SUMMARY
Post-transcriptional regulation of gene expression is an important mechanism for modulating protein levels in eukaryotes, especially in developmental pathways. The highly conserved homodimeric STAR/GSG proteins play a key role in regulating translation by binding bipartite consensus sequences in the untranslated regions of target mRNAs, but the exact mechanism remains unknown. Structures of STAR protein RNA binding subdomains have been determined, but structural information is lacking for the homodimerization subdomain. Here, we present the structure of the C. elegans GLD-1 homodimerization domain dimer, determined by a combination of X-ray crystallography and NMR spectroscopy, revealing a helix-turn-helix monomeric fold with the two protomers stacked perpendicularly. Structure-based mutagenesis demonstrates that the dimer interface is not easily disrupted, but the structural integrity of the monomer is crucial for GLD-1 dimerization. Finally, an improved model for STAR-mediated translational regulation of mRNA, based on the GLD-1 homodimerization domain structure, is presented.
INTRODUCTION
Post-transcriptional regulation of gene expression is a conserved mechanism that ensures the proper localization and timing of developmental processes in eukaryotes and often involves specific protein-RNA complexes with the 5'- or 3'-untranslated region (UTR) of mRNAs (Curtis et al., 1995). The Signal Transduction and Activation of RNA / GRP33, Sam68, GLD-1 (STAR/GSG) family, conserved in higher eukaryotes, consists of RNA-binding proteins that act as post-transcriptional regulators. Many STAR proteins play key roles in the coordination of several developmental processes such as mammalian spermatogenesis, metazoan central nervous system development, and Drosophila wing development (Matter et al., 2002; Larocque & Richard, 2005; Zaffran et al., 1997; Fyrberg et al., 1997).
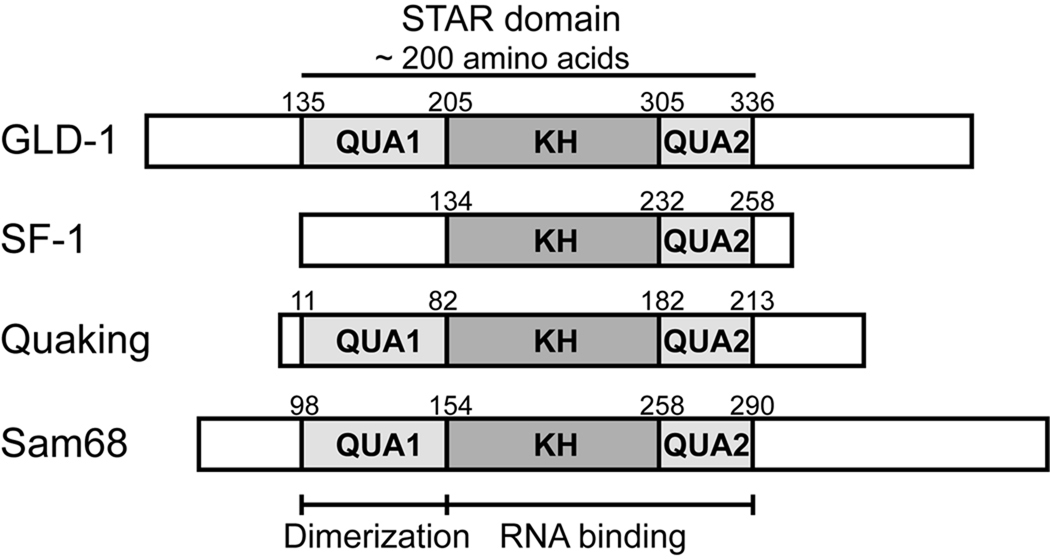
The highly conserved STAR domain consists of a central KH domain flanked by two homologous regions, termed Qua1 and Qua2 (Fig. 1; Vernet & Artzt, 1997). STAR proteins bind RNA via the KH and Qua2 regions (Ryder et al., 2004). In addition, most STAR family members homodimerize through interactions in the Qua1 subdomain (Chen et al., 1997; Chen & Richard, 1998). Current models for STAR dimer RNA binding suggests a bipartite consensus sequence in the target mRNA, composed of a 6 nucleotide UA-rich element bound by one STAR protomer, and an additional shorter half-site in close proximity, recognized by the second protomer (Ryder et al., 2004; Galarneau & Richard 2009).
Figure 1. The STAR family of RNA-binding proteins.
Domain structure of representative members of the STAR/GSG protein family. The Qua1, KH and Qua2 subdomains are shaded.
In Caenorhabditis elegans, the GLD-1 STAR protein controls germ cell differentiation and regulates the sperm-to-oogenesis switch in hermaphrodites (Jones et al., 1996; Jan et al., 1999). GLD-1 is a prototypical STAR protein that exerts developmental control by repressing translation of certain germline genes through directly binding conserved 28 nucleotide TGE (Tra-2 & GLI response Element) repeats in the 3'-UTR of target mRNAs (Jan et al., 1999). Although the KH-Qua2 subdomain is sufficient for GLD-1 RNA binding, the absence of Qua1 reduces the affinity for TGE by about one order of magnitude (Ryder et al., 2004). The lethal phenotype observed for Quaking mutants that abolish dimerization (Chen & Richard, 1998) though suggests that STAR domain dimerization serves other important functions in addition to facilitating RNA binding. The structural basis for GLD-1 RNA binding is not known, but high sequence conservation of the STAR domain (Vernet & Artzt, 1997) suggests the mode of RNA recognition is conserved within the STAR family. Solution structures of the KH-Qua2 domains of SF-1 in complex with RNA (Liu et al., 2001) and of the KH-Qua2 region of Xenopus Quaking (XQua) without RNA (Maguire et al., 2005) provide information about the RNA-binding subdomain organization of STAR proteins. Specifically, the SF-1 structure details the interaction of a monomeric KH-Qua2 domain with one single-stranded hexamer RNA consensus site. However, SF-1 is the only known STAR protein involved in splicing regulation and it also lacks the Qua1 dimerization subdomain (Berglund et al., 1997). Because there are no known structures of a complete STAR domain or the Qua1 dimerization region, it is not clear how homodimeric STAR proteins bind bipartite, asymmetric RNA target sequences. Previous sequence-based structure predictions suggest that Qua1 contains two α-helical segments and may form a coiled-coil dimer (Chen & Richard, 1998).
In order to better understand the role of homodimerization in STAR family functions such as RNA binding, we have determined the 2.0 Å resolution crystal structure of the Qua1 homodimerization subdomain from GLD-1. Overall, the Qua1 structure and structure-based mutational studies establish the molecular basis for STAR domain organization and further detail the biological model for STAR-mediated repression of translation.
RESULTS
NMR of GLD-1 Qua1 identifies the structural core comprising two α-helices connected by a short ordered linker
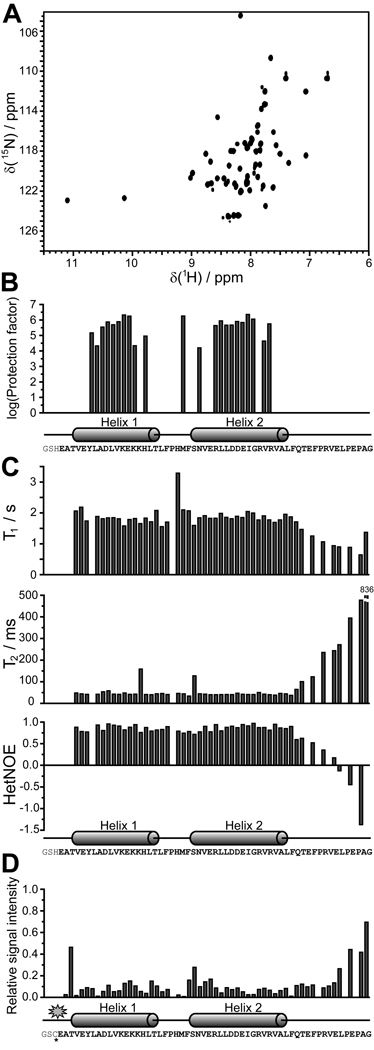
The GLD-1 Qua1 subdomain (residues 144–200) gave sharp lines and good dispersion in the 1H,15N-HSQC spectrum (Fig. 2A), where each resonance originates from an individual backbone amide. To identify structured regions, the backbone (HN, N, C, Cα, Hα) and Cβ resonances were assigned using standard triple resonance NMR experiments (Sattler et al., 1999). However, five N-terminal amino acids not present in the HSQC spectrum could not be assigned. Secondary structure predictions using the backbone chemical shifts (Berjanskii et al., 2006; Wishart & Sykes, 1994) suggest Qua1 is composed of two α-helices connected by a short linker, with an unfolded C-terminus.
Figure 2. Identification of the structured region in the Qua1 region of GLD-1 by NMR spectroscopy.
(A) 1H,15N-HSQC spectrum for the Qua1 region. (B) The H/D exchange experiment confirms that the two predicted helices are structured and thus protected against proton exchange with the solvent. (C) 15N-relaxation results suggest that the structured core of the Qua1 region includes both helices and a short, not very flexible linker. (D) Paramagnetic relaxation enhancement of a spin-labeled Qua1 region shows strong signal attenuation toward the C-terminal end of helix 2, suggesting either an antiparallel orientation of two monomers or a helix-turn-helix monomer.
Hydrogen-Deuterium exchange experiments monitored by NMR give site-specific information about secondary structure elements and their stability (Wildes & Marqusee, 2004). Amide protons in secondary structure elements are protected and exhibit a dramatic slowing of the proton exchange relative to those in random coil regions. For Qua1, protection factors in the range of 104 – 106 (Fig. 2B), indicative of structured regions, are observed for residues within both predicted helices and for F167, which is directly adjacent to the N-terminus of helix 2. For all other residues, H/D exchange was too fast to be measured, indicating that these residues are not in stable secondary structure elements. 15N-relaxation experiments (Fig. 2C) also define the ordered core region of Qua1, which includes both helices. The short linker between helices 1 and 2 exhibits relaxation times and heteronuclear NOE values similar to the adjacent helices, indicating that it is ordered. In contrast, the drastic increase in T2 and decrease of heteronuclear NOE values for the C-terminal 12 residues demonstrate the flexibility of this region.
The Qua1 regions of STAR proteins Qk1, XQua and HOW were predicted to form a coiled-coil (Chen & Richard, 1998), and the NMR measurements for GLD-1 Qua1 are consistent with this hypothesis. To assess whether Qua1 monomers adopt a parallel or antiparallel orientation in the dimer, a paramagnetic MTSL spin label was placed at the Qua1 N-terminus. Paramagnetic centers broaden NMR resonances of atoms in close proximity. A single Cys residue introduced at the Qua1 N-terminus was coupled to an MTSL spin label, and 1H,15N-HSQC spectra of both spin-labeled and unlabeled Qua1 were recorded (Fig. S1). The ratio of signal intensities between the labeled and unlabeled samples was attenuated for almost all resonances (Fig. 2D). Except for the residues adjacent to the spin label, the largest effect was observed towards the end of helix 2, implying that the C-terminus of helix 2 is spatially close to the N-terminus of helix 1 in the Qua1 dimer. Furthermore, the electron paramagnetic resonance (EPR) spectrum of spin-labeled Qua1 shows no sign of spin-spin coupling, suggesting the distance between the two spin labels within the dimer is larger than 20 Å. The spin labeling data is consistent with an antiparallel coiled-coil model because signal attenuation for a parallel coiled-coil would be expected only at the N-terminus, and spin-spin coupling would be observed in the EPR spectrum because of close proximity of the two spin labels. However, a helix-turn-helix monomeric fold, in which the N-terminus of helix 1 and the C-terminus of helix 2 are spatially close within the same monomer, could not be excluded.
A CS-Rosetta generated Qua1 monomer structural model used for crystallographic phasing
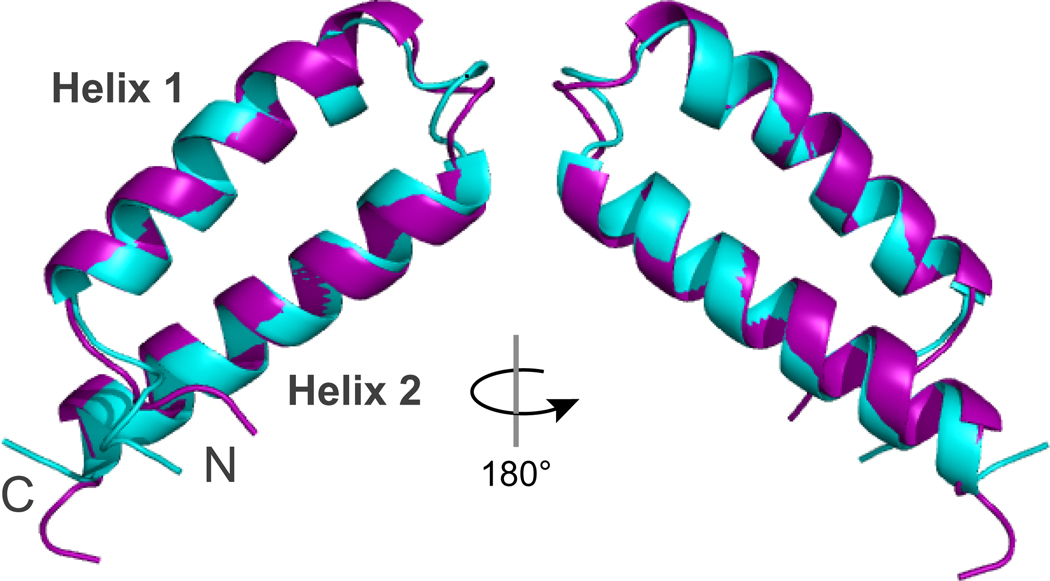
Chemical shift data for the backbone (HN, N, C, Cα, Hα) and Cβ atoms were used as input to the CS-Rosetta de novo structure prediction software (Shen et al., 2008a,b). C-terminal residues (190–200) identified as flexible by the 15N-relaxation experiments were omitted from the calculation which improved convergence (Fig. S2). The CS-Rosetta models converged on a well-defined structural model for the Qua1 monomer with a helix-turn-helix fold (Fig. 3). The average Cα-RMSD of the ten lowest-energy structures is 1.1 ± 0.2 Å (Fig. S2) and RMSD values for most Qua1 models cluster within 2 Å relative to the lowest-energy model. Since CS-Rosetta is designed for structure prediction of monomeric proteins, the calculated model does not provide information about the dimer interface and topology, and it was not clear if the predicted Qua1 structure was accurate or if potential dimer interface residues were forced into artificial contacts within the monomer.
Figure 3. Comparison of the CS-Rosetta model (cyan) and the crystal structure (purple) of the Qua1 region.
Alignment of the CS-Rosetta model and the lowest-rmsd crystal structure model reveals a Cα RMSD of 0.8 Å for the structured core region (residues 146–187).
Crystallization trials yielded well diffracting crystals of both wildtype Qua1 and an N169A mutant, but suitable search models for molecular replacement (MR) are lacking. However, recently a CS-Rosetta calculated model was successfully used in molecular replacement for a monomeric protein (Szymczyna et al. 2009). In following, the Qua1 monomer CS-Rosetta model was used as search model in MR. Phasing solutions were obtained for both data sets and the density-modified electron density maps show clear solvent boundaries and side chain density. Five N-terminal residues differ in position between the CS-Rosetta model and the crystallographic maps, likely because chemical shift information was unavailable for these residues. The N169A mutant structure was refined at 2.28 Å (R = 19.5%, Rfree = 26.4%; see Fig. S3 for the N169A mutant structure and Table S1 for crystallographic statistics).
X-ray crystal structure of the GLD-1 Qua1 region reveals a unique dimer topology
To confirm the MR solution obtained from the CS-Rosetta model, the Qua1 structure was also solved at 2.04 Å with R = 19.9% and Rfree = 24.5% from selenomethionine labeled wildtype Qua1 crystals using of MAD phasing (Table 1). The resulting structure adopts a helix-turn-helix fold nearly identical to the model predicted by CS-Rosetta. The Qua1 region is dimeric, as expected based on previous biophysical and biochemical studies (Ryder et al., 2004), but unexpectedly, the two protomers are stacked almost perpendicularly in the dimer (Fig. 4A).
Table 1.
Data collection and refinement statistics for wildtype Qua1. Values in parenthesis indicate statistics for the highest resolution shell.
| SeMet1 | native2 | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P41212 | P212121 | ||
| Cell dimensions (Å) | a = b = 37.91 | a = 33.89, | ||
| c = 158.33 | b = 42.96, | |||
| c = 154.64 | ||||
|
SeMet (peak) |
SeMet (remote) |
SeMet (inflection) |
native | |
| Wavelength (Å) | 0.9787 | 0.9184 | 0.9792 | 0.9795 |
| Resolution (Å) | 50-2.50 (2.54-2.50) |
50-2.50 (2.54-2.50) |
50-2.50 (2.54-2.50) |
50-2.04 (2.08-2.04) |
| Total reflections | 66156 | 66714 | 67549 | 111657 |
| Unique reflections | 4516 (221) | 4543 (214) | 4604 (233) | 15152 (717) |
| Rsym (%) | 4.4 (10.0) | 4.0 (9.8) | 3.8 (9.9) | 4.8 (34.2) |
| I/σ(I) | 41.4 (28.0) | 47.0 (28.6) | 54.2 (28.9) | 33.7 (5.7) |
| Completeness (%) | 99.5 (100.0) | 99.4 (100.0) | 99.5 (100.0) | 99.9 (100.0) |
| Redundancy | 14.6 (15.2) | 14.7 (15.1) | 14.7 (15.4) | 7.4 (7.5) |
| Refinement | ||||
| Resolution (Å) | 37.55-2.04 | |||
| (2.20-2.04) | ||||
| Unique reflections | 15092 (2802) | |||
| Rwork | 19.9 (20.7) | |||
| Rfree | 24.5 (24.7) | |||
| Stereochemistry | ||||
| Res. in favored region (%) | 92.4 | |||
| Res. in allowed region (%) | 7.6 | |||
| Number of atoms | ||||
| Protein | 1685 | |||
| Water | 103 | |||
| B factors | ||||
| Protein | 40.88 | |||
| Water | 48.22 | |||
| Rmsd | ||||
| Bond lengths (Å) | 0.005 | |||
| Bond angles (°) | 0.751 | |||
| Residues not modeled | ||||
| A | −2, 196–200 | |||
| B | −2, 191–200 | |||
| C | −2, 196–200 | |||
| D | −2, 188–200 | |||
used for phasing and initial model building
used for structure refinement after molecular replacement with initial model from MAD data.
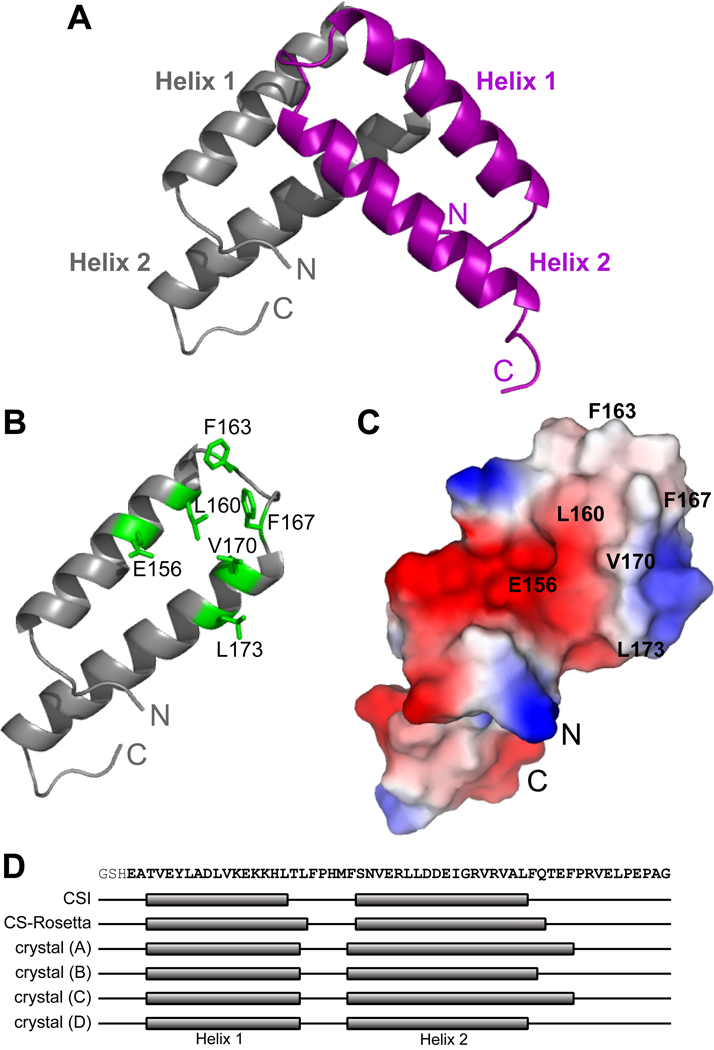
Figure 4. X-ray crystal structure of the homodimeric Qua1 region.
(A) Structure of the Qua1 homodimer. Two helix-turn-helix monomers are stacked in a perpendicular orientation. The first N-terminal and the C-terminal 5–13 residues are disordered in the electron density map. (B) The dimer interface consists of a hydrophobic patch (highlighted in green). (C) Surface charge potential of the Qua1 monomer. Dimer interface residues are labeled. (D) Mapping of the secondary structure determined from chemical shift indices (CSI), the CS-Rosetta model and all four monomers within the asymmetric unit of the crystal structure onto the Qua1 sequence.
The asymmetric unit contains two Qua1 dimers with essentially identical structures, Cα-RMSD between the monomers ranging from 0.35 Å to 1.01 Å for the structured core (residues 146–187), and the Cα-RMSD for the structured core of the CS-Rosetta model (Fig. 3) relative to the crystal structure (0.80 Å– 1.12 Å) is comparable to the deviation observed between the 4 Qua1 monomers in the asymmetric unit (for a complete list of RMSD comparisons see Table S2).
The Qua1 monomer adopts a helix-turn-helix fold that coincides with the structured region identified by NMR relaxation. The helix-turn-helix motif is stabilized by a hydrophobic "zipper" and a hydrogen bond between Y149 and E177 (Fig. 6C). Helix 2 extends past the zipper region and is more solvent exposed, which explains the lack of protection for these residues in the H/D exchange experiment, despite adopting an α-helical structure. Both protomers are very similar in the structured core region (Cα-RMSD 0.55 Å – 1.01 Å; Table S2), but more pronounced variations are observed C-terminal to the core region due to differences in crystal environment. For instance, helix 2 extends for an additional turn in monomers A and C compared to B and D, whereas its length is the average of these two in the CS-Rosetta solution model (Fig. 4D). The C-terminal tail that is flexible in solution is also unstructured and largely disordered in the crystal, adopting different positions depending on the crystal contacts.
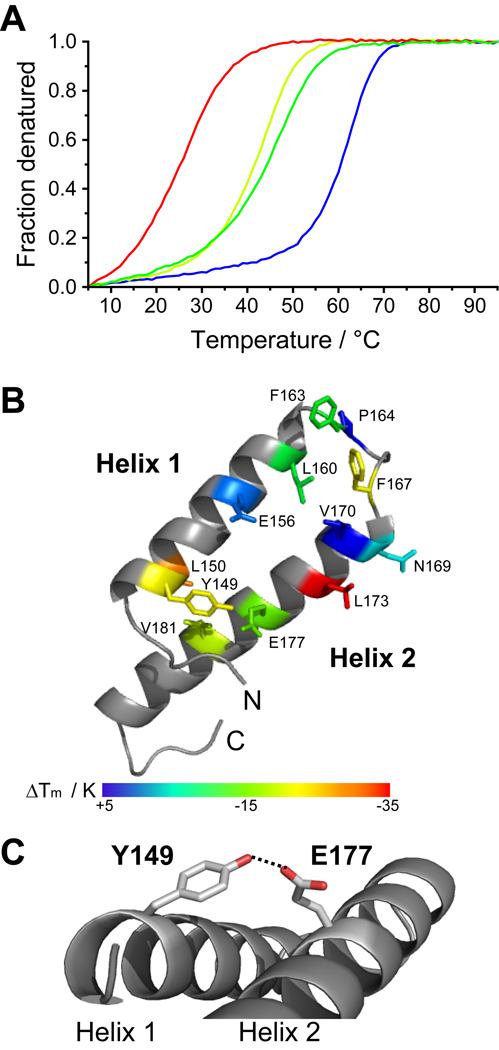
Figure 6. The stability of the dimer is more affected by mutations in the monomer zipper than by mutations in the dimer interface itself.
(A) Representative CD melting curves for wildtype Qua1 (blue), and the mutants E177A (dark green), Y149F (light green) and L173A (red). (B) The effect of Qua1 mutations on the melting temperature is mapped onto the crystal structure. (C) Y149 and E177 form a hydrogen bond that stabilizes the monomer helix-turn-helix fold, but is not part of the dimer interface.
The dimer interface (Fig. 4B,C & S4) comprises roughly 1400 Å2 of buried surface area and is formed by a hydrophobic patch (L160, F163, F167, V170, and L173) close to the turn between helices of each protomer. Further stabilization arises from two hydrogen bonds formed between the side chain of E156 and the backbone amides of N169' and V170' (the prime denotes residues in the other protomer).
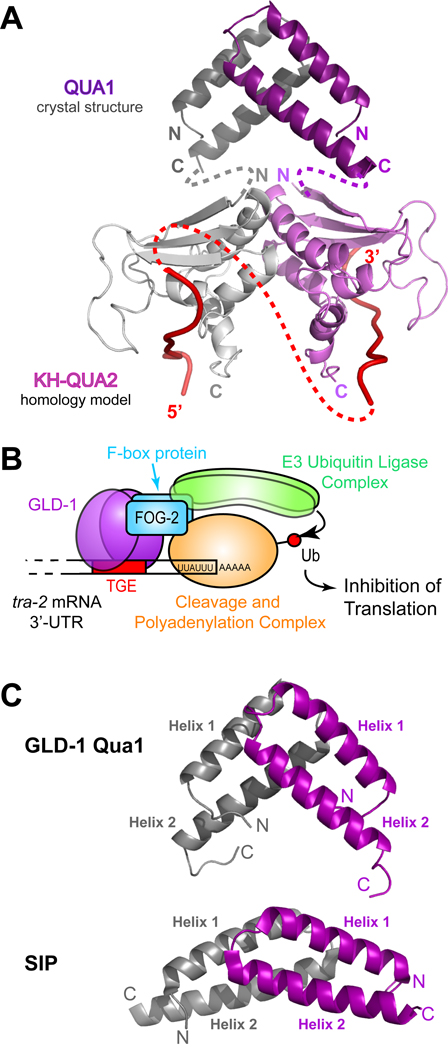
Structural similarity searches using DALI (Holm et al., 2008), 3D-BLAST (Tung et al., 2007) and PISA (Krissinel and Henrick, 2007) identified Qua1 as part of a superfamily that uses helix-turn-helix motifs to form four-helix bundle dimers. Most members have a classical antiparallel orientation like the synthetic metal binding peptide DF1 (Maglio et al., 2003), or a parallel orientation like some ROP variants (Willis et al., 2000). However, Qua1 is unique because of its almost perpendicular geometry. The most similar monomer arrangement is found in the Siah-binding protein dimer (SIP; Santelli et al., 2005), which also has two monomers stacked at an angle via a hydrophobic patch near the turn of the hairpin fold, but the angle between the monomers is much wider than for Qua1 (Fig. 8C & Discussion).
Figure 8. Model of a GLD-1 STAR dimer bound to RNA and model of the GLD-1 translation repression complex.
(A) Model of a GLD-1 STAR dimer. The Qua1 crystal structure is represented as dark gray and purple ribbons. The KH and Qua2 regions are a homology model based on the SF-1 NMR structure (Liu et al., 2001) and are shown as light gray and purple ribbons. The interface between the KH-Qua2 domains was modeled based on the Nova KH3 dimer (Ramos et al., 2002). The connectivity between Qua1 and the KH-Qua2 model is shown schematically as dashed gray and purple lines. Red ribbons represent RNA, and the connectivity of RNA between protomers is depicted as a dashed red line. (B) Model of the GLD-1 translation repression complex on the RNA. GLD-1 recruits an E3 ubiquitin ligase via its interaction with the F-box protein FOG-2. The cleavage and polyadenylation complex may be a potential target for ubiquitination. (C) Comparison of the GLD-1 Qua1 (top) and SIP (bottom) homodimerization domains. The two protomers within the homodimers are colored in gray and purple.
Residual Dipolar Couplings reveal that Qua1 dimer topology in the crystal and in solution are identical
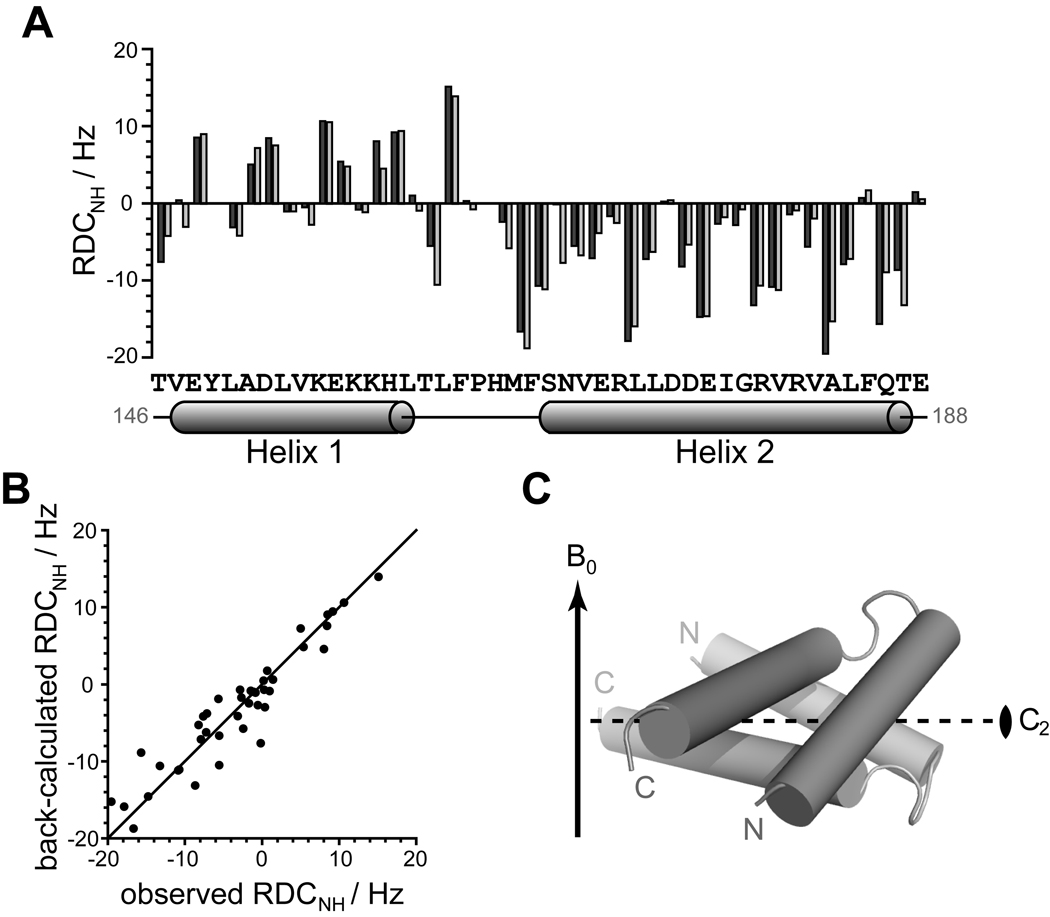
The agreement between the CS-Rosetta model and the crystal structure confirms that Qua1 monomers adopt the same fold in solution and in crystals. However, this does not provide direct evidence that the quaternary structure of the dimer in solution is identical to that observed in the crystal. NMR residual dipolar coupling (RDC) measurements were used to address this question. RDC values are determined in partially aligned media and provide direct information about the orientation of atomic bonds relative to the magnetic field and thus their orientation relative to each other (Bax, 2003). If a crystal structure is available, experimental RDC values can easily be compared to back-calculated values derived from rigid-body fitting of the crystal structure.
1H,15N-RDC data for Qua1 were obtained by orienting the sample with Pf1 phage. Since crystal contacts break the symmetry of the dimer, a symmetric structural model was generated by aligning the structured core (residues 146–188) of the CS-Rosetta model with both protomers A and B in the crystal structure. PALES (Zweckstetter and Bax, 2000) was then used to back-calculate the RDC values for this symmetric model and compare them to the experimental values. The calculated and experimental RDC values agree for the Qua1 dimer (Fig. 5). Furthermore, the symmetry axis of the dimer aligns perpendicular to the phage and magnetic field (Fig. 5C) such that each amide adopts an equivalent position in both protomers. Each monomer alone, in the same orientation, would yield identical RDC values because of the inherent symmetry, but analytical gel filtration studies (Ryder et al., 2004) confirm that Qua1 is a dimer in solution. In conclusion, the dimer structure in the crystal is consistent with the RDC data in solution.
Figure 5. Experimental 1H-15N residual dipolar couplings (RDC) agree well with the Qua1 homodimer crystal structure.
(A) Experimental RDC obtained from aligning 15N-labeled Qua1 with Pf1 phage (dark gray) and back-calculated 1H-15N-RDCs from the crystal structure (light gray) plotted against protein sequence for the structural core of Qua1. The secondary structure according to the CS-Rosetta model is mapped onto the sequence. (B) Plotting of the back-calculated RDC from the crystal structure against the experimental values shows good correlation. (C) Alignment of the dimer model relative to the magnetic field B0 (indicated by an arrow) as determined by fitting the orientation of the Qua1 dimer crystal structure to the experimental RDC. The 2-fold symmetry axis (C2) of the homodimer is indicated by a dashed line.
Structure-based mutational analysis identifies determinants of Qua1 dimer stability
Key dimer interface residues were mutated in order to investigate their role in Qua1 dimerization, including the highly conserved residues E177 and Y149, and other residues in the monomer zipper. All thermal melting curves monitored by CD spectroscopy (Fig. 6) show a single transition and similar amplitudes, indicating that the constructs are folded at low temperatures. These data also suggest that Qua1 forms obligate dimers, in which dimer dissociation and disruption of monomer secondary structure occur simultaneously in a two-state unfolding transition. However, the midpoint of the melting transition (TM) is mutation-dependent (Table 2). Most mutations within the dimer interface have negligible to moderate effects, the one exception being L173A which confers the largest observed destabilization. L173 is located at the inner edge of the interface close to the dimer symmetry axis and packs against itself (L173') in the adjacent protomer. Removal of this pair of side chains results in a large void at one position in the interface while the other mutants are spatially separated.
Table 2.
Stability of Qua1 point mutants / CD melting. (H6) indicates constructs with a C-terminal hexahistidine tag that was not removed.
| Mutant | TM / °C | ΔTM / K |
|---|---|---|
| wt | 63 | 0 |
| wt (H6) | 63 | 0 |
| Dimer interface: | ||
| E156A | 62 | −1 |
| L160A | 53 | −10 |
| F163A | 52 | −11 |
| P164A | 65 | +2 |
| F167A | 44 | −19 |
| N169A | 58 | −5 |
| V170A | 67 | +4 |
| L173A (H6) | 28 | −35 |
| Monomer Zipper: | ||
| Y149F (H6) | 44 | −19 |
| L150A (H6) | 37 | −26 |
| E177A (H6) | 48 | −15 |
| V181A | 46 | −17 |
E177 is also interesting because it is highly conserved among STAR proteins and shows an embryonic lethal phenotype in mouse Quaking (E48) when mutated to Gly (Ebersole et al., 1996). In an in vivo immunoprecipitation assay, mouse Quaking E48G fails to pull down wild type Quaking, suggesting it prevents homodimerization (Chen & Richard, 1998). Surprisingly, our structure reveals that E177 is not a key residue in the GLD-1 dimer interface, instead forming a Van-der-Waals contact to L174'. However, the structure suggests this is only a minor stabilization compared to the hydrophobic interactions. The most prominent structural feature of E177 is a hydrogen bond to the highly conserved Y149, which forms a clamp between both helices in the monomer. Therefore, E177 appears to play a more important role in stabilizing the monomeric fold rather than the dimer interface, suggesting that dimerization may be affected more by mutations that disrupt the structural integrity of the monomer.
To test this hypothesis, E177, Y149 and additional residues in the zipper were mutated, resulting in a significantly greater destabilization relative to the dimer interface mutants (Table 2). Removal of the hydrogen bond between E177 and Y149 (E177A and Y149F) yielded a similar degree of destabilization. Because the E177G mutant only expressed in very small amounts and degraded during purification, it seemed likely that replacing E177 with a Gly instead of Ala has an even more pronounced effect on the monomer stability, and thus dimerization. Because glycine often acts as a helix breaker, this may explain why no coiled-coil was predicted for the corresponding Quaking E48G mutant (Chen & Richard 1998). The significant effects observed for the Qua1 L150A and V181A mutants further underscores the importance of monomer stability. In conclusion, in the absence of any individual interface contacts required for dimerization, monomer integrity seems key because it drives the correct presentation of the multiple weak interface contacts that combine to form a stable Qua1 dimer interface.
DISCUSSION
The STAR protein family is highly conserved among higher eukaryotes (Vernet & Artzt, 1997), and many members regulate gene expression post-transcriptionally by forming specific protein-RNA complexes within the 3'-UTR of target mRNAs (Saccomano et al., 1999; Jan et al., 1999). The STAR protein core consists of a KH RNA-binding domain flanked by two regions homologous to the mouse Quaking protein: the Qua2 extension, which forms part of the interface with RNA, and the Qua1 homodimerization domain (Ryder et al., 2004). The structure of splicing factor SF-1 in complex with branch point RNA (Liu et al., 2001) provides a model for the interaction of a single KH-Qua2 domain with one hexamer RNA consensus site. However, our understanding of how dimerization affects RNA binding and STAR protein function has been limited due to the lack of structural information available on the Qua1 region.
The structure of the GLD-1 Qua1 region presented here provides a more complete model of how the GLD-1 STAR domain homodimer may associate with its asymmetric RNA target sequence. Translational regulation by GLD-1 also involves additional proteins, like FOG-2, that directly interact with a GLD-1 RNA complex. By studying the genetically well-characterized germline developmental pathways in the model organism C. elegans, we aim to define the molecular basis for translational regulation by protein-RNA complexes in the 3'-UTR.
The GLD-1 Qua1 region is a helix-turn-helix dimer
The structure of the GLD-1 Qua1 region presented here represents the first structure of a STAR/GSG homodimerization domain. In contrast to previous predictions (Chen & Richard, 1998), Qua1 does not form a coiled-coil, but adopts a helix-turn-helix fold. The monomeric fold is stabilized by a hydrophobic zipper between the two helices and a hydrogen bond between the conserved residues Y149 and E177, while the C-terminal 13 residues are unstructured and flexible. The dimer interface consists of a hydrophobic patch and is located near the loop region of the monomer hairpin. Although helix-turn-helix dimers are common, Qua1 is distinct because of the unique, almost perpendicular angle between protomers.
Furthermore, residual dipolar coupling experiments and structure prediction using NMR chemical shifts with CS-Rosetta both reveal that the structures of Qua1 in solution and in the crystal are consistent. The successful combination of experimental chemical shifts and computational structure prediction using CS-Rosetta to obtain a molecular replacement model for crystallography corroborates previous work (Szymczyna et al., 2009) and also represents the first example of using this approach to solve the structure of a homodimeric protein. Therefore, this method may be broadly applicable, even for more complex dimeric structures. Interestingly, side chains in the monomer zipper are predicted with high precision relative to the crystal structure and among the 10 lowest-energy CS-Rosetta structures, while the side chains in the dimer interface show considerably larger deviations. Consistently, the monomer zipper is more important for the stability of the overall fold, which may have contributed to the success of the CS-Rosetta prediction that does not consider the presence of the second protomer.
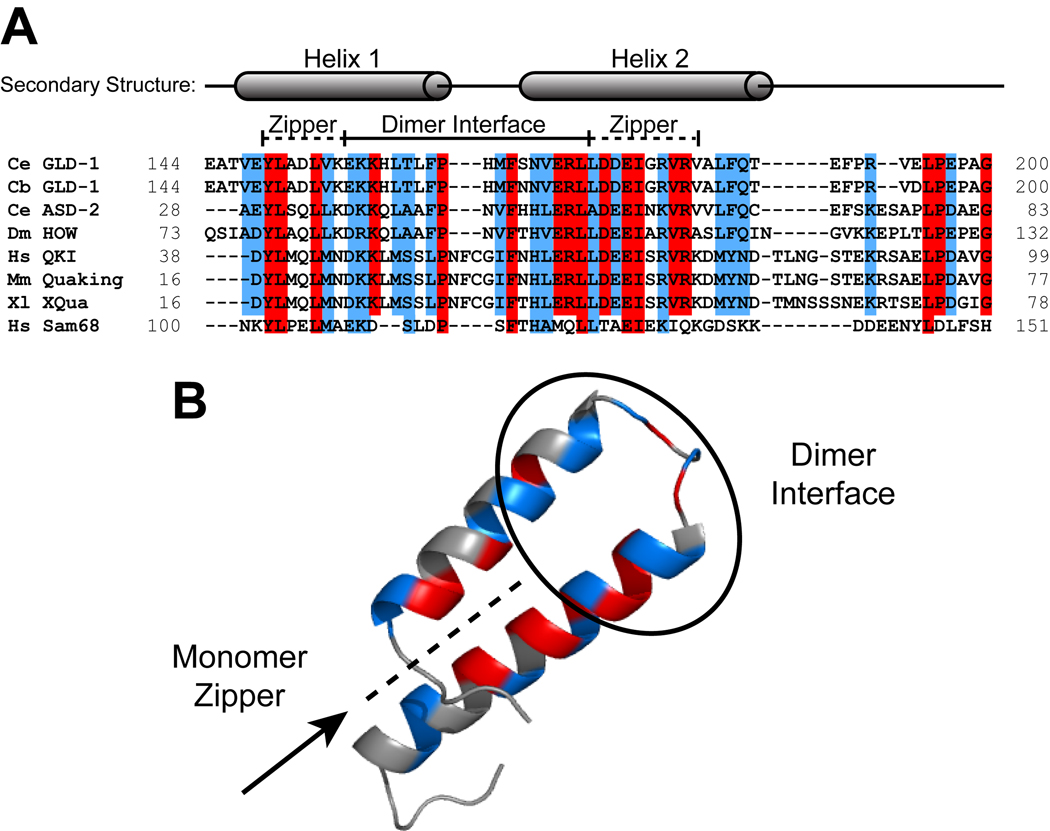
The Qua1 dimerization domain is highly conserved among STAR family members
The Qua1 region of the STAR domain is highly conserved among most STAR family members (Fig. 7), implying that the structure is also conserved. The residues within the monomer zipper are the most conserved. Within the dimer interface, similar hydrophobic residues are generally tolerated and may serve to discriminate between paralogs within the same organism. The Qua1 region of Sam68 is less conserved relative to the Quaking homologs, and may represent a different subfamily (Chen & Richard, 1998; Chen et al., 1997).
Figure 7. Sequence alignment of the Qua1 region for representative members of the STAR/GSG family.
(A) Sequence alignment. The secondary structure is mapped on top of the sequences. Identical residues are highlighted in red and similar residues in blue. The dimer interface and zipper regions are marked by a bar. (B) Identical (red) and similar (blue) residues mapped onto the GLD-1 Qua1 crystal structure.
Previous studies demonstrated that a lethal point mutation of E48 to glycine in the Qua1 region of mouse Quaking prevents homodimerization (Chen & Richard, 1998), suggesting that this residue may be part of the dimer interface. However, the GLD-1 Qua1 structure shows the corresponding E177 is part of the monomer zipper. Single mutations of this residue and others located in the monomer zipper destabilize Qua1 more than single mutations in the dimer interface. The importance of the zipper residues for Qua1 dimerization and their high degree of conservation imply that, in the absence of any individual interface contacts required for dimerization, monomer integrity is crucial because it ensures the correct arrangement of the multiple weak interface contacts that combine to form a stable Qua1 dimer interface.
A model for STAR domain organization and RNA binding
The SF-1 structure in complex with branch point RNA (Liu et al., 2001) allowed construction of a homology model for the GLD-1 KH-Qua2 region bound to RNA (Ryder et al., 2004; Fig. 8A). Since SF-1 lacks the Qua1 region and does not dimerize, it only serves as a model of RNA binding by a single STAR monomer. As such, this model does not explain how a symmetric STAR protein homodimer binds its target mRNAs with asymmetrical binding sites.
Although KH-Qua2 in GLD-1 does not dimerize on its own, it binds RNA cooperatively, indicating that two KH-Qua2 protomers might interact within the context of a full STAR domain bound to RNA (Ryder et al., 2004). This model is further supported by Sam68, which requires parts of the KH region and the presence of RNA in addition to Qua1 to dimerize (Chen et al., 1997). The model depicting KH domain interactions in the dimer was based on the interface in the Nova crystal structure (Lewis et al., 2000). The Nova-1 KH3 domain is in a monomer/dimer equilibrium in solution, mediated by a specific interface. Nova-1 binds to RNA as a dimer and is implicated in regulation of alternative splicing in neurons (Ramos et al. 2002). It remains a possibility that the presence of the main dimerization domain Qua1 and Qua2 could significantly affect KH dimerization in GLD-1, but the Nova dimer remains the best available model. If the KH-Qua2 region extends the dimer interface, it seems logical that the Qua1 dimer and the KH-Qua2 dimer model share the same symmetry axis. The linker between Qua1 and KH-Qua2 (residues 190–205) is unstructured and flexible in Qua1. Considering the length of the linker, Qua1 could potentially reach the top or the bottom of the modeled KH-Qua2 dimer. The high proline density in this region suggests that a portion of this linker is unstructured, but the many hydrophobic residues may indicate they play a role in the structure.
One possible model for RNA binding involves one GLD-1 protomer recognizing the 5'-UACUCA-3' hexamer consensus within the TGE, while the second protomer binds the upstream UA element. The upstream dinucleotide increases the affinity of GLD-1 RNA binding by about 10-fold relative to a single hexameric site (Ryder et al., 2004; Carmel et al., submitted). The RNA sequence separating the two specificity elements in the TGE comprises only 10 nucleotides, versus at least 16–20 required in the symmetric model. To bind this bipartite target sequence efficiently, the RNA binding sites need to be positioned close enough. In the symmetric model (Fig. 8A), direct connections between the RNA pieces appear too long to fulfill this condition. Therefore, the KH domains are likely oriented in a different way with respect to the RNA, bringing the RNA-binding interfaces closer together to accommodate a single TGE. Interestingly, the tra-2 3'-UTR contains two identical TGE repeats plus an additional relaxed consensus site in between the two TGEs, suggesting that GLD-1 may bind two hexamers in different TGE elements. Relative to a single TGE site, GLD-1 binds 2.5-fold tighter to RNA sequences with two hexamer consensus sites allowing somewhat variable spacing (Carmel et al., submitted). It is therefore possible that STAR dimers bind two consensus sites in a symmetric fashion, provided the linker RNA sequence in between is sufficiently long. Our current efforts to determine the structure of the full GLD-1 STAR domain in complex with RNA will provide more insight into the arrangement of the subdomains within the complex and the path of the asymmetric TGE RNA through the GLD-1 dimer.
Implications for the role of GLD-1 in translational repression
The mechanism of GLD-1-mediated mRNA translational repression remains poorly understood. GLD-1 interacts with FOG-2 in a yeast two-hybrid assay (Clifford et al., 2000), and both FOG-2 and GLD-1 deletion mutants in C. elegans result in germline feminization (Francis et al., 1995; Schedl and Kimble, 1988). FOG-2 contains an F-box domain commonly found in SCF (Skp1 – Cul1 – F-box) E3 ubiquitin ligase complexes (Ho et al., 2008). The SCF complex contains numerous proteins and binds to ubiquitin-conjugating enzymes (E2) to form the active ligase complex (Petroski & Deshaies, 2005). F-box proteins confer specificity to SCF ubiquitin ligases by binding both the ubiquitination target and other E3 ligase components (Najak et al., 2005; Yamanaka et al., 2002). FOG-2 also interacts with the Skp1 related (SKR) protein, which is a known component of an SCF complex (Yamanaka et al., 2002). Therefore, FOG-2 may be part of an E3 ubiquitin ligase that is targeted to the mRNA via GLD-1 (Fig. 8B). It is unlikely that GLD-1 is targeted for ubiquitin-mediated protein degradation because both GLD-1 and FOG-2 are necessary for promoting male germ line development in C. elegans (Clifford et al., 2000).
Since the poly-A tail of some GLD-1 targets was found to be relatively short (Jan et al., 1997), the cleavage and polyadenylation complex may be tagged for ubiquitination, and translational repression may be achieved by inhibition of poly-A tail elongation. However, a biological target for the putative FOG-2 containing ubiquitin ligase and its role in translational repression has yet to be determined. Furthermore, multiple distinct mechanisms of GLD-1 mediated translational control exist dependent on the mRNA target (Lee & Schedl, 2001).
Interestingly, the human Siah-interacting protein (SIP) has a dimer topology similar to Qua1 (Fig. 8C) and is also part of an SCF-like E3 ubiquitin ligase complex (Santelli et al., 2005). It functions as an adapter protein by binding both Skp1 and Siah1, and could act as a scaffold that mediates E3 ligase assembly. GLD-1 might also function as an adapter protein that assembles an E3 ligase complex on its RNA target. Interestingly, both SIP and the Qua1 region of GLD-1 have a proline-rich region in the unstructured C-terminus. In SIP, this region is important for SIAH binding (Santelli et al., 2005), and the C-terminal domain present in the full-length isoform of SIP contains the binding determinants for Skp1 (Matsuzawa & Reed, 2001). It is possible that in GLD-1 the linker between Qua1 and KH-Qua2 is also important for protein-protein interactions. Our current efforts strive to determine the FOG-2 binding site within the GLD-1 STAR domain and characterize the GLD-1/FOG-2 interaction.
C. elegans possesses a large family of FOG-2-related (FTR) proteins with over 100 members (Nayak et al., 2005), implying that specificity in STAR protein-mediated translational repression may be achieved by modular assembly of GLD-1 (in the germline) or STAR2/ASD-2 (in somatic tissue) with different FTR proteins. Because STAR proteins are highly conserved and F-box-containing SCF ubiquitin ligases are ubiquitous, recruiting ubiquitin ligases to mRNAs via modular multiprotein complexes might be a general mechanism of post-transcriptional regulation.
EXPERIMENTAL PROCEDURES
Protein constructs and Purification
Hexahistidine-tagged GLD-1 Qua1 (residues 144–200) was cloned into a pET22b-based expression vector and expressed in E. coli BL21 Gold (DE3). Single point mutations were introduced using Quikchange (Stratagene). The constructs were purified on a Nickel-NTA affinity column followed by a HiTrap Q anion exchange column (GE Healthcare). For details on protein expression and purification see Supplementary Material.
NMR Spectroscopy
NMR samples were prepared in 20 mM sodium phosphate (pH 7.5), 0.02% NaN3 containing 10% D2O / 90% H2O with a final protein concentration of 0.5 mM. NMR spectra were recorded at 35°C on Bruker spectrometers (500, 750 and 800 MHz) with 5 mm TXI/HCN triple resonance probes. Backbone assignments of 15N,13C-Qua1 were accomplished using classical triple-resonance (Sattler et al., 1999) and HCCH-COSY (Gehring & Ekiel, 1998) experiments. Details and references for all experiments are available in the Supplementary Material. For NMR data processing and analysis, NMRPipe (Delaglio et al., 1995) and CARA (Keller, 2005) were used. All chemical shifts are relative to 2,2-dimethyl-2-silapentane-5-sulfonate (Wishart et al., 1995).
H/D Exchange Experiments
The amide H/D exchange rates for 15N-Qua1 were measured at 20°C on a 500 MHz Bruker Avance spectrometer with a TXI 5 mm probe. The sample was exchanged into D2O by lyophilizing the protein in aqueous 20 mM sodium phosphate (pH 7.5) to dryness and quickly dissolving the pellet in D2O. The exchange of the amide protons with deuterium was monitored by collecting a series of successive 1H,15N-HSQC spectra for a total exchange time of 16.5 h. The first spectrum was initiated 15 min after adding D2O and the remaining spectra were acquired at 18 min intervals. Protection factors were calculated as kint/kex where kex is the exchange rate constant obtained by fitting a monoexponential function to the amide signal intensities, and kint is the intrinsic exchange rate estimated using SPHERE (Bai et al., 1993).
15N-Relaxation Measurements
Nitrogen T1, T2 and steady-state heteronuclear NOE relaxation experiments for 15N-Qua1 were collected at 35.0°C using sensitivity-enhanced pulse programs (Farrow et al., 1994) on a shielded 800 MHz Bruker Avance spectrometer with a TXI/HCN 5 mm probe. T1 and T2 relaxation delay times ranged from 10 ms to 3200 ms and 6 ms to 130 ms, respectively, and were collected in random order. The saturated and unsaturated heteronuclear NOE experiments were collected in an interleaved manner, and the ratio of signal intensities was calculated for each resonance. T1 and T2 were fit to a monoexponential function with offset using Curvefit (Mandel et al., 1995). Signal intensities were analyzed using NMR View (Johnson and Blevins, 1994).
Spin labeling and Paramagnetic Relaxation Experiments
(1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) Methanethiosulfonate (MTSL) spin label was obtained from Toronto Research Chemicals, Inc. and used as a 100 mM stock solution in acetonitrile. 15N-C-Qua1 with an N-terminal Cys was exchanged into 20 mM sodium phosphate, pH 7.5 without reducing agent using a PD-10 desalting column (GE Healthcare). The protein (90 µM) was incubated at room temperature with 100-fold molar excess of spin labeling reagent over night. Excess spin label was removed with a PD-10 column before concentrating the protein. 10% D2O was added to the NMR sample with a final protein concentration of 0.5 mM. 15N-C-Qua1 (0.5 mM) without spin label in 20 mM sodium phosphate, 2 mM DTT, 10% D2O / 90% H2O was used as the diamagnetic sample. 1H,15N-HSQC spectra were collected for both samples at 35°C on an 800 MHz Bruker DRX spectrometer with TCI/HCN 5 mm cryo-probe, and the ratio of signal intensities was calculated for each residue. Since the spectra were nearly identical, amide assignments were inferred from the backbone assignments of the Qua1 construct.
Residual Dipolar Couplings (RDC)
The isotropic sample contains 0.5 mM 15N-Qua1 in the previously described standard NMR buffer with 10% D2O. The aligned sample was prepared by adding Pf1 phage (Alsa Biotech) at a final concentration of 12 mg/ml to a second sample identical to the isotropic one. Two-dimensional IPAP 1H,15N-HSQC spectra using Watergate water suppression (Ottinger et al., 1998) were collected in an interleaved manner on a 750 MHz Bruker Avance spectrometer with a TXI/HCN ATMM 5 mm probe at 35.0°C. In total, 48 experimental N-H RDC were obtained. PALES (Zweckstetter & Bax, 2000) was used to fit the experimental RDC to the X-ray crystal structure and back-calculate RDC from the structure. Only the structured core comprising residues 146–188 could be fit and RDC for the highly flexible C-terminal tail (189 – 200) and N-terminus (−2 – 145) were ignored.
Secondary and tertiary structure prediction
Secondary structure prediction from chemical shifts was based upon the Chemical Shift Index (CSI) and PREDITOR secondary structure prediction programs (Wishart and Sykes, 1994; Berjanskii et al., 2006). The tertiary structure of Qua1 was predicted using CS-Rosetta (Shen et al., 2008a&b) with the backbone (HN, N, C, Cα, Hα) and Cβ chemical shifts as input. The flexible C-terminal residues 190–200 were omitted from the calculations. CS-Rosetta calculations were run in parallel on a local 64-bit Linux cluster with 3936 CPUs, generating 1200 structures in total. The 500 lowest-energy structural models were extracted to assess convergence. The molecular replacement search model for the N169A mutant was obtained by changing residue 169 in the CS-Rosetta model to Ala.
EPR spectroscopy
EPR spectra were recorded at room temperature on a Varian E-109 spectrometer fitted with a two-loop one-gap resonator (Hubbell et al., 1987). Protein samples of 5 uL (100 µM without, 50 µM with 30% w/v sucrose) were loaded in Pyrex capillaries (0.84 mm o.d. × 0.6 mm i.d.) sealed on one end. Spectra were acquired using a 2 mW incident microwave power and ca. 1 G field modulation amplitude at 100 kHz. All spectra were normalized to the same area.
X-ray Crystallography
Crystallization
Qua1 crystals were crystallized by vapor diffusion in 24-well hanging drop plates, 2µl drop volume, at 22°C. For detailed crystallization and cryo-protection conditions see Supplementary Material.
Data collection and processing
Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) BL 11-1 processed with HKL2000 (Otwinowski et al., 1997). The PHENIX package (Adams et al., 2002) was used for MAD phasing of SeMet-Qua1, molecular replacement of native Qua1 using the SeMet-Qua1 model, and molecular replacement of native Qua1 wildtype and N169A using the CS-Rosetta model (log likelihood gain (LLG)/ translation function Z score (TFZ) = 284 / 10.4 for wildtype and 180 / 8.7 for N169A). Initial model building (SeMet-Qua1) or model rebuilding (native Qua1, N169A) after density modification was carried out in PHENIX AutoBuild, followed by alternating rounds of refinement with PHENIX Refine and model rebuilding and evaluation in Coot (Emsley and Cowtan, 2004). In later rounds of refinement, TLS refinement using each monomer as an independent TLS group was performed in addition to coordinate and atomic B-factor refinement. Iterative rebuilding and refinement converged on a final model with R = 19.9% and Rfree = 24.5% (wildtype Qua1); R = 19.5 % and Rfree = 26.4% (N169A mutant) and good stereochemistry (92.4% / 7.6% of residues in the most favored / additionally allowed region for wildtype and 92.9% / 7.1% for N169A). For complete crystallographic statistics see Table 1 & Table S1.
CD Spectroscopy
Thermal melting experiments were performed using an Aviv Circular Dichroism Spectrometer Model 202SF and a 1 mm quartz cuvette. The samples consist of 50 µM protein in 20 mM sodium phosphate (pH 7.5). Melting curves were recorded between 5°C and 95°C in 1°C-steps monitoring the signal at 222 nm.
Supplementary Material
ACKNOWLEDGEMENTS
We thank I. Wilson for access to X-ray equipment; J. Kelly for access to the CD spectrometer; G. Kroon, X. Dai, S. Kwan, G. Bhaba, B. Koehntop, X. Zhu, P. Verdino and B. Chapados for fruitful discussions and assistance.
X-ray data were collected at SSRL, a national user facility operated by Stanford University on behalf of the U.S. Dept. of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE, BES, and by the NIH, NCRR, Biomedical Technology Program, and the NIGMS.
This work was supported by the NIH grants GM53320 (J.R.W.) and 1F32GM068286 (L.C.), as well as DFG (C.B), and CIHR (B.R.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates have been deposited in the Protein Data Bank with the accession code 3K6T (wildtype) and 3KBL (N169A mutant).
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UAUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berjanskii MV, Neal S, Wishart DS. PREDITOR: A web server for predicting protein torsion angle restraints. Nucleic Acid Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: Role of the KH domain. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Richard S. Structure-Function Analysis of Qk1: A lethal point mutation in mouse quaking prevents homodimerization. Mol. Cell. Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Curtis D, Lehmann R, Zamore PD. Translational control in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeiffer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene unites signal transduction and RNA binding in the developing nervous system. Nat. Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg C, Becker J, Barthmaier P, Mahaffey J, Fyrberg E. A Drosophila muscle-specific gene related to the mouse quaking locus. Gene. 1997;197:315–323. doi: 10.1016/s0378-1119(97)00278-3. [DOI] [PubMed] [Google Scholar]
- Galarneau A, Richard S. The STAR binding proteins GLD-1, OKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol. Biol. 2009;10:47–58. doi: 10.1186/1471-2199-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring K, Ekiel I. H(C)CH-COSY and (H)CCH-COSY experiments for 13C-labeled proteins in H2O solution. J. Magn. Reson. 1998;135:185–193. doi: 10.1006/jmre.1998.1543. [DOI] [PubMed] [Google Scholar]
- Ho MS, Ou C, Chan Y-R, Chien C-T, Pi H. The utility F-box for protein destruction. Cell. Mol. Life Sci. 2008;65:1977–2000. doi: 10.1007/s00018-008-7592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WH, Froncisz W, Hyde JS. Continuous and stopped flow EPR Spectrometer based on a loop gap resonator. Rev. Sci. Instrum. 1987;58:1879–1886. [Google Scholar]
- Jan E, Yoon JW, Walterhouse D, Iannaccone P, Goodwin EB. Conservation of the C. elegans tra-2 3'UTR translational control. EMBO J. 1997;16:6301–6313. doi: 10.1093/emboj/16.20.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR view: A computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Keller RLJ. PhD thesis. Switzerland: ETH Zürich, Zürich; 2005. Optimizing the process of NMR spectrum analysis and computer aided resonance assignment. [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Larocque D, Richard S. Quaking KH domain proteins as regulators of glial cell fate and myelination. RNA Biol. 2005;2:37–40. doi: 10.4161/rna.2.2.1603. [DOI] [PubMed] [Google Scholar]
- Lee M-H, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley M, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Krämer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- Maglio O, Nastri F, Pavone V, Lombardi A, DeGrado WF. Preorganization of molecular binding sites in designed diiron proteins. Proc.Natl.Acad.Sci.USA. 2003;100:3772–3777. doi: 10.1073/pnas.0730771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire ML, Guler-Gane G, Nietlispach D, Raine ARC, Zorn AM, Standart N, Broadhurst RW. Solution structure and backbone dynamics of the KH-QUA2 region of the Xenopus STAR/GSG Quaking protein. J. Mol. Biol. 2005;348:265–279. doi: 10.1016/j.jmb.2005.02.058. [DOI] [PubMed] [Google Scholar]
- Mandel AM, Akke M, Palmer AG., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: Correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- Matsusawa S-I, Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:E6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottinger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Ramos A, Hollingworth D, Major SA, Adinolfi S, Kelly G, Muskett FW, Pastore A. Role of dimerization in KH/RNA complexes: The example of Nova KH3. Biochemistry. 2002;41:4193–4201. doi: 10.1021/bi011994o. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Frater LA, Abramowitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nature Struct. Mol. Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- Saccomanno L, Loushin C, Punkay E, Artzt K, Goodwin EB. The STAR protein QKI-6 is a translational repressor. Proc. Natl. Acad. Sci. USA. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E, Leone M, Li C, Fukushima T, Preece NE, Olson AJ, Ely KR, Reed JC, Pellecchia M, Liddington RC, Matsuzawa S-I. Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J. Biol. Chem. 2005;280:34278–34287. doi: 10.1074/jbc.M506707200. [DOI] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletzky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008a;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR. 2008b;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna BR, Taurog RE, Young MJ, Snyder JC, Johnson JE, Williamson JR. Synergy of NMR, computation, and X-ray crystallography for structural biology. Structure. 2009;17:499–507. doi: 10.1016/j.str.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C-H, Huang J-W, Yang J-M. Kappa-alpha plot derived structural alphabet and BLOSUM-like substitution matrix for fast protein structure database search. Genome Biology. 2007;8:R31.1–R31.16. doi: 10.1186/gb-2007-8-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- Wildes D, Marqusee S. Hydrogen-exchange strategies applied to energetics of intermediate processes in protein folding. Methods in Enzymology. 2004;380:328–349. doi: 10.1016/S0076-6879(04)80015-6. [DOI] [PubMed] [Google Scholar]
- Willis MA, Bishop B, Regan L, Brunger AT. Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core. Structure Fold.Des. 2000;8:1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Masayoshi Y, Imaki H, Koga M, Oshima Y, Nakayaman K-I. Multiple Skp1-Related Proteins in Caenorhabditis elegans: Diverse Patterns of Interaction with Cullins and F-Box Proteins. Current Biology. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Astier M, Gratecos D, Sémériva M. The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development. 1997;124:2087–2098. doi: 10.1242/dev.124.10.2087. [DOI] [PubMed] [Google Scholar]
- Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.