Abstract
Background
Accumulation of amyloid β (Aβ) in the brain is believed to represent one of the earliest events in the Alzheimer disease process. Aβ is generated from amyloid precursor protein after sequential cleavage by β- and γ-secretase. Alternatively, α-secretase cleaves within the Aβ sequence, thus, precluding the formation of Aβ. A lot of research has focused on Aβ production, while less is known about the non-amyloidogenic pathway. We have previously shown that Aβ is present in human cerebrospinal fluid (CSF) as several shorter C-terminal truncated isoforms (e.g. Aβ1–15 and Aβ1–16), and that the levels of these shorter isoforms are elevated in media from cells that have been treated with γ-secretase inhibitors. Objective:To explore the effect of N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester (DAPT), a γ-secretase-inhibitor, treatment on the Aβ isoform pattern in brