Abstract
Alzheimer's disease (AD) is characterized by extensive neuronal death in distinct brain regions, including the frontal cortex and hippocampus, although the specific mechanisms of neuronal degeneration in AD remain a topic of intense scientific pursuit. One model for cell death in AD postulates that abortive cell cycle events in neurons, including tetraploidy, precede neuronal death, and novel therapeutics based on suppressing cell cycle re-entry are being pursued. Using DNA content fluorescence-activated cell sorting combined with fluorescence in situ hybridization and immunostaining, we analyzed neuronal nuclei from postmortem human brain samples from the frontal cortex and hippocampus of nondiseased and AD patients for evidence of tetraploidy. Here, we show that tetraploid nuclei are similarly prevalent in AD and control brains and are exclusively non-neuronal, contrasting with an absence of tetraploid neurons. Our findings demonstrate that neuronal tetraploidy is nonexistent in the AD brain and intimate a reevaluation of neuronal cell cycle re-entry as a therapeutic target for AD.
Key Words: Alzheimer's disease, Cell cycle, Fluorescence in situ hybridization, Tetraploidy, Fluorescence-activated cell sorting, Neurons, Cerebral cortex, Hippocampus
Introduction
Neurodegenerative diseases are marked by neuronal cell death and loss [1]. In Alzheimer's disease (AD), the selective loss of neurons, particularly in the hippocampus and frontal cortical associative areas, is a feature of the disease [2]. Although a unifying model for neurodegeneration in AD is incomplete, abnormal accumulation of β-amyloid and tau represent leading pathogenic mechanisms [3]. In addition, novel paradigms for cell death in AD, ranging from mitochondrial dysfunction to metal toxicity, may also contribute to the development and progression of AD [4,5]. One current hypothesis for AD pathogenesis postulates that neuronal death is causally linked to aberrant cell-cycle reentry, through a process termed ‘cycle-related neuronal death’ (CRND) [6]. Evidence for CRND includes the observation that some neurons in AD brains are immunoreactive for cell cycle proteins such as PCNA [7,8], and Ki67 [9], as well as several cyclins [10,11,12] and cyclin-dependent kinases (CDKs) [13,14].
An important consequence of the proposed cell cycle re-entry is the existence of tetraploid neurons, which has also been inferred through the observation of tetrasomic nuclei in AD brains without the benefit of neuronal markers [15]. Tetrasomy refers to a chromosomal duplication, usually of a single chromosome pair, contrasting with tetraploidy, which refers to a complete duplication of all chromosomes (2N), resulting in a 4N karyotype. When identified using single chromosome pair fluorescence in situ hybridization (FISH), four signals would be identified for both a tetrasomic and tetraploid cell, yet total DNA content would differ vastly between these two states. This distinction is important in view of the presence of aneuploid cells in the normal brain [16,17], raising the possibility that the inferred tetraploid neurons may actually be tetrasomic. In addition, the neuronal identity of such cells remains unclear. Two corollaries stemming from CRND are that: (1) truly tetraploid neurons must exist in the AD brain, and (2) they should be strikingly more prevalent than in control brains that do not have the same degree of neuronal death. Here we have addressed these corollaries by combining DNA fluorescence-activated cell sorting (FACS), FISH and neuronal immunolabeling on nuclei from human AD brains.
Materials and Methods
Tissue Processing
All human protocols were approved by the Human Subjects Committee at The Scripps Research Institute and conform to National Institutes of Health guidelines and public law. Brain tissue was obtained from the University of Maryland Brain and Tissue Bank (National Institute of Child Health and Human Development NO1-HD-8-3284) and stored at −80°C until use. For this study, we isolated nuclei from 7 (4 cortical and 3 hippocampal) AD and 7 control patients (mean age = 66.3 years old for control and 80.0 for AD). Cortical samples were obtained from the frontal cortex (Broadman's areas 9, 10 and 11), while CA1, CA3 and dentate gyrus fields were present in sagittal sections of the hippocampal samples. AD brain samples were pathologist verified and had Braak scores of V or VI, and control samples had Braak scores of II or less. Samples were thawed at −20°C prior to incubation in Ca2+/Mg2+-free PBS (CMF-PBS) supplemented with 2 mM EGTA for 60 min on ice. Following this, samples were gently triturated using a series of filtered pipet tips with decreasing bore diameter and filtered through 40-μm nylon mesh (BD Biosciences, Bedford, Mass., USA). After centrifugation at 500 g for 5 min at 4°C, nuclei were isolated from the cell pellet as described previously, and fixed for 10 min with ice-cold 1% paraformaldehyde at 4°C. After removal of the fixative, the isolated nuclei were washed twice in CMF-PBS and stored overnight in CMF-PBS with propidium iodide (50 μg/ml). Prior to FACS, nuclei were treated with 50 μg/ml RNAse A for 20 min at 37°C.
FISH Probes
FISH centromere enumeration probes (CEP) against the centromeric sequence of chromosomes 4 or 6 were obtained from Abbott Molecular (Des Plaines, Ill., USA). Locus-specific FISH probes for chromosomes 4, 6, and 21 were generated by nick translation. Briefly, BAC clones containing DNA sequences for human chromosome 4 (RP11-11P12, RP11-7G22, and RP11-185B5), chromosome 6 (RP11-24F12, RP11-91C23), and chromosome 21 (RP11-30D19, RP11-50N20, RP11-48C23, RP11-66H5, CTC-82515) were grown in LB medium supplemented with 12.5 μg/ml chloramphenicol (Sigma Aldrich) overnight at 37°C. BAC DNA was isolated using NucleoBond BAC kits (Clontech, Mountain View, Calif., USA). Amino-allyl dUTP (Sigma Aldrich) was incorporated into template DNA using standard nick translation, to which Alexa Flour 488 was then covalently attached. After labeling and purification using QuiaPrep PCR Purification Kits (Qiagen, Valencia, Calif., USA), a 20-fold excess of human Cot-1 DNA (Invitrogen, Carlsbad, Calif., USA) was added prior to ethanol precipitation. The purified, labeled DNA probe was resuspended in hybridization buffer (2× SSC/50% formamide/10% low-molecular-weight dextran sulfate), and stored at −20°C until use. The high-sequence homology of the chromosome 21 and chromosome 13 centromeres necessitated generation of two locus-specific FISH probes for the q-arm of chromosome 21 (image not shown).
FACS and Immunostaining
Isolated nuclei were sorted using a FACS Aria (Becton Dickinson) flow cytometer after machine calibration with chick erythrocyte nuclei (CEN) (Biosure, Grass Valley, Calif., USA). Unsorted, fraction N (typically composed of 90–95% of all nuclei) and fraction C (typically composed of 5–10% of all nuclei) were sorted into CMF-PBS and then dropped onto precleaned glass slides to allow the liquid to evaporate. Slides were blocked overnight in 2.5% BSA/1% Triton-X 100 PBS at pH 7.4. The primary antibodies NeuN (used at 1:100 dilution; Chemicon, Temecula, Calif., USA) and HuC/D (used at 10 μg/ml; Invitrogen) were detected with an Alexa Fluor 488-labeled donkey anti-mouse secondary antibody (used at 1:500 dilution; Invitrogen).
Fluorescence in situ Hybridization
After sorting and immunostaining, slides were immersed in 2× SSC for 10 min at room temperature. The slides were dehydrated through ethanol, denatured at 75°C for 2 min in denaturation solution (2× SSC/70% formamide), and passed through an ice-cold ethanol dehydration series. DNA probes were denatured at 80°C for 7 min and allowed to reanneal for 60 min before hybridization. After overnight hybridization at 37°C, slides were rinsed in 4× SSC to remove the coverslips, and passed through 2× SSC/50% formamide pH 7.0 for 5 min, 4× SSC 0.1% (v/v) Tween-20 for 2 min, and 2× SSC for 2 min. After counterstaining with DAPI, slides were dehydrated through ethanol and coverslipped with Vectashield antifade solution (Vector Labs, Burlingame, Calif., USA). Slides were stored at −20°C until analysis. For sequential hybridizations, a series of FISH images was acquired using an Olympus IX70 microscope equipped with an automated stage (Applied Precision, Issaquah, Wash., USA), capable of marking the nuclear position on the slide. After removing the first probe set by immersion in 2× SSC/70% formamide pH 7.0 for 2 min at 60°C, the next probe set was applied and FISH images were acquired from the same nuclei. This process was repeated for the third hybridization. For experiments where neuronal immunolabeling was followed by chromosome-specific FISH, a minimum of 500 neuronal (NeuN or HuC/D positive) fraction C nuclei per sample were scored for FISH signals.
Results
Isolation and Characterization of Nuclei from Postmortem Human Brain Samples
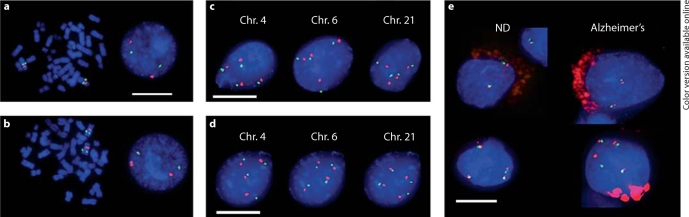
To test the possibility that tetraploid neurons exist in the adult brain and are more prevalent in the AD brain, nuclei from pathologist-verified and sex-matched sporadic AD and nondemented control frontal cortices and hippocampi were isolated, blind to their identity, then sorted based on DNA content to identify tetraploid cells. Nuclei prepared with an optimized propidium iodide staining protocol (including RNAse pretreatment) were sorted by FACS to isolate two pools of nuclei: putative nonmitotic nuclei (fraction ‘N’) with near-diploid DNA contents, and putative S-phase cycling nuclei (fraction ‘C’) with greater-than-diploid DNA contents (fig. 1a, b; N vs. C). To provide a very conservative assessment of potentially tetraploid neurons, fraction C included both the traditional 4N peak as well as populations flanking the 4N peak (including the region of the 2N peak exhibiting the greatest fluorescence), to account for hypothetical technical artifacts that may have missed 4N neurons. Fraction C nuclei were then dropped onto glass slides for further analysis by immunolabeling and FISH. We detected neurons by immunolabeling for the neuronal antigens NeuN (fig. 1c) and HuC/D (fig. 1d), which comprised between 17 and 48% of fraction C nuclei, depending on the sample (fig. 1e). Despite variations in neuronal number between individual brain samples, the percentages of neurons within each sample were similar for NeuN or HuC/D (fig. 2). A truly tetraploid neuron must be present in fraction C, based upon its DNA content. In order to test this possibility, we generated FISH probes for three selected autosomes, chromosomes 4, 6 and 21. For chromosomes 4 and 6, we generated a locus-specific q-arm FISH probe for use in conjunction with a commercially available centromeric probe (fig. 3a, b). The probe set for chromosome 21 consisted of two locus-specific q-arm probes (data not shown). FISH probes were validated for binding specificity by hybridization to interphase and metaphase human lymphocytes (fig. 3a, b). Additional verification of probe specificity was performed by hybridization of locus/centromeric probes in combination with the corresponding whole chromosome paint (data not shown) [16]. FISH analyses of chromosomal copy number in fraction C revealed many nuclei with four signals in all samples analyzed (fig. 3c). Although the percentages of tetrasomic nuclei varied between samples, we did not detect an increase or decrease in the percentage of tetrasomic nuclei in AD brain samples compared to controls for either the frontal cortex or hippocampus (fig. 4). Remarkably, up to 35% of fraction C nuclei were tetrasomic (4–35% range); however, because fraction C comprised around 10% of all brain nuclei (propidium iodide positive and ungated), the overall level of tetrasomy in the brain based on these data would range between 0.4 and 3.5%, consistent with previous reports [16,17]. In order to determine if these nuclei were truly tetraploid (rather than tetrasomic for the chromosomes assayed), we sorted fraction C nuclei by FACS and then sequentially hybridized the same nuclei with probes for these three different chromosomes. We applied a first set (chromosome 4 telomeric and centromeric) of FISH probes to the target nuclei and used deconvolution microscopy to obtain images of tetrasomic nuclei. We next marked their position on the slide, stripped off the first set of probes and applied the second set (chromosome 6), using their marked locations to return to the same nucleus for imaging. The process was repeated for the third set (chromosome 21). In every instance, we found tetrasomic signals for all chromosomes analyzed, in both control and AD brain samples. These data demonstrated that fraction C nuclei were likely tetraploid, based on their DNA content and assayed FISH signals (fig. 3d). By comparison, fraction N contained the expected prevalence of eusomic and aneusomic neurons and nonneuronal cells observed in prior reports [16,17] along with an absence of tetraploid cells (data not shown).
Fig. 1.
FACS and immunolabeling of human brain nuclei. a Human brain nuclei were distributed into nonmitotic (N) and cycling (C, red shading) fractions compared to an internal control population, CEN, using flow cytometry. b An orthogonal scatter plot of the sorted N and C fractions, plotted as cell size (forward scatter width, FSC-W) against DNA content. Neuronal nuclei are detected in fraction C by immunolabeling. Fraction C nuclei stained with DAPI (blue) were immunopositive for NeuN (c) or HuC/D (d; green). e In the frontal cortex (CTX) and hippocampus (HP) of nondiseased (control) and AD brain samples, NeuN and HuC/D were present in between 17 and 48% of all fraction C nuclei depending on the sample. Error bars reflect standard deviations of neuronal nuclei percentages within sample groups. c, d Scale bar = 20 μm. Note: colors only in online version.
Fig. 2.
Percentages of NeuN- and HuC/D-positive nuclei in fraction C of control and AD brains. After FACS and immunolabeling of control (a, b) and AD (c, d) samples, between 300 and 600 nuclei were scored for both NeuN and HuC/D labeling for hippocampal (a, c) or cortical (b, d) samples. Only single nuclei were included. Similar percentages of neurons were detected within each brain sample using each antibody.
Fig. 3.
FISH probes used in this study. Centromeric (red) and locus-specific (green) probes applied to human interphase and metaphase lymphocytes stained with DAPI (blue) for chromosomes 4 (a) and 6 (b; chromosome 21 probes not shown). Apparent ‘split signals’ for telomeric loci are seen in metaphase spreads owing to the resolution of maternal and paternal alleles. c Fraction C contains nuclei that are enriched for tetrasomies for chromosomes 4, 6, and 21 as identified by single-chromosome FISH (representative examples from a control cortex are shown). d Sequential hybridizations for chromosomes 4, 6, and 21 on the same nuclei (from fraction C) supported their identification as tetraploid combined with DNA content analysis. A cortical nucleus from a control sample is shown with the same DAPI layer in each image for clarity. e In brain sections, tetrasomic nuclei (shown for chromosome 21, bottom row) are found in neuron-rich (grey matter) and neuron-poor (white matter) regions of both nondiseased control (ND) and AD brains with morphologies similar to those of neurons (top row, as identified by neuroanatomical position and chromatin structure). ae Scale bar = 10 μm. Note: colors only in online version.
Fig. 4.
Tetrasomy levels in fraction C nuclei. The percentage of tetrasomies in fraction C from individual brains ranged from 4 to 35%. The percentage of tetrasomies within each sample was similar among chromosomes 4, 6, and 21 in control (a, b) and AD (c, d) brains. No difference in tetrasomy levels were detected between control and AD brain regions (hippocampus in a and c, frontal cortex in b and d).
We next asked if tetraploid nuclei had a stereotyped distribution in the nondiseased brain as compared to AD. To determine the neuroanatomical position of these apparent tetraploid nuclei in the brain, we performed FISH in tissue sections, where the detection of tetraploid cells is less likely to be limited by a tissue-sectioning artifact. In the nondiseased cortex, we found tetrasomic nuclei in all cortical layers (1–5/6) as well as in white matter tracts below the ventricular surface. Tetrasomic nuclei showed a similar random pattern of distribution in the nondiseased hippocampus. However, the prevalence and distribution of these cells were indistinguishable between control and AD samples in cortical and hippocampal areas (fig. 3e). The intense red/green fluorescence along the nuclear periphery (fig. 3e) comes from lipofuscin, a broadly autofluorescent metabolic byproduct that accumulates with age in neurons and glial cells of the human brain [18,19].
Sequential Immunolabeling and FISH of Tetraploid Nuclei from Postmortem Human Brain Samples
The finding that neurons and tetraploid cells were present in fraction C prompted us to address whether there was some overlap between these two populations. The possible neuronal identity of tetraploid nuclei was assessed by immunolabeling for the neuron-specific nuclear markers NeuN and HuC/D, combined with FISH to produce nuclei double-labeled for neuronal and chromosomal markers [20,21]. After sorting fraction C nuclei, we performed NeuN or HuC/D immunolabeling as described previously, marking the location of neuronal nuclei using a microscope equipped with an automated stage. These samples were then processed for FISH analysis to identify any tetraploid neurons. Using this approach, we were able to double label fraction C nuclei for a neuronal marker and a chromosome probe. After identity-blind analyses of over 3,500 NeuN or HuC/D immunolabeled nuclei from 14 individuals (control and AD), no tetraploid neuronal nuclei were identified in the cerebral cortex or hippocampus. This result contrasted with hundreds of nonneuronal (NeuN or HuC/D negative) nuclei showing 4 FISH signals in all brain samples analyzed (fig. 5). Thus, despite the high prevalence of tetraploid nuclei in the AD brain, none were demonstrably tetraploid neurons.
Fig. 5.
Tetrasomy is restricted to NeuN- or HuC/D-negative populations in the control and AD brain. Fraction C nuclei (n = 3,655) labeled with the neuronal markers NeuN or HuC/D (green, shown for NeuN) prior to FISH analysis for selected chromosomes (shown for chromosome 6 in red, with white arrows in a). All tetrasomic nuclei were NeuN and HuC/D negative in control (a) or AD (c) frontal cortices as well as control (b) or AD (d) hippocampi. Larger chromosomes (such as shown for chromosome 6) tended to cluster around the nuclear periphery; however, all FISH signals were within the nuclear regions defined by DAPI staining (inset in b). Similar results were seen with FISH for the autosomes 4 and 21 (data not shown). ad Scale bar = 10 μM. Note: colors only in online version.
Discussion
CRND predicts that the AD brain should have vastly more tetraploid neurons. Our data are incompatible with this prediction. All tetraploid nuclei observed, assessed by not only FISH but DNA content FACS and neuronal marker immunolabeling, were nonneuronal along with being indistinguishable with respect to prevalence and distribution between control and AD brains. The absence of tetraploid neurons in fraction C is unlikely to be explained by antigen loss associated with cell death, as NeuN remains detectable even in apoptotic cells [22], while complementary results were obtained using the distinct neuronal antigen, HuC/D. Virtually all neurons in the cortex and hippocampus reportedly express NeuN [21]; however, it is formally possible that some proportion of tetraploid nuclei may be neuronal without expressing this antigen. Nevertheless, even if this unlikely scenario were to exist, our data indicate that such hypothetical tetraploid neurons would not occur specifically in AD.
These data do not address the role of cell cycle proteins reported in postmitotic neurons [reviewed in [6]], although the data do not support their canonical cell cycle function in CRND. This begs the question of what roles cell cycle proteins play in postmitotic neurons. Novel functions for PCNA, cyclins and CDKs have been described beyond their roles in cell cycle progression, notably in DNA damage responses [23,24,25,26]. Given the extent of DNA damage incurred by aging and degenerating neurons in AD, it seems likely that the observed re-expression of many cell cycle proteins in this setting may reflect a DNA repair and maintenance response of vulnerable neurons [27,28,29], or perhaps other as yet unidentified functions distinct from CRND.
