 and Vanderbilt-Ingram Cancer Center Support Grant P30 CA68485.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Buckley BJ, Li S, Whorton AR. Keap1 modification and nuclear accumulation in response to S-nitrosocysteine. Free Radic Biol Med. 2008;44:692–698. doi: 10.1016/j.freeradbiomed.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple IM, Goldring CE, Jenkins RE, Chia AJ, Randle LE, Hayes JD, Kitteringham NR, Park BK. The hepatotoxic metabolite of acetaminophen directly activates the Keap1-Nrf2 cell defense system. Hepatology. 2008;48:1292–1301. doi: 10.1002/hep.22472. [DOI] [PubMed] [Google Scholar]
- Dietz BM, Liu D, Hagos GK, Yao P, Schinkovitz A, Pro SM, Deng S, Farnsworth NR, Pauli GF, van Breemen RB, Bolton JL. Angelica sinensis and its alkylphthalides induce the detoxification enzyme NAD(P)H: quinone oxidoreductase 1 by alkylating Keap1. Chem Res Toxicol. 2008;21:1939–1948. doi: 10.1021/tx8001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem Res Toxicol. 2007;20:1878–1884. doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau LV, Pickett CB. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J Biol Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou G. How to flip the (redox) switch. Cell. 2002;111:607–610. doi: 10.1016/s0092-8674(02)01165-0. [DOI] [PubMed] [Google Scholar]
- Holland R, Hawkins AE, Eggler AL, Mesecar AD, Fabris D, Fishbein JC. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem Res Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzclaw WD, Dinkova-Kostova AT, Talalay P. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv Enzyme Regul. 2004;44:335–367. doi: 10.1016/j.advenzreg.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005a;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005b;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Maruyama A, Kang MI, Kawatani Y, Shibata T, Uchida K, Warabi E, Noguchi N, Itoh K, Yamamoto M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J Biol Chem. 2005;280:27244–27250. doi: 10.1074/jbc.M502551200. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Jiao D, Conaway CC, Wang MH, Yang CS, Koehl W, Chung FL. Inhibition of N-nitrosodimethylamine demethylase in rat and human liver microsomes by isothiocyanates and their glutathione, L-cysteine, and N-acetyl-L-cysteine conjugates. Chem Res Toxicol. 1996;9:932–938. doi: 10.1021/tx9502094. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- Liebler DC, Hong F, Sekhar KR, Freeman ML. Site-specific modification of teh electrophile sensor protein Keap1 and activation of Nrf2-dependent gene expression. Adv. Mol. Toxicol. 2006;1:55–71. [Google Scholar]
- Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Prestera T, Holtzclaw WD, Zhang Y, Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci U S A. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee NH, Zheng N. A ubiquitin-like protein unleashes ubiquitin ligases. Cell. 2008;135:209–211. doi: 10.1016/j.cell.2008.09.049. [DOI] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- Satoh T, Kosaka K, Itoh K, Kobayashi A, Yamamoto M, Shimojo Y, Kitajima C, Cui J, Kamins J, Okamoto S, Izumi M, Shirasawa T, Lipton SA. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. J Neurochem. 2008;104:1116–1131. doi: 10.1111/j.1471-4159.2007.05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa T, Zaki MH, Okamoto T, Akuta T, Tokutomi Y, Kim-Mitsuyama S, Ihara H, Kobayashi A, Yamamoto M, Fujii S, Arimoto H, Akaike T. Protein S-guanylation by the biological signal 8-nitroguanosine 3',5'-cyclic monophosphate. Nat Chem Biol. 2007;3:727–735. doi: 10.1038/nchembio.2007.33. [DOI] [PubMed] [Google Scholar]
- Sekhar KR, Yan XX, Freeman ML. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene. 2002;21:6829–6834. doi: 10.1038/sj.onc.1205905. [DOI] [PubMed] [Google Scholar]
- Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006a;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006b;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]