Abstract
Oxidative stress plays an important part in the pathogenesis of a variety of diseases. The ability to mount an efficient response against the continuous threat posed by exogenous and endogenous oxidants is essential for cellular homeostasis and survival. Oxidative stress activates transcription of a variety of antioxidant genes through cis-acting sequence known as antioxidant response element (ARE). Members of the Cap-N-Collar family of transcription factors, including Nrf1 and Nrf2, have been identified that bind ARE. Nrf1 and Nrf2 are expressed in a wide range of tissues and cell types, and both bind the ARE as heterodimers with small-Maf proteins. Numerous studies indicate a pivotal role of Nrf2 in ARE function. Herein, we review data derived from cell-based studies and knockout mice in an attempt to define the role and regulation of Nrf1 in oxidative stress response and other functions.
Introduction
Expression of genes encoding antioxidant enzymes is regulated at the transcriptional level through cis-active sequences known as the antioxidant response element (ARE). The nuclear factor erythroid-derived 2-related factor 2 (Nrf2) protein plays a central role in mediating activation through the ARE. Nrf2 belongs to the Cap-N-Collar family of regulatory proteins that also includes Nrf1, Nrf3, and p45NFE2. This review focuses on Nrf1 and highlights some of the knowledge that is beginning to emerge about specific roles of Nrf1 and regulation of Nrf1 function. Although considered originally for its role in beta-globin gene expression in erythroid cells, Nrf1 has been shown to bind the ARE and regulate the expression of a number genes involved in oxidative stress. However, the spectrum of Nrf1-dependent genes has expanded to include genes involved in cellular and tissue differentiation, inflammation, and other cellular processes. Nrf1 knockout studies in mice indicated that it is an essential gene. A role for Nrf1 in maintaining hepatocyte homeostasis and in the pathogenesis of steatohepatitis and cancer was demonstrated by tissue-specific knockout studies. There are two major protein isoforms of Nrf1. Recent evidence indicates that full length Nrf1 protein is targeted to the ER membrane, while the N-terminally deleted Nrf1 variant is constitutively nuclear and functions as dominant negative in ARE-mediated gene activation. Future studies using genetic and biochemical approaches are necessary to provide further understanding about the functions of Nrf1 in other tissues and cell types, and uncover the networks of partner proteins, and target genes that regulate various aspects of stress response and cellular functions.
Structure and Properties of Nrf1
Nrf1 (nuclear factor erythroid-derived 2-related factor 1), which is also known as NFE2L1/LCRF1/TCF11, is a member of the CNC subfamily of basic-leucine zipper (bZIP) transcription factors (CNC-bZIP) (Chan et al., 1993a). CNC-bZIP factors are characterized by a highly conserved 43 amino acid homology region that lies immediately N-terminal to the basic-DNA-binding domain and is referred to as the “CNC” domain after the drosophila cap-n-collar protein. CNC-bZIP factors were isolated as a result to identify regulators of beta-globin gene expression, and this family of proteins in human and mouse include p45 NF-E2, Nrf2, Nrf3, Bach1 and Bach2. Available evidence indicates that CNC-bZIP factors function as obligate heterodimers by forming dimers with small Maf proteins (Maf G, Maf K and Maf F) for DNA binding (Itoh et al., 1995; Johnsen et al., 1996; Johnsen et al., 1998; Kobayashi et al., 1999). In addition to small Maf proteins, a comprehensive analysis using protein arrays to identify bZIP protein interactions indicates ATF/CREB proteins and c-Maf are potential partners of Nrf1 (Newman and Keating, 2003).
Nrf1 maps to chromosome 17q21.3 in human and chromosome 11 in mouse (Chan et al., 1995; McKie et al., 1995). In humans, the Nrf1 gene which spans 15 kb of genomic DNA, and has 9 exons with alternate first exons (1a and 1b) and terminal exons (6 and 6a), and 2 polyadenylation sites (Luna et al., 1994). Alternative first exons, differential splicing, and alternate polyadenylation sites give rise to at least four different transcripts of Nrf1. The full-length human Nrf1 transcript is predicted to code for a protein made up of 772 amino acids, whereas the mouse transcript codes for a 741 amino acid protein (Luna et al., 1994; Luna et al., 1995). The Nrf1 protein is highly conserved throughout evolution, and several different Nrf1 proteins are detected in cells. Western blot experiments show a doublet band at 120-kDa and a smaller band migrating at 65-kDA (Wang et al., 2007). The doublet corresponds to the full-length Nrf1, and the upper band may represent a post-translationally modified form of the protein. The faster migrating band at 65-kDA presumably correspond to a protein that it is produced via alternative translation initiation (Chan et al., 1993b). In support of this is that the Nrf1 open reading frame contains multiple AUG codons downstream of the 5′ initiation codon that are in the context of a Kozak sequence, while the 5′ AUG codon is in a suboptimal context for initiation of protein synthesis. The relative positions of these AUGs in the open reading frame and their overall sequence context are evolutionarily conserved.
The 120-kDA isoform of Nrf1 is localized primarily in the ER as an integral membrane protein, whereas the 65-kDA isoform is nuclear (Wang and Chan, 2006). Studies indicate that endoplasmic reticulum (ER) localization is mediated through the N-terminus region of Nrf1. The deletion of 30 amino acids from the amino-terminus of Nrf1 resulted in nuclear distribution of the protein as determined by both immunofluorescent staining and Western blotting. Examination of the N-terminal protein sequence of Nrf1 revealed a hydrophobic domain between amino acids 7 and 24, and secondary structure analysis of this domain predicts an amphipathic-helical structure that is observed in other membrane-associated proteins. A fusion protein of a heterologous protein plus the amino-terminus of the Nrf1 protein also targeted to the ER. Thus, the amino-terminal amino acids of Nrf1, which contain a putative transmembrane domain, are both necessary and sufficient for targeting to the ER (Wang and Chan, 2006; Zhang et al., 2006). Nrf1 has also been reported to be glycosylated. Thus glycosylation may also play a role in controlling the localization of Nrf1 in the cell (Zhang et al., 2009). These findings raise the issue of how Nrf1 gains access to the nucleus, as well as whether Nrf1 has any functional role in the ER. One possibility is that Nrf1 is sequestered in the ER to mediate stress response emanating from this compartment. In addition to the transmembrane domain, Nrf1 contains a Neh2-like domain located between amino acids 171-244 that shows 72% homology to the Neh2 domain of Nrf2 with complete conservation of the DLG and ETGE motifs important for Keap1 binding (Wang and Chan, 2006). The Neh2 domain of Nrf2 binds Keap1, which regulates Nrf2 stability. Although Nrf1 has also been shown to interact with Keap1, the biologic significance of this interaction remains to be determined.
Gene targets of Nrf1
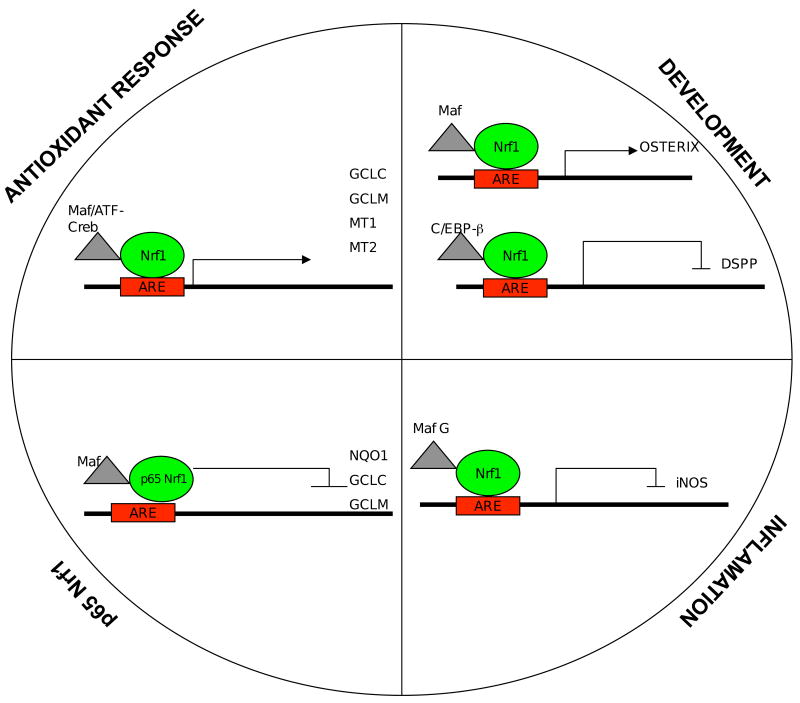
Binding-site selection experiments have shown that Nrf1-MafG heterodimers bind preferentially to a consensus sequence of TGCTGAGTCAT that is identical to the antioxidant response element sequence (Johnsen et al., 1998). The antioxidant response element (ARE), which is also referred to as electrophile responsive element, has been shown to regulate various genes involved in oxidative stress response. Examples of these genes include heme oxygenase, glutathione peroxidase, heavy and light chains of ferritin, and superoxide dismutase that function to protect cells from ROS-mediated injury. In addition, a number of phase II enzymes involved with xenobiotic metabolism are also under the control of the ARE. These include NAD(P)H:quinone oxidoreductase, and various isoforms of glutathione-S-transferases, and UDP-glucoronyltransferases that functions to catalyze the conversion of electrophilic compounds to less toxic and more excretable metabolites in the cell.
Although Nrf2 has been shown to be crucial in activation of genes regulated by the antioxidant response element, involvement of Nrf1 in ARE function has also been demonstrated through transfection studies and gene expression analysis in knockout cells and animals. These targets of Nrf1 include genes encoding enzymes involved in glutathione (GSH) biosynthesis and other oxidative defense enzymes. GSH is a major antioxidant in the cell, and it is an important cofactor for a number of enzymes involved in the metabolism and elimination of endogenous and xenobiotic compounds (Wu et al., 2004). GSH synthesis is catalyzed by two enzymes–gamma-glutamylcysteine ligase, which is composed of a catalytic (GCLC) and a regulatory subunit (GCLM), and glutathione synthetase (GSS). Fibroblasts derived from Nrf1 null embryos showed reduced expression of GCLM and GSS, and Nrf1 has been shown to bind and activate the GCLM promoter (Kwong et al., 1999). Cell-based studies also implicate GCLC and GSS as targets of Nrf1. Enforced expression of Nrf1 in cells upregulates luciferase gene expression driven by GCLC or GSS promoter, and these promoters have been shown to bind Nrf1 by gel-shift and chromatin immunoprecipitation experiments (Myhrstad et al., 2001; Lee et al., 2005). Basal expression of genes including NQO1, ferritin-H, and MT1, was essentially abolished by loss of both Nrf1 and Nrf2 (Leung et al., 2003). However, induction of ARE-regulated genes was significantly blunted in Nrf2-/- compared to Nrf1-/- fibroblasts. These results indicate that Nrf1 and Nrf2 have overlapping roles in regulating basal expression of ARE-containing antioxidant genes, whereas inducible expression is largely dependent on Nrf2. Recent studies suggest that a subset of oxidative stress response genes may be preferentially activated by Nrf1. Metallothionein-1 and -2 (MT-1 and MT-2), which protects cells from heavy metal-induced damage, are down-regulated in Nrf1-deficient livers (Ohtsuji et al., 2008). While both Nrf1 and Nrf2 bind with equal affinity to the ARE in the MT1 gene promoter, only Nrf1 is able to up-regulate reporter gene expression driven by the MT-1 promoter. Interestingly, expression of Nrf2 target genes such NQO1, GSTP1, GCLC and HO-1 is increased in the Nrf1 knockout livers (Ohtsuji et al., 2008).
Aside from antioxidant genes, Nrf1 has been shown to regulate genes involved in development and various cellular processes. Microarray analysis of bones from a mutant mouse strain (sfx) that develops spontaneous fractures has identified osterix as another Nrf1 target gene (Xing et al., 2007). Osterix is a zinc finger transcription factor that plays an important role in the differentiation of osteoblast and bone formation. The promoter region of osterix has a well-conserved ARE that mediates ascorbic acid-induced expression, and is bound by Nrf1 as indicated by gel-shift and chromatin immunoprecipitation assays (Xing et al., 2007).
Nrf1 has also been reported to function as a repressor of transcription. The dentin sialophosphoprotein (DSPP) gene encodes DSP and DPP, which are expressed in terminally differentiated odontoblast. In undifferentiated odontoblast, Nrf1 interacts with C/EBP-β to repress DSPP expression. In fully differentiated odontoblasts, the loss of interaction between C/EBP-β and Nrf1 results in increased expression of DSPP gene (Narayanan et al., 2004). These results suggest that Nrf1 is a negative regulator of odontoblast differentiation. In addition, Nrf1 may also play a role in modulating the inflammatory response. The inducible form of nitric oxide synthase (iNOS) plays a critical role in regulating vascular response in inflammation and injury, and TGF-β has been shown to be a negative regulator of iNOS expression. TGF-β treatment of human smooth muscle cells leads to increased expression and binding of Nrf1/MafG heterodimers to an ARE-like site in the iNOS promoter (Berg et al., 2007). The knockdown of Nrf1 blocks the suppression of iNOS expression by TGF-β. Other potential Nrf1 regulated genes identified by gene expression profiling studies done on Nrf1 deficient mouse livers include glycosylation-related proteins, chaperones, metabolic enzymes, transporters, signal transduction proteins, cell cycle and differentiation regulatory proteins, cytoskeletal organization and nuclear proteins, immunological proteins etc. However, it is yet to be determined whether these genes are direct Nrf1 targets (Ohtsuji et al., 2008).
As described above, there are two distinct isoforms of Nrf1 and they appear to have different abilities to activate gene expression in transient transfection experiments. Enforced expression of the shorter Nrf1 protein suppresses Nrf2 mediated activation of ARE dependent reporter genes in cells, and it also suppresses electrophilic induction of endogenous ARE genes in cells leading to hypersensitivity to oxidative stress (Wang et al., 2007). The ability of p65 to modulate stress response is demonstrated by its ability to inhibit oxidative preconditioning in cells. The shorter isoform of Nrf1 is capable of competing with Nrf2 for interaction with small Maf proteins and binding to ARE in vivo. Together, these findings suggest that the short isoform of Nrf1 act as a dominant negative factor, and raises the interesting possibility that one of its functions in the cell is to modulate oxidative stress response. Given that p65Nrf1 is widely distributed and is abundantly present in the nucleus, p65Nrf1 may serve as another mechanism to limit oxidative stress response mediated by Nrf2, as well as a mechanism to further fine-tune expression of ARE-responsive genes in response to different physiological conditions (Wang et al., 2007). While p65Nrf1 does not appear to activate gene expression, it remains to be determined whether under certain circumstances p65Nrf1 functions as an activator through interactions with other transcription factors or co-activator proteins. Additional studies are required to examine these possibilities.
Physiologic functions of Nrf1
During early stages of development, in situ hybridizations done on 7 to 7.5 post coitum (dpc) mice showed Nrf1 is uniformly expressed in embryonic as well as extra embryonic tissues (Murphy and Kolsto, 2000). At around 9 dpc, there appears to be dorsoventral gradient in expression of Nrf1 with increased levels in the posterior embryo where development is less advanced compared to the anterior part of the animal. At this stage, there is also localized increase in expression of Nrf1 in the heart, presumptive midbrain, head mesenchyme and migrating neural crest cells (Murphy and Kolsto, 2000). In adults, Nrf1 mRNA is present in all tissues examined thus far, but high levels are found in the heart, muscle, liver, kidney and various compartments with high secretory capacity including salivary glands, prostate, and bronchio-epithelium (JYC unpublished data). Various cell-lines including HeLa, HepG2, D551 and MCF7 have been shown to express Nrf1 transcripts. Thus Nrf1 is widely expressed, but physiological functions of the protein are not well understood.
Nrf1 is an essential gene during development. Loss of Nrf1 function in mice results in late gestational embryonic lethality (Chan et al., 1998). Mutant embryos suffer from anemia secondary to abnormal fetal liver erythropoiesis that is non-cell autonomous. Although Nrf1 has been suggested to have a role in globin gene regulation, no defect in expression of globin genes was detected in erythrocytes. Fibroblasts derived from Nrf1 mutant embryos showed enhanced sensitivity to the toxic effects of oxidant compounds and decreased glutathione levels (Kwong et al., 1999). Consistent with this defect, genes involved in glutathione biosynthetic pathway are downregulated in Nrf1 deficient fibroblast. In contrast to the late embryonic lethality in Nrf1 mutants, a combined deficiency in both Nrf1 and Nrf2 leads to lethality between embryonic days 9 and 10 (Leung et al., 2003). Compound mutants exhibit extensive apoptosis that is not observed in the single Nrf1 knockout mutants. Cells derived from compound mutants showed exaggerated oxidative stress and apoptosis under ambient air that can be rescued by culturing under reduced oxygen tension or by addition of antioxidants. Correspondingly, expression of antioxidant defense genes was severely impaired in compound mutant cells compared with single mutant cells. These findings indicate that Nrf1 and Nrf2 have overlapping functions in regulating antioxidant gene expression during early embryogenesis.
However, it is also becoming clear that the functions of Nrf1 and Nrf2 are not entirely redundant. The different phenotypes of Nrf1 and Nrf2 knockouts indicate that Nrf1 must have roles distinct from that of Nrf2. Analyses of genetic chimeras made by injecting normal blastocysts with Nrf1-deficient ES cells indicate that Nrf1 is essential for the hepatocyte lineage. Mouse ES cells deficient in Nrf1 function developed normally and contributed to most tissues in adult chimeras except the liver. While Nrf1 function is dispensable during early stages of liver formation, chimeric livers from late gestational embryos showed severe degeneration (Chen et al., 2003). Consistent with this finding, hepatocyte-specific knockout of Nrf1 leads to high levels of apoptosis in the liver (Xu et al., 2005). Together, these findings indicate a cell-autonomous role for Nrf1 in survival of hepatocytes. Even though there are evidences of impaired antioxidant gene expression, it is not known if cell death is primarily caused by oxidative stress in chimeras and liver-specific knockouts.
Loss of Nrf1 function in hepatocytes also leads to steatohepatitis and hepatic neoplasia, which is not observed in Nrf2 knockouts (Xu et al., 2005). The underlying mechanisms for these pathologies, and whether Nrf1 functions as a tumor suppressor in hepatocytes remain to be determined. Proliferation of smooth endoplasmic reticulum and induction of microsomal genes indicates activation of fatty acid oxidation pathways. However, it is not known whether Nrf1 directly regulates genes involved in maintaining lipid homeostasis in the liver. Although inflammation may be secondary to increased cell death in the knockouts, it is interesting in this regard that Nrf1 has been implicated as a negative regulator of inducible nitric oxide synthase (iNOS) expression. As iNOS plays a role in inflammatory responses by increasing the production of nitric oxide, it is possible that inflammation in Nrf1 liver knockouts occurs in part as a result of dysregulated production of nitric oxide through sustained activation of iNOS. Regardless of the mechanisms, steatosis is likely to contribute to oxidative stress as a result of increased ROS production from metabolism of fatty acid, and inflammation is also an important source of ROS. In addition, steatosis can also cause direct toxicity to cells and induce apoptosis. Together, these defects provide a synergistic combination in promoting apoptosis and cell proliferation that contributes to neoplastic development in the knockouts.
Nrf1 also plays a role in bone development
Osteoblast-specific Nrf1 knockout mice have reduced bone mineral content and bone area (S. Mohan and JYC unpublished data). The mechanism for impaired bone formation in these animals may be related to the reduction in osterix levels, which is a target gene involved in bone differentiation that's activated by Nrf1 (Xing et al., 2007). Nrf1 has also been implicated in neuronal survival after acute brain injury. An up-regulation of Nrf1 and HO-1 expression is observed in the hippocampus of mice injected with the excitioxin, kainic acid (Hertel et al., 2002). Whether Nrf1 plays a role in protecting the brain against oxidative stress remains to be determined.
Concluding Remarks
Nrf1 and Nrf2 are members of the CNC-bZIP family of transcription factors that share similar amino acid sequences and patterns of tissue expression. While the field of Nrf2 in stress response has been evolving rapidly, our understanding of Nrf1 in this pathway is not as extensive. As outlined above, genetic and biochemical evidence support a partially overlapping role between Nrf1 and Nrf2 in regulating ARE-driven genes in the oxidative stress response in cells. However, given the divergent phenotypes exhibited by knockouts of Nrf1 and Nrf2, Nrf1 is likely to have functions separate from Nrf2. Thus one important unresolved issue is to establish these unique functions related to cell survival, hepatic function, oncogenesis and development. In addition, it is still unclear whether Nrf1 possess unique functions in the context of stress response. This will require identification of unique target genes, and a comparison of the repertoires of genes regulated by each protein under different conditions. The molecular mechanisms responsible for the fundamental differences in the transcriptional activities of Nrf1 and Nrf2 have yet to be determined. In addition, Nrf1 can function as a transcriptional activator, as well as a repressor. Defining the coactivators, corepressors and binding partners of Nrf1 should shed light on this network of gene regulation. Another important unresolved issue is to establish the relationship between the different Nrf1 isoforms. Apart from the liver, Nrf1 is also highly expressed in several other tissue compartments in the body. The development of additional tissue-specific mouse mutants should facilitate our understanding of its physiologic functions. These additional studies should reveal exciting aspects regarding the function and regulation of Nrf1.
Fig 1. Gene Targets of Nrf1.
Details are provided in the text.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berg DT, Gupta A, Richardson MA, O'Brien LA, Calnek D, Grinnell BW. Negative regulation of inducible nitric-oxide synthase expression mediated through transforming growth factor-beta-dependent modulation of transcription factor TCF11. J Biol Chem. 2007;282:36837–36844. doi: 10.1074/jbc.M706909200. [DOI] [PubMed] [Google Scholar]
- Chan JY, Cheung MC, Moi P, Chan K, Kan YW. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum Genet. 1995;95:265–269. doi: 10.1007/BF00225191. [DOI] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A. 1993a;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. Isolation of cDNA encoding the human NF-E2 protein. Proc Natl Acad Sci USA. 1993b;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY. Nrf1 is critical for redox balance and survival of liver cells during development. Mol Cell Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel M, Braun S, Durka S, Alzheimer C, Werner S. Upregulation and activation of the Nrf-1 transcription factor in the lesioned hippocampus. Eur J Neurosci. 2002;15:1707–1711. doi: 10.1046/j.1460-9568.2002.01992.x. [DOI] [PubMed] [Google Scholar]
- Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen O, Skammelsrud N, Luna L, Nishizawa M, Prydz H, Kolsto AB. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996;24:4289–4297. doi: 10.1093/nar/24.21.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap‘n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- Kwong M, Kan YW, Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J Biol Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- Lee TD, Yang H, Whang J, Lu SC. Cloning and characterization of the human glutathione synthetase 5′-flanking region. Biochem J. 2005;390:521–528. doi: 10.1042/BJ20050439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- Luna L, Johnsen O, Skartlien AH, Pedeutour F, Turc-Carel C, Prydz H, Kolsto AB. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics. 1994;22:553–562. doi: 10.1006/geno.1994.1428. [DOI] [PubMed] [Google Scholar]
- Luna L, Skammelsrud N, Johnsen O, Abel KJ, Weber BL, Prydz H, Kolsto AB. Structural organization and mapping of the human TCF11 gene. Genomics. 1995;27:237–244. doi: 10.1006/geno.1995.1037. [DOI] [PubMed] [Google Scholar]
- Marini MG, Chan K, Casula L, Kan YW, Cao A, Moi P. hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2. J Biol Chem. 1997;272:16490–16497. doi: 10.1074/jbc.272.26.16490. [DOI] [PubMed] [Google Scholar]
- McKie J, Johnstone K, Mattel MG, Scambler P. Cloning and mapping of murine Nfe211. Genomics. 1995;25:716–719. doi: 10.1016/0888-7543(95)80015-e. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap‘n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Murphy P, Kolsto A. Expression of the bZIP transcription factor TCF11 and its potential dimerization partners during development. Mech Dev. 2000;97:141–148. doi: 10.1016/s0925-4773(00)00413-5. [DOI] [PubMed] [Google Scholar]
- Myhrstad MC, Husberg C, Murphy P, Nordstrom O, Blomhoff R, Moskaug JO, Kolsto AB. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochim Biophys Acta. 2001;1517:212–219. doi: 10.1016/s0167-4781(00)00276-1. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Ramachandran A, Peterson MC, Hao J, Kolsto AB, Friedman AD, George A. The CCAAT enhancer-binding protein (C/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J Biol Chem. 2004;279:45423–45432. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- Wang W, Kwok AM, Chan JY. The p65 isoform of Nrfl is a dominant negative inhibitor of ARE-mediated transcription. J Biol Chem. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J Biol Chem. 2007;282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci USA. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. Nrfl and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1. Mol Cell Biol. 2005;25:5933–5946. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrfl transcription factor by its N-terminal domain is independent of Keap1: Nrfl, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lucocq JM, Hayes JD. The Nrfl CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem J. 2009;418:293–310. doi: 10.1042/BJ20081575. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lucocq JM, Yamamoto M, Hayes JD. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrfl is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem J. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]