AD is a slowly progressing disease, often taking decades to manifest in humans. Estimates regarding the rate of neuronal clearance in AD vary [2,30], and it is conceivable that our analysis failed to detect tetraploidy in neurons because of their rarity. Recent studies using BrdU injections and NeuN FACS have estimated that the maximum number of adult generated neocortical neurons is <0.07% of all neocortical cells [31,32]. These studies did not actually detect any tetraploid neurons, but arrived at this maximum rate by assuming that a theoretical ‘next cell’ in their analysis was a true positive, and then calculating the maximum rate of observable events. A similar analytical treatment of data can be used to determine a detection limit for tetraploid neurons in our study. As a representative example, FACS-sorted fraction C nuclei from the AD hippocampus made up around 5% of the total input nuclei (all propidium iodide-positive non-CEN events). NeuN immunolabeling of fraction C AD hippocampal nuclei revealed that 35% of this population was neuronal (fig. 2). For each sample, we counted a minimum of 500 fraction C neurons [500/0.35 = 1,428 fraction C nuclei (neuronal and nonneuronal) which is 5% of the total number of unsorted nuclei or 1,428/.05 = 28,560 total nuclei]. If the next nucleus analyzed represented a hypothetical tetraploid neuron in fraction C, then this would also represent a maximum observable rate of neuronal tetraploidy at any given time of 1 in 28,571, or 0.003% of all nuclei. A calculation for the AD cortex reveals a similarly low hypothetical maximum rate of neuronal tetraploidy of 0.003%. Therefore, while it remains formally possible that tetraploid neurons were missed in our analysis, these cells would be (1) exceedingly rare and (2) a priori equally probable in both AD and control cases, rendering this possibility nonspecific for AD. There remains a formal possibility that the rate of cell death through a neuronal tetraploid intermediate could be increased early in the disease and cleared by apoptotic/necrotic mechanisms. Our approach of using samples between 63 and 88 years of age might miss these events, although it is notable that this situation is common to many if not most studies using typical AD brain samples. Moreover, it would indicate that even a 25-year time span for our sporadic AD samples is insufficient to capture any of the postulated tetrapolid neurons.
Alternative functions for cell cycle proteins are further suggested by the singular expression of either cyclins or CDKs in the adult AD brain, which may not be sufficient for cell-cycle re-entry since the expression of a cyclin/CDK complex is required for biological activity [33]. Recent studies have suggested that in rodent models of injury-induced neuronal apoptosis and human presenile dementia, DNA synthesis as detected by BrdU incorporation occurs predominately in microglial and endothelial cells, but not in neurons [34,35]. Combined with data presented here, it appears that nonneuronal cells and/or tetrasomic rather than tetraploid neurons could account for prior data supporting CRND. New therapies to treat AD based upon CRND may be potentially viable [36,37]; however, their mechanisms of action would most likely involve nonneuronal cells.
Acknowledgements
This research was supported by a Neuroplasticity of Aging fellowship (J.W.W.) and the NIH (grant MH076145, J.C.). We thank C. Paczkowski for helpful discussion in formulating the manuscript, and we thank Gregory Cox for supplying the FISHTag kits used to generate the fluorescent probes. We also thank Alex Ilic and Cody Fine at The Scripps Research Institute Flow Cytometry Core Facility for help with FACS.
References
- 1.Price DL, Tanzi RE, Borchelt DR, Sisodia SS. Alzheimer's disease: genetic studies and transgenic models. Annu Rev Genet. 1998;32:461–493. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- 2.Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 3.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Mancuso M, Siciliano G, Filosto M, Murri L. Mitochondrial dysfunction and Alzheimer's disease: new developments. J Alzheimers Dis. 2006;9:111–117. doi: 10.3233/jad-2006-9203. [DOI] [PubMed] [Google Scholar]
- 5.Shcherbatykh I, Carpenter DO. The role of metals in the etiology of Alzheimer's disease. J Alzheimers Dis. 2007;11:191–205. doi: 10.3233/jad-2007-11207. [DOI] [PubMed] [Google Scholar]
- 6.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 7.Nagy Z, Esiri MM, Smith AD. Expression of cell division markers in the hippocampus in Alzheimer's disease and other neurodegenerative conditions. Acta Neuropathol. 1997;93:294–300. doi: 10.1007/s004010050617. [DOI] [PubMed] [Google Scholar]
- 8.Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer's disease brain. J Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith MZ, Nagy Z, Esiri MM. Cell cycle-related protein expression in vascular dementia and Alzheimer's disease. Neurosci Lett. 1999;271:45–48. doi: 10.1016/s0304-3940(99)00509-1. [DOI] [PubMed] [Google Scholar]
- 10.Nagy Z, Esiri MM, Cato AM, Smith AD. Cell cycle markers in the hippocampus in Alzheimer's disease. Acta Neuropathol. 1997;94:6–15. doi: 10.1007/s004010050665. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J Neurosci. 2003;23:2557–2563. doi: 10.1523/JNEUROSCI.23-07-02557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoozemans JJ, Bruckner MK, Rozemuller AJ, Veerhuis R, Eikelenboom P, Arendt T. Cyclin D1 and cyclin E are co-localized with cyclo-oxygenase 2 (COX-2) in pyramidal neurons in Alzheimer disease temporal cortex. J Neuropathol Exp Neurol. 2002;61:678–688. doi: 10.1093/jnen/61.8.678. [DOI] [PubMed] [Google Scholar]
- 13.Vincent I, Jicha G, Rosado M, Dickson DW. Aberrant expression of mitotic Cdc2/cyclin B1 kinase in degenerating neurons of Alzheimer's disease brain. J Neurosci. 1997;17:3588–3598. doi: 10.1523/JNEUROSCI.17-10-03588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McShea A, Harris PL, Webster KR, Wahl AF, Smith MA. Abnormal expression of the cell cycle regulators P16 and CDK4 in Alzheimer's disease. Am J Pathol. 1997;150:1933–1939. [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer's disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, Monakhov VV, Soloviev IV. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS ONE. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riga D, Riga S. Lipofuscin and ceroid pigments in aging and brain pathology. A review. I. Biochemical and morphological properties. Rom J Neurol Psychiatry. 1995;33:121–136. [PubMed] [Google Scholar]
- 19.Braak E. On the fine structure of the external glial layer in the isocortex of man. Cell Tissue Res. 1975;157:367–390. doi: 10.1007/BF00225527. [DOI] [PubMed] [Google Scholar]
- 20.Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 21.Mullen RJ, Buck CR, Smith AM. Neun, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 22.Bengzon J, Kokaia Z, Elmer E, Nanobashvili A, Kokaia M, Lindvall O. Apoptosis and proliferation of dentate gyrus neurons after single and intermittent limbic seizures. Proc Natl Acad Sci U S A. 1997;94:10432–10437. doi: 10.1073/pnas.94.19.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 24.Park DS, Morris EJ, Bremner R, Keramaris E, Padmanabhan J, Rosenbaum M, Shelanski ML, Geller HM, Greene LA. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons evoked by DNA damage. J Neurosci. 2000;20:3104–3114. doi: 10.1523/JNEUROSCI.20-09-03104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazumder S, Plesca D, Almasan A. A Jekyll and Hyde role of cyclin E in the genotoxic stress response: switching from cell cycle control to apoptosis regulation. Cell Cycle. 2007;6:1437–1442. doi: 10.4161/cc.6.12.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, Carrington M, Koeffler HP, Restle A, Wiesmuller L, Sobczak-Thepot J, Berdel WE, Serve H. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer's disease. Nucleic Acids Res. 2007;35:7497–7504. doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subba Rao K. Mechanisms of disease: DNA repair defects and neurological disease. Nat Clin Pract Neurol. 2007;3:162–172. doi: 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- 29.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 30.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease. Lancet. 1994;344:769–772. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 31.Bhardwaj RD, Curtis MA, Spalding KL, Buchholz BA, Fink D, Bjork-Eriksson T, Nordborg C, Gage FH, Druid H, Eriksson PS, Frisen J. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 33.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 34.Bauer S, Patterson PH. The cell cycle-apoptosis connection revisited in the adult brain. J Cell Biol. 2005;171:641–650. doi: 10.1083/jcb.200505072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boekhoorn K, Joels M, Lucassen PJ. Increased proliferation reflects glial and vascular-associated changes, but not neurogenesis in the presenile Alzheimer hippocampus. Neurobiol Dis. 2006;24:1–14. doi: 10.1016/j.nbd.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Lu KP, Zhou XZ. The prolyl isomerase pin1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 37.Arendt T, Holzer M, Stobe A, Gartner U, Luth HJ, Bruckner MK, Ueberham U. Activated mitogenic signaling induces a process of dedifferentiation in Alzheimer's disease that eventually results in cell death. Ann N Y Acad Sci. 2000;920:249–255. doi: 10.1111/j.1749-6632.2000.tb06931.x. [DOI] [PubMed] [Google Scholar]