Abstract
Quinolinic acid phosphoribosyltransferase (QAPRTase, EC 2.4.2.19) forms nicotinate mononucleotide (NAMN) from quinolinic acid (QA) and 5-phosphoribosyl 1-pyrophosphate (PRPP). Previously determined crystal structures of QAPRTase•QA and QAPRTase•PA•PRPP complexes show positively charged residues (Arg118, Arg152, Arg175, Lys185 and His188) lining the QA binding site. To assess the roles of these residues in the Salmonella typhimurium QAPRTase reaction, they were individually mutated to alanine and the recombinant proteins overexpressed and purified from a recombineered E. coli strain that lacks the QAPRTase gene. Gel filtration indicated that the mutations did not affect the dimeric aggregation state of the enzymes. Arg175 is critical for the QAPRTase reaction, and its mutation to alanine produced an inactive enzyme. The kcat values for R152A and K185A were reduced by 33-fold and 625-fold, and binding affinity of PRPP and QA to the enzymes decreased. R152A and K185A mutants displayed 116-fold and 83-fold increases in activity towards the normally inactive QA analog, nicotinic acid (NA), indicating roles for these residues in defining the substrate specificity of QAPRTase. Moreover, K185A QAPRTase displayed a 300-fold higher kcat/Km for NA over the natural substrate QA. Pre-steady state analysis of K185A with QA revealed a burst of nucleotide formation followed by a slower steady-state rate, unlike the linear kinetics of WT. Intriguingly, pre-steady state analysis of K185A with NA produced a rapid but linear rate for NAMN formation. The result implies a critical role for Lys185 in the chemistry of the QAPRTase intermediate. Arg118 is an essential residue that reaches across the dimer interface. Mutation of Arg118 to alanine resulted in 5000-fold decrease in kcat value, and a decrease in the binding affinity of QA and PRPP to R152A. Equimolar mixtures of R118A with inactive or virtually inactive mutants produced approximately 50% of the enzymatic activity of WT, establishing an interfacial role for Arg118 during catalysis.
Quinolinate phosphoribosyltransferase (QAPRTase, EC 2.4.2.19) is a type II phosphoribosyltransferase (PRTase) that participates in the de novo biosynthesis of the pyridine coenzyme, NAD (1, 2). Recently, nicotinate phosphoribosyltransferase (NAPRTase) and nicotinamide phosphoribosyltransferase (NMPRTase), involved in the salvage pathways of NAD biosynthesis, have been classified as type II PRTases (3–5). The type II PRTases catalyze the transfer of a phosphoribosyl moiety from 5-phosphoribosyl-1-pyrophosphate (PRPP) to their specific substrates. The phosphoribosyl transfer reaction catalyzed by QAPRTase is linked to an irreversible decarboxylation reaction at position 2 of the quinolinic acid (QA) ring, with no cofactor requirement (2, 6).
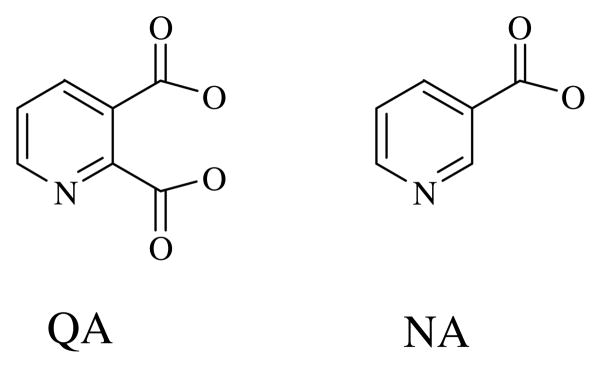
The nadC gene of Salmonella typhimurium which encodes QAPRTase is one of the three nonessential genes involved in de novo NAD biosynthesis (7). Unlike the other two genes (nadA (L-aspartate oxidase) and nadB (quinolinate synthase)), which are known to be regulated by the NadI repressor, the nadC gene is not genetically controlled (8). However, QAPRTase is specific for QA and cannot use its analog nicotinic acid (NA). QA differs from NA by the presence of the negatively charged carboxylate group at position 2 of the molecule (Figure 1). In the salvage pathway of NAD biosynthesis, NAPRTase converts NA to NAMN. Like QAPRTase, NAPRTase shows a high degree of specificity for its substrate, as indicated in genetic and enzymological studies (9, 10). Appropriate bacterial strains carrying nadC (to block QAPRTase activity) or pncB mutations (to block NAPRTase) show auxotrophies that are not relieved by NA and QA respectively (9). Additionally, NA does not detectably inhibit QAPRTase, whereas several pyridine analogs with negative charges at the 2 position have been shown to be effective inhibitors of the enzyme (11, 12). Although crystal structures are available for all the type II PRTases, it remains unknown how the enzymes discriminate among their respective substrates.
Figure 1.
Chemical structures of quinolinate and nicotinate.
The reaction catalyzed by QAPRTase follows an ordered-sequential mechanism, in which QA binds first, followed by PRPP, releasing NAMN and then PPi (13). The phosphoribosyl transfer reaction that forms the putative quinolinate mononucleotide (QAMN) intermediate is thought to precede the decarboxylation of the QAMN to form NAMN (14; a recent theoretical study (15) has reviewed this idea). QAMN has never been isolated or synthesized, and its chemical stability and properties are unknown. Thus, it is not clear if the decarboxylation is enzymatic or spontaneous. Based on inhibition studies, the putative QAMN intermediate was proposed to decarboxylate via an ylide mechanism (12, 16–17). The ylide mechanism has also been proposed for the decarboxylation of orotidine 5′-monophosphate by OMP decarboxylase, which, like QAPRTase, is an α/β barrel enzyme, and has no cofactor requirement (18). Among cofactorless decarboxylases, a lysine amino group was recently proposed to act as the initial carboxyl receptor in a bacterial oxaloacetate decarboxylase (19). Studies with model compounds showed that non-enzymatic decarboxylation of pyridine 2-carboxylic acids substituted with a carboxyl group at position 3 occurred spontaneously (16, 17). However, the rate of the QAPRTase reaction is 5 orders of magnitude faster than the spontaneous decarboxylation of the model compounds. QAPRTase may thus be involved in the decarboxylation step. Positively charged residues in the actives site of the enzyme might directly stabilize the negative charge at the 2-position of QA after decarboxylation of the putative QAMN intermediate (2, 6).
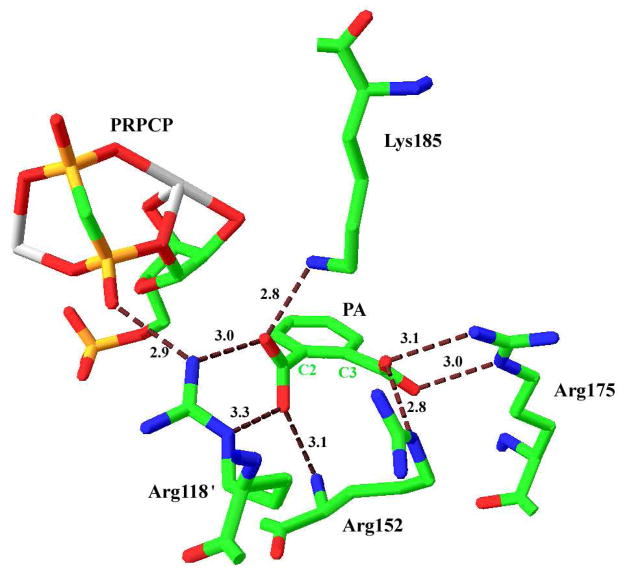
Crystal structures of substrate-bound complexes of QAPRTase from various organisms provide insights into the structure-function relationship of the enzyme (2, 6, 20–22). The active site of the enzyme is located at the C-terminal α/β-barrel domain of one subunit, which is bordered by the N-terminal domain of the adjacent subunit, forming a head-to-tail, domain-swapped dimer (2). In the crystal structure of S. typhimurium QAPRTase, the QA binding site (Figure 2) is composed of the following positively charged residues; Arg118′ (sequence numbering is based on the mature Salmonella enzyme; the prime designates a residue from the adjacent subunit), Arg152, Arg175, Lys185 and His188. One of the oxygen atoms of the 2-carboxylate of PA (Figure 2) forms hydrogen bonds with the amino nitrogen of Lys185 (2.8 Å) and it is 3 Å away from NH1 of Arg118′. Arg118′ is involved in an extensive hydrogen bonding. The Mycobacterium tuberculosis QAPRTase•PA•PRPPC ternary analog complex shown here demonstrated that in addition to interacting with QA, NH1 of Arg118′ is within hydrogen-bonding distance of the α-phosphate of PRPP (6). The second oxygen atom of the 2-carboxylate of PA is 3.3 Å away from the Nε of Arg118′, and forms a hydrogen bond with the mainchain N of Arg152. These residues may be important for binding of QA or exclusion of NA. Additionally, the negative charge at the 2-position of the QA ring after decarboxylation of QAMN intermediate might be stabilized indirectly, by inductive electron withdrawal through interactions of Arg152 and Arg175 (2, 6). Arg175 makes a bidentate hydrogen bond with the oxygen atoms of the C3 carboxylate of PA, while the Nε of Arg152 is seen to interact with one of the oxygen atoms of PA. His188 (not shown) is also in the wall of the active site, but its closest approach to QA (via the C2 carboxylate) is 8.3 Å.
Figure 2.
Binding site for QA in the M. tuberculosis QAPRTase•PA•PRPCP•Mn2+ Michaelis complex (PDB entry 1QPR). Residues that interact with QA are shown and their hydrogen-bonding distances are represented in Å. The primes on Arg118′ designates residue from the adjacent subunit. C2 and C3 are designations of the homologous positions on QA. The Two Mn2+ are coordinated by PRPC are shown in gray. This figure was generated using SwissPDB Viewer (23).
The origin of the striking base specificity of QAPRTase remains a major issue. It is easiest to imagine that NA simply fails to form the panoply of hydrogen bonds that QA can. In line with this, NA is a particularly poor inhibitor of QAPRTase (11). That poor binding may extend to poor catalysis; in particular a dual interaction of Arg118′ with both the 2-carboxylate of QA and the pyrophosphate portion of PRPP may allow QA to orient for catalysis, whereas NA may not. Alternatives would include the proposal that NA may be able to undergo weak and nonproductive “wrong way” binding in which the C3 carboxylate occupies the site normally bound by the 2-carboxylate of QA, with the N1 of NA now occupying the site normally occupied by the C4 of QA. In thymidylate synthase, exclusion of dCMP from the dUMP site appeared to arise by steric exclusion mediated by Asn229, but study of indiscriminant D229N mutants suggests a more complex mechanism including rearrangement of active site residues. The potential mobility of Lys185, and its somewhat “kinked” sidechain conformation, makes a role for exclusion of NA a possibility. However, examination of structures does not seem to support an alternative position of the Lys185 sidechain that blocks access to the QA site by NA.
The positively charged residues that line the QA binding site are thus base binding residues, candidates for base specificity and might act directly or indirectly stabilize the QAMN intermediate. In a recent characterization of the human QAPRTase the homologous residues were identified and preliminary characterization of mutants was reported (21). In this paper, the residues were mutated individually to alanine, in order to explore their contribution to catalysis. The kinetic characterization of the mutant enzymes with respect to WT is presented, allowing conclusions about the origin of the base specificity of the enzyme.
Materials and Methods
Materials
Materials used have been previously described (24). SDS-PAGE (14–20%) gels were from Lonza Biosciences. [14C]NA was obtained from Sigma.
Over-expression and Purification of Enzymes
Construction of ZB100, the deleted nadC strain of BL21(DE3) has been described (24). The QuikChange site-directed mutagenesis kit (Stratagene) was used to carry out the mutations. The following primers were used to construct the mutants: R118A (forward, G CTG ACC GGC GAG GCG ACG GCG CTA AAC TTT GTC; reverse, GAC AAA GTT TAG CGC CGT CGC CTC GCC GGT CAG C), R152A (forward, C CAG TTG CTC GAC ACG GCG AAA ACG CTG CCG GGT C; reverse G ACC CGG CAG CGT TTT CGC CGT GTC GAG CAA CTG G), K153A (forward CAG TTG CTC GAC ACG CGT GCG ACG CTG CCG GGT CTG CGC; reverse GCG CAG ACC CGG CAG CGT CGC ACG CGT GTC GAG CAA CTG), R175A (forward, C GGC GGC GCC AAT CAT GCG CTG GGC CTC ACT GAC G; reverse, C GTC AGT GAG GCC CAG CGC ATG ATT GGC GCC GCC G), K185A (forward, CTC ACT GAC GCG TTC CTG ATT GCG GAA AAC CAT ATT ATC GCC TCC; reverse, GGA GGC GAT AAT ATG GTT TTC CGC AAT CAG GAA CGC GTC AGT GAG) and H188A (forward, C GCG TTC CTG ATT AAA GAA AAC GCG ATT ATC GCC TCC GGT TCG GTT C; reverse, G AAC CGA ACC GGA GGC GAT AAT CGC GTT TTC TTT AAT CAG GAA CGC G) with the mutated codon in bold. The ZB100 strain used for protein expression was transformed with the mutant plasmids following standard procedure. The mutant enzymes were over-expressed and purified as previously described (24).
Characterization of Mutant QAPRTases
The aggregation state, equilibrium binding of [14C]PRPP and [3H]QA and QAPRTase activity of WT and mutants enzyme forms was assayed as described (24).
Pre-steady State Measurements
Pre-steady state analysis for the formation of [3H]NAMN and [14C]NAMN using [3H]QA and [14C]NA as substrates, respectively, were carried out. Rapid quench experiments were carried out at 25°C in buffer QA using RQF-3 Chemical-Quench-Flow apparatus (KinTek Instruments). Reactions were initiated when 15 μL enzyme (80 μM) pre-equilibrated with 2 mM [3H]QA or [14C]NA was mixed with 15 μL PRPP (8 mM) in buffer QA. Reaction times ranged from 1 s to 10 min. Reactions were quenched with 2 M HCl and collected in 1.5 ml Eppendorf tubes. After all the samples were collected, the tubes were centrifuged at 3000 x g for 2 min to remove the denatured protein. Aliquots of 10 μL of the quenched sample were spotted and chromatographed on 3MM Whatman paper with a 4:6 mixture of 1M ammonium acetate and 95% ethanol. Under the given conditions, QA and NAMN have Rf values of 0.25 and 0.4, respectively. The [3H]QA and [3H]NAMN were quantitated as described previously (24). Control experiments in the absence of enzyme in which the reaction was quenched with HCl determined the background activity. Formation of NAMN by R118A and K185A using [3H]QA were performed by manual mixing. The data for the product formed by the reaction of K185A with [3H]QA was fitted to the burst equation 1 (25).
| (1) |
where A is the burst amplitude, k1 the pre-steady-state rate constant for product formation, t, time, and kss the steady-state rate of product formation.
Alternative Substrates for Mutant Enzymes
2,4-pyridinedicarboxylic acid, 2,5-pyridinedicarboxylic acid, 2,6-pyridinedicarboxylic acid, 3,4-pyridinedicarboxylic acid, 3,5-pyridinedicarboxylic acid, nicotinamide, pyridine-4-carboxylic acid, 6-aminonicotinic acid and NA were incubated individually, with WT and mutant enzymes under standard radiolabel assay conditions (24). Reaction products were visualized under 254 nm UV light after chromatographic separation of products from the transfer assay. The reaction mixture contained 500 μg of mutant enzyme, 0.5 mM of alternative substrate and 2mM PRPP in a 200 μL reaction volume. The reaction mixtures were incubated for 3 h at 30°C, inactivated with 2M HCl and centrifuged for 5 min (14 800 × g) in a microcentrifuge. The supernatant (25 μL) was applied to 3MM Whatman paper and developed as described (24). The formation of [14C]NAMN by WT and R152A using [14C]NA as a substrate was measured by the radiolabel transfer assay.
Restoration of Activities of by Mutant Enzymes by R118A Complementation
Heterodimers of R118A with mutant enzymes (R152A, K153A, R175A and K185A) were formed individually by mixing equal concentrations (2 mg/mL) of the enzymes in buffer QA containing 1 mM DTT, and incubating them for 3 h in ice. The enzymatic activities of the heterodimers were determined by the standard spectrophotometric assay.
RESULTS
The Hydrogen Bond Network in the QA Binding Site
The cationic nature of the quinolinate binding sites of all known QAPRTases has been noted (2, 6, 21–22). Close examination of X-ray structures reveals an intricate web of deduced hydrogen bonding, exemplified by the structure of the Mycobacterium ternary substrates analog complex in Figure 2, which is similar to a recent structure of the yeast enzyme (22; bond lengths are for the complex shown here, and vary somewhat with other complexes, analogs and species). The sidechain of Lys185 forms a 2.8 Å hydrogen bond to one C2 carboxylate oxygen, which also interacts (2.95 Å) with NH1 of Arg118′. Arg118 also interacts through its NE with the other C2 carboxylate oxygen (3.3 Å) and, via NH1, with an α-phosphate oxygen of PRPP (2.9 Å). The C2 carboxylate forms a fourth deduced hydrogen bond (3.1 Å) with Arg152 through the mainchain N of the latter. At the C3 carboxylate, three apparent hydrogen bonds, from NE of Arg152 (2.8 Å), and from NE (3.0 Å) and NH1 (3.1 Å) are formed. Thus, each of the two carboxylates and each of the Arg residues makes multidentate hydrogen bonds that could serve to read substrate identity. Lys185, which forms a single hydrogen bond, appears more free to move, and a recent structure of the binary complex of QA with the yeast enzyme shows Lys185 interacting via a water molecule with a C3 carboxylate oxygen (22).
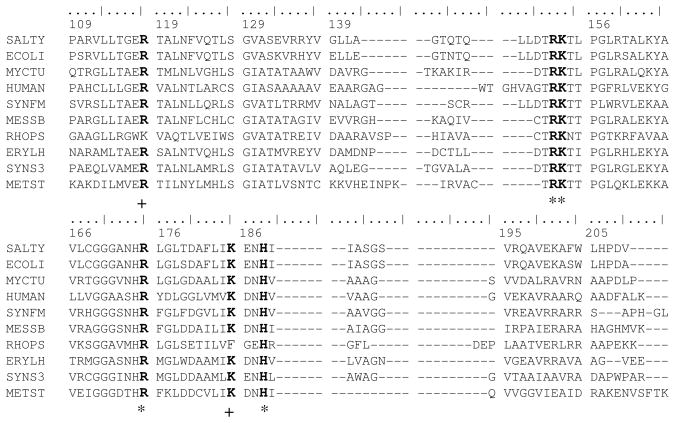
Examination of a large sequence library (Materials and Methods) showed that the residues lining the QA site were highly conserved (Figure 3). Arg152, Lys153, Arg175 and His188 were all absolutely conserved. Arg118 and Lys185 were conserved, except for replacement by Lys and Phe, respectively in Rhodopseudomonas palustris. The extreme sequence conservation is in keeping with important functional roles.
Figure 3.
Partial sequence alignment of QAPRTase from various organisms. 10 of the 230 sequences aligned as described in Methods are displayed. Sequences are: SALTY, Salmonella typhimurium LT2; ECOLI, Escherichia coli K-12; MYCTU, Mycobacterium tuberculosis H37Rv; HUMAN, Homo sapiens; SYNFM, Syntrophobacter fumaroxidans; MESSB, Mesorhizobium sp. BNC1; RHOPS, Rhodopseudomonas palustris BisB5; ERYLH, Erythrobacter litoralis; SYNS3, Synechococcus sp. CC9311; METST, Methanosphaera stadtmanae. Residue numbering at top is from the mature S. typhimurium protein sequence. Conserved residues are indicated in bold, absolutely conserved residues are marked with *, and highly conserved residues are marked with + underneath the alignments.
Expression and Purification of Mutant QAPRTases
The over-expression and purification of the mutant enzymes produced 200–250 mg of protein per 6 L of culture, and were similar to that of the WT enzyme. The mutant enzymes appeared homogeneous as assessed by SDS-PAGE.
Aggregation State of Mutant QAPRTases
Gel filtration of the mutant enzymes under native conditions was performed on a Superdex 200 column. All mutant enzymes eluted at the same position as WT (data not shown), with similar peak-widths at half height, indicating that the enzymes were stably dimeric with molecular mass of 65 kDa, as predicted for WT (64 856 (26)).
Steady-state Kinetic Measurement for Mutants
Mutation of the residues lining the QA binding site had large impacts on catalytic activity. The steady-state kinetic constants for WT and mutant enzymes are summarized in Table 1. The mutation of Arg118 diminished kcat/Km,PRPP for R118A by 281 000-fold compared to WT, indicating the importance of Arg118 in catalysis. This reduction was a result of a 5 000-fold lower kcat value and approximately 55-fold elevated Km,PRPP value. The kcat/Km,QA for R118A is reduced by 25 000-fold, due to the 5 000-fold reduction in kcat and a 5-fold increase in Km,QA value.
Table 1.
Steady state kinetic parameters for WT and mutant QAPRTases
| kinetic constant | WT | R118A | K185A | R152A | H188A |
|---|---|---|---|---|---|
| kcat, (s−1) | 1.0 ± 0.10 | 2.4 × 10−4 | 1.6 × 10−3 | 0.03 | 1.0 ± 0.02 |
| KM, QA (μM) | 27 ± 6 | 136 ± 20 | 1478 ± 441 | 278 ± 23 | 67 ± 5 |
| kcat/KM, QA (μM−1 s−1) | 3.7 × 10−2 | 1.5 × 10−6 | 1.1 × 10−6 | 1.0 × 10−4 | 1.6 × 10−2 |
| KM, PRPP (μM) | 57 ± 7 | 3138 ± 800 | 2181 ± 500 | 832 ± 180 | 547 ± 23 |
| kcat/KM, PRPP (μM−1 s−1) | 1.8 × 10−2 | 6.4 × 10−8 | 7.3 × 10−7 | 3.5 × 10−5 | 1.8 × 10−3 |
| KD, QA (μM) | 25 ± 7 | 128 ± 14 | ND | 261 ± 38 | 90 ± 15 |
| N | 2.0 ± 0.1 | 1.9 ± 0.1 | 1.8 ± 0.3 | ||
| KD,PRPP (μM) | 53 ± 11 | ND | ND | ~ 1000 + | 480 ± 75 |
| n = 1.7 ± 0.3 |
ND, not detected
Lys185 and Arg152, which interact with the 2-carboxylate and 3-carboxylate of QA, respectively, gave, as expected, decreases in activity for their mutations. Removal of the side-chain of Lys185 resulted in a 625-fold decrease in kcat, with a 38-fold higher Km,PRPP and a 55-fold increase in Km,QA causing 34 000-fold decrease in kcat/Km,QA and 25 000-fold reduction in kcat/Km,PRPP value. For R152A, kcat decreased by 33-fold, with a 15-fold increase in Km,PRPP and 10-fold increase in Km,QA leading to a 500-fold and 370-fold reduction in kcat/Km,PRPP and kcat/Km,QA, respectively.
No measurable activity was detected for R175A, indicating that Arg175 is essential for catalysis. The undetectable activity of R175A like that of K153A (24) suggests that activities of other extremely sluggish mutants described here and in the preceding paper (24) are intrinsic to those mutant enzymes and not a result of contamination by active enzymes.
Mutation of the absolutely conserved His188 to alanine did not affect the ability of the enzyme to catalyze the QAPRTase reaction, as the kcat was similar to that of WT. The Km,PRPP value increased by 10-fold, leading to a 10-fold reduction in kcat/Km,PRPP H188A. With the exception of His188, the steady state results indicate that all the residues at the QA binding site of QAPRTase are important for catalysis.
Binding of PRPP and QA to the Mutant Enzymes
The effect on formation of binary complexes of mutant QAPRTase was determined by equilibrium binding experiments with [3H]QA and [14C]PRPP by centrifugal dialysis (Table 1). Surprisingly, a reduced binding affinity for PRPP was observed for all the mutant enzymes, though most of their mutated residues do not directly interact with PRPP as demonstrated from the crystal structures. A 10-fold increase was measured for KD,PRPP for H188A, in agreement with its Km value. The KD,PRPP for R152A was approximately 1mM, almost 18-fold higher than WT and in agreement with its Km,PRPP value. The binding of [14C]PRPP to R118A, R175A and K185A mutant enzymes was not detected by centrifugal dialysis, indicating KD,PRPP values that are greater than 1mM.
The binding of PRPP to the mutants in the presence of the QA analog, PA, which tightens binding of PRPP in WT, had no effect on the KD values of PRPP (Data not shown).
The KD value of QA for R152A was 260 μM, 10-fold higher than that of WT and consistent with its Km value. For the binding of [3H]QA to R118A, the KD value measured is represented by Scatchard analysis shown in Fig 4. Consistent with the Km,QA, a 5-fold higher KD value for QA was measured for R118A. Binding of [3H]QA was not detected for R175A and K185A mutant enzymes, indicating KD values greater than 1 mM.
Figure 4.
Scatchard plot for binding of R118A to [3H]QA. The value of KD was 128 μM, with an n value of 1.9 mol of [3H]QA/mol QAPRTase dimer.
Alternative substrates
Alternative substrates were employed to test whether there was a change in base specificity of mutant QAPRTase. To assess the contribution of Arg152 and Lys185 in substrate discrimination, the ability of their respective alanine mutants to carry out the QAPRTase reaction was tested using various pyridine analogs. With the exception of NA and 6-aminonicotinic acid, all other QA analogs investigated were not detectably utilized by the mutant enzymes. Formation of NAMN by WT enzyme using NA as a substrate was detected by the radiolabel transfer assay. The specific activity of WT with NA at 1 mM and 2 mM PRPP was 0.0006 Umg−1. This rate represents a 2000-fold reduction in comparison to its activity of 1.2 Umg−1 with 1 mM QA as the substrate. The specificity of R152A for QA and NA is more relaxed than that of WT. A specific activity of 0.07 Umg−1 was measured for the reaction of R152A with NA versus 0.1 Umg−1 using the natural QA substrate. R152A uses NA 116-fold better than WT does. Detailed steady state kinetic analysis for WT or R152A with NA was not performed.
K185A exhibited an 83-fold higher specific activity for its ability to utilize NA than the WT enzyme did with NA as a substrate. Remarkably, K185A showed a strong preference for NA over QA. Its activity with NA was sufficient to allow measurement of kinetic parameters (Table 2) by the spectrophotometric assay. A 31-fold elevation was observed in kcat,NA compared to the kcat,QA value for the mutant enzyme. The Km,NA value for K185A decreased by 10-fold and Km,PRPP(NA) decreased by 6-fold in comparison to the respective Km values with the natural QA substrate. The values of kcat/Km,NA and kcat/Km,PRPP(NA) are greater by 300-fold and 178-fold, respectively, in comparison to the values with QA as the substrate. K185A was the only mutant to detectably utilize 6-aminonicotinic acid as a substrate.
Table 2.
Comparison of kinetic parameters of K185A using NA as an alternate substrate
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| QA | 1.6 × 10−3 | 1478 ± 441 | 1.1 × 10−6 |
| NA | 4.8 × 10−2 | 151 ± 13 | 3.2 × 10−4 |
Pre-steady State Kinetics
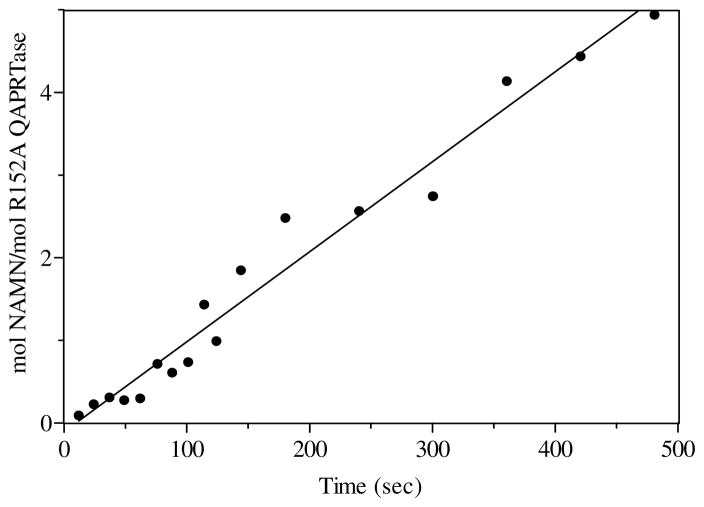
It has previously been shown that the chemistry step in the QAPRTase reaction is rate limiting as evidenced from the linear rate in NAMN formation in the pre-steady state (13, 24). The pre-steady state rate of NAMN formation by R118A using QA was measured by the manually quenched radiolabel transfer method. The initial rate of NAMN formation was linear, at 2 × 10−5 s−1, consistent with the steady state kcat value (2.4 × 10−4 s−1). For R152A using QA, the initial rate of NAMN formation was also linear at 0.1 s−1, consistent with its steady state kcat value (Figure 5).
Figure 5.
Pre-steady state kinetics of [3H]NAMN formation by R152A with [3H]QA. Rapid mixing was performed in a Kintek Chemical-Quench-Flow apparatus as described in Materials and Methods. The solid line represents a linear least square fit to the data points. The kcat of the reaction was 0.1 s−1.
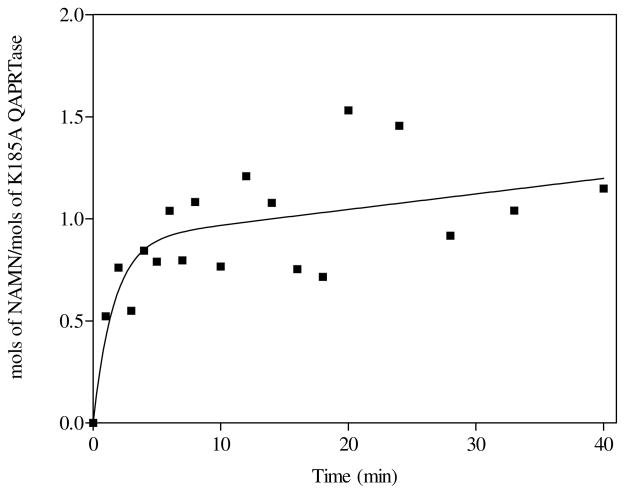
For the K185A mutant, chemical-quench experiments performed with [3H]QA as the substrate revealed a pre-steady state burst of [3H]NAMN formation, followed by a slow linear rate (Figure 6). The scatter in the data is due to the low activity of the mutant enzyme. Nevertheless, the existence of a pre-steady state burst was reproducible, with an amplitude of 0.9 ± 0.1 mol [3H]NAMN/mol of K185A. A relatively fast pre-steady state rate of 0.01s−1, and a slower steady state rate of 0.00013 s−1 (0.008 min−1) provided a good fit to the data. In contrast to WT, the presence of a burst of [3H]NAMN formation by K185A when utilizing [3H]QA indicates a relatively fast chemistry step for the mutant enzyme, with either product release or further processing providing the rate-limitation.
Figure 6.
[3H]NAMN formation by K185A with [3H]QA in the pre-steady state. Rapid mixing was performed manually as described in Materials and Methods. The curve represents the best fit of the data to equation 1, with rate constants equal to 0.01s−1 and 0.00013 s−1, for the pre-steady and steady states, respectively.
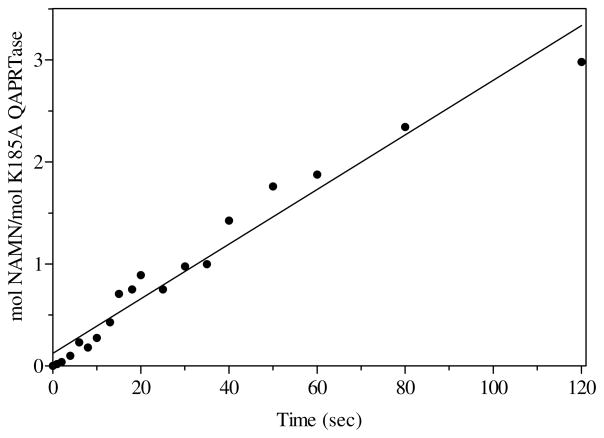
To confirm this implication, rapid quench experiments were performed with K185A, using [14C]NA as the substrate. In direct contrast to the reaction of K185A with [3H]QA, the pre-steady state result of K185A with [14C]NA showed no burst of NAMN formation (Figure 7). The initial rate was linear at 0.03 s−1, in agreement with the steady state kcat value for NA (0.05 s−1), and in particular, in rough agreement with the pre-steady burst state rate (0.01s−1) of product formation for the reaction of K185A with QA. The result of the reaction of K185A with NA suggests that the release of NAMN as observed for WT is not rate-limiting in the use of QA by this mutant. Therefore, it must be that for the reaction of K185A with QA, an intermediate was formed, whose subsequent reaction or release was rate-limiting.
Figure 7.
Pre-steady state kinetics of [14C]NAMN formation by K185A with [14C]NA. Rapid mixing was performed in the Kintek Chemical-Quench-Flow apparatus as described in Materials and Methods. The solid line represents a linear least square fit to the data points. The kcat of the reaction was 0.03 s−1.
The burst of on-enzyme product formation by K185A with [3H]QA suggests the existence of an intermediate whose further reaction may limit the turnover. On paper chromatography, the behavior of the products from the reaction of K185A with [3H]QA was similar to that of NAMN.
Restoration of enzymatic activity by mutant complementation
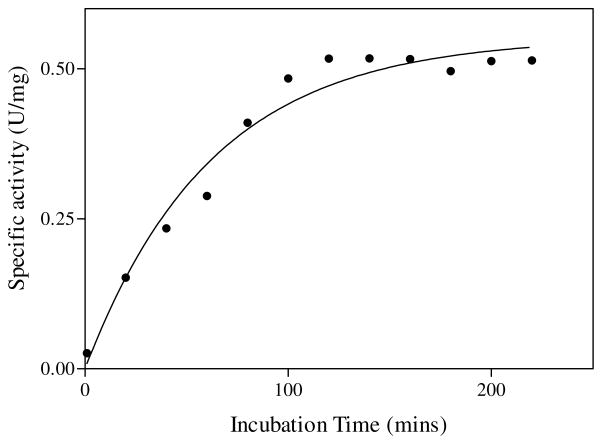
In the S. typhimurium QAPRTase dimer, Arg118′ interacts with QA across the subunit interface. R118A was used as a probe in complementation experiments with other debilitated or inactive mutant enzymes, to test the importance of this interfacial participation. Incubation of equal molar concentrations of R118A with R175A resulted in restoration of enzymatic activity (Figure 8). Similar results were observed with R152A and K185A. The restoration of activity was maximal within about 2–3 hrs. The activities of individual mutants and the activities of equimolar mixtures of R118A with mutant enzymes are presented in Table 3, as percentages of the activity of WT enzyme. The general observation is that mixing R118A with all poorly active or with inactive mutants restored activity. The R118A mutant enzyme also complemented other poorly active mutants (E214A, E214D, K153A and K284) described in the preceding paper. However, mixing binary combinations from among R152A, K153A, R175A, E214A, E214D and K284A did not restore enzyme activity (Unpublished results). Because the enhanced activity was observed only in the presence of R118A, it is most likely associated with formation of heterodimers between the two mutants. Because the activities of the equilibrated heterodimeric mutants were about 50% of WT, the most logical explanation is that one of the two active sites of the dimer can function at full activity with the other in an inactive state, and that heterodimers are preferred in mixtures of R118A with the other mutants.
Figure 8.
Time dependent complementation of inactive R175A mutant. Poorly active R118A mutant was mixed with equimolar R175A as described in Materials and Methods. The line represents a fit of data to a single exponential function.
Table 3.
Activity of complementation mixtures containing R118A
| Complementing Enzyme | Specific Activity (U mg−1) | Mixture (1:1) Tested | Specific Activity (U mg−1) |
|---|---|---|---|
| WT | 1 | R118A/WT | 1.1 |
| R152A | 0.03 | R118A/R152A | 0.5 |
| K153A | ND | R118A/K153A | 0.4 |
| R175A | ND | R118A/R175A | 0.5 |
| K185A | 1.6 × 10−3 | R118A/K185A | 0.5 |
ND, not detected
Activity of R118A was 2.4 × 10−4 U mg−1
DISCUSSION
The site-directed mutagenic investigation of the conserved residues at the QA binding site of QAPRTase showed that the interactions made by the residues are important for binding and catalysis. Lys185 most likely plays a direct chemical role in the stability of the QAMN ylide intermediate in the decarboxylation step, and it is a substrate specificity determinant for QAPRTase. Arg152 functions in QA binding and substrate discrimination. The very low activities of the enzyme mutated at Arg175 implied that the residue is essential and might play a role in both QA and PRPP binding. The importance of head to tail domain swapping (3) in QAPRTase is highlighted by Arg118′ as one of the critical participants across the dimer interface.
The hydrogen bond provided to the 2-carboxylate of QA by Lys185 appears by design to bind QA and confer base specificity for QAPRTase. The Lys185 sidechain is one that makes a notable conformational change between different complexes. In the Mycobacterium apoenzyme structure, the sidechain amino group lies close to the carboxylates of Asp186 and Asn187. In the Salmonella QA binary complex, the sidechain has repositioned and compacted to bind to the QA 2-carboxylate. In neither state does the sidechain position appear to preclude base NA binding. The interaction at C2 could potentially stabilize the negative charge at the 2 position on the putative QAMN ylide intermediate that results from decarboxylation. The major catalytic role of Lys185 is demonstrated by the significant decrease in catalysis by K185A. Ablation of the Lys185 sidechain to eliminate its interactions with the 2-carboxylate of QA resulted in a weakened binding affinity for QA, an inability to discriminate against NA, and therefore a lack of QA specificity. The base specificity of K185A is the reverse of WT, with WT favoring QA over NA by 2000-fold and K185A exhibiting a 300-fold increase in catalytic efficiency for NA over QA. The reverse base discrimination by K185A enzyme clearly supports a major role for Arg152 in QA binding and specificity. The role of Lys185 in substrate determination reconciles with the change in conformation of Lys185, as observed in the crystal structure of the Mycobacterium enzyme upon binding of QA (6).
The weak binding affinities of R152A and K185A enzymes for QA suggests roles for these residues in binding QA. The role of Arg152 and Lys185 in catalytic substrate discrimination was addressed from the ability of R152A and K185A to utilize NA as a substrate.
The pre-steady state burst observed for K185A when QA is utilized as the substrate indicated that formation of an initial product was fast, and either its release or further modification was slow. However, the rapid linear reaction of K185A using NA as the substrate indicated a difference in product formation. If the initial-burst product of K185A reaction with QA were NAMN, its further kinetic behavior would be similar to the product of the K185A reaction with NA, which is without question NAMN. The presence of a slower steady state rate for the reaction of K185A with QA implied that the product that formed was not NAMN. An explanation of the burst of product formation for the reaction of K185A with QA is that the putative QAMN intermediate was formed, and either the decarboxylation of QAMN or its release into solution is the rate limiting step. Attempts to detect the QAMN intermediate were unsuccessful. The unstable nature of the putative QAMN intermediate might not have permitted its capture under the current isolation procedure. The pre-steady state experiment with K185A strongly suggests that Lys185 participates in the decarboxylation step of QAPRTase, and most likely uses its positive charge to stabilize the negative charge of QAMN ylide intermediate. Lysine residues are known to be carbamoylated by carbon dioxide, usually to form a catalytically stable part of a metal-ion binding site. We note with interest the possibility that a lysine residue acts catalytically covalently accepts the initial carbon dioxide of Vibrio cholerae oxaloacetate decarboxylase (19). A water molecule found associated with the amino of Lys185 noted in some complexes of the yeast QAPRTase (22) could potentially represent a carboxy-lysine.
The importance of Arg118′ in catalysis is supported by several results. First, the location of Arg118 within the active site, forming hydrogen bonds to both substrates, but stretching across the subunit in crystal structures suggested a catalytic role. Second, sequence analysis showed absolute conservation of Arg118′. Third, mutation to alanine resulted in a dramatic decrease in the catalytic efficiency for PRPP due to the reduced kcat and a higher Km value for PRPP. Finally, the undetectable binding of PRPP to R118A strongly indicates an important role for Arg118′ in PRPP binding.
The observed restoration of the activity of inactive and poorly active mutants when mixed with R118A supports the structural studies in the interfacial participation of Arg118′ in catalysis. Dimerization facilitates proper structural alignment of the otherwise distant and essential Arg118′ residue from the adjacent subunit in the active site of QAPRTase. Earlier work by Ozturk et al. (27) has also shown that a type I PRTase, OPRTase, possesses this property of active site sharing of residues from the adjacent subunit. OPRTase has a flexible loop containing the Lys103 residue that is required for activity. Like QAPRTase in capping the enzyme active site, the loop in OPRTase acts as a lid for the active site formed by the adjacent subunit of the homodimer. In the OPRTase relative HGPRTase, the homologous loop is provided by the same subunit (28).
The very weak binding of R152A and K185A mutant enzymes to PRPP was unexpected, for neither of the residues interact directly with PRPP. The likeliest explanation for this observation is that the web of hydrogen bonds formed by residues interacting with QA (Figure 2) was disrupted, possibly via Arg118. Additionally, other residue-residue interactions appear important. In the M. tuberculosis QAPRTase•PA•PRPPC ternary complex, Arg152, which does not interact with PRPP is sequence adjacent to Lys153, which does. Arg152 is also seen to interact with the main-chain of Asn122′, which in turn, forms a hydrogen bond with main chain of Arg118′. Lys185, which does not interact with PRPP, is adjacent to Asp173, which does.
Abbreviations
- QA
quinolinic acid
- PA
phthalic acid
- PRPP
5-phosphoribosyl-1-pyrophosphate
- NAMN
nicotinate mononucleotide
- PPi
pyrophosphate
- NAPRTase
nicotinate phosphoribosyltransferase
- QAMN
quinolinate mononucleotide
- PRPCP
5-phosphoribosyl-1-(β-methylene)pyrophosphate
Footnotes
Supported by NIH grant GM48623 to CG
References
- 1.Musick WD. Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. CRC Crit Rev Biochem. 1981;11:1–34. doi: 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- 2.Eads JC, Ozturk D, Wexler TB, Grubmeyer C, Sacchettini JC. A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase. Structure. 1997;5:47–58. doi: 10.1016/s0969-2126(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 3.Chappie JS, Canaves JM, Han GW, Rife CL, Xu Q, Stevens RC. The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure. 2005;13:1385–1396. doi: 10.1016/j.str.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J Biol Chem. 2005;280:18326–18335. doi: 10.1074/jbc.M501622200. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Zhang X, Bheda P, Revollo JR, Imai S, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 6.Sharma V, Grubmeyer C, Sacchettini JC. Crystal structure of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis: a potential TB drug target. Structure. 1998;6:1587–1599. doi: 10.1016/s0969-2126(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 7.Foster JW, Moat AG. Mapping and characterization of the nad genes in Salmonella typhimurium LT-2. J Bacteriol. 1978;133:775–779. doi: 10.1128/jb.133.2.775-779.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holley EA, Foster JW. Bacteriophage P22 as a vector for Mu mutagenesis in Salmonella typhimurium: isolation of nad-lac and pnc-lac gene fusions. J Bacteriol. 1982;152:959–962. doi: 10.1128/jb.152.2.959-962.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster JW, Kinney DM, Moat AG. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol. 1979;137:1165–1175. doi: 10.1128/jb.137.3.1165-1175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster JW, Moat AG. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980;44:83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann DF, Byerrum RU. Quinolinic acid phosphoribosyltransferase from castor bean endosperm. I. Purification and characterization. J Biol Chem. 1974;249:6817–6823. [PubMed] [Google Scholar]
- 12.Kalikin L, Calvo KC. Inhibition of quinolinate phosphoribosyl transferase by pyridine analogs of quinolinic acid. Biochem Biophys Res Commun. 1988;152:559–564. doi: 10.1016/s0006-291x(88)80074-3. [DOI] [PubMed] [Google Scholar]
- 13.Cao H, Pietrak BL, Grubmeyer C. Quinolinate phosphoribosyltransferase: kinetic mechanism for a type II PRTase. Biochemistry. 2002;41:3520–3528. doi: 10.1021/bi012148g. [DOI] [PubMed] [Google Scholar]
- 14.Bhatia R, Calvo KC. The sequencing expression, purification, and steady-state kinetic analysis of quinolinate phosphoribosyl transferase from Escherichia coli. Arch Biochem Biophys. 1996;325:270–278. doi: 10.1006/abbi.1996.0034. [DOI] [PubMed] [Google Scholar]
- 15.Rozenberg A, Lee JK. Theoretical studies of the quinolinic acid to nicotinic acid mononucleotide transformation. J Org Chem. 2008;73:9314–9319. doi: 10.1021/jo8012379. [DOI] [PubMed] [Google Scholar]
- 16.Brown EV, Moser RJ. Further evidence as to the nature of the transition state leading to decarboxylation of 2-pyridinecarboxylic acids. Electrical effects in the transition state. J Org Chem. 1971;36:454–457. [Google Scholar]
- 17.Dunn GE, Thimm HF. Kinetics and mechanism of decarboxylation of some pyridine carboxylic acids in aqueous solution. Can J Chem. 1977;55:1342–1347. [Google Scholar]
- 18.Harris P, Navarro Poulsen JC, Jensen KF, Larsen S. Structural basis for the catalytic mechanism of a proficient enzyme: orotidine 5′-monophosphate decarboxylase. Biochemistry. 2000;39:4217–4224. doi: 10.1021/bi992952r. [DOI] [PubMed] [Google Scholar]
- 19.Studer R, Dahinden P, Wang WW, Auchli Y, Li X-D, Dimroth P. Crystal structure of the carboxyltransferase domain of the oxaloacetate decarboxylase Na+ pump from Vibrio cholerae. J Mol Biol. 2007;367:547–557. doi: 10.1016/j.jmb.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Kim MK, Im YJ, Lee JH, Eom SH. Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori. Proteins. 2006;63:252–255. doi: 10.1002/prot.20834. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Woznica K, Catton G, Crawford A, Botting N, Naismith JH. Structural and kinetic characterization of quinolinate phosphoribosyltransferase (hQPRTase) from Homo sapiens. J Mol Biol. 2007;373:755–763. doi: 10.1016/j.jmb.2007.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.di Luccio E, Wilson DK. Comprehensive X-ray structural studies of the quinolinate phosphoribosyl transferase (BNA6) from Saccharomyces cerevisiae. Biochemistry. 2008;47:4039–4050. doi: 10.1021/bi7020475. [DOI] [PubMed] [Google Scholar]
- 23.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 24.Bello ZI, Stitt BL, Grubmeyer C. Interactions at the 2 and 5 positions of 5-phosphoribosyl pyrophosphate are essential in Salmonella typhimurium quinolinate phosphoribosyltransferase. Biochemistry. 2009 doi: 10.1021/bi9018219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson KA. The Enzymes. Academic Press, Inc; New York: 1992. Transient-state kinetic analysis of enzyme reaction pathways; pp. 1–61. [Google Scholar]
- 26.Hughes KT, Dessen A, Gray JP, Grubmeyer C. The Salmonella typhimurium nadC gene: sequence determination by use of Mud-P22 and purification of quinolinate phosphoribosyltransferase, J. Bacteriol. 1993;175:479–486. doi: 10.1128/jb.175.2.479-486.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozturk DH, Dorfman RH, Scapin G, Sacchettini JC, Grubmeyer C. Locations and functional roles of conserved lysine residues in Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1995;34:10755–10763. doi: 10.1021/bi00034a007. [DOI] [PubMed] [Google Scholar]
- 28.Eads JC, Scapin G, Xu Y, Grubmeyer C, Sacchettini JC. The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP. Cell. 1994;78:325–334. doi: 10.1016/0092-8674(94)90301-8. [DOI] [PubMed] [Google Scholar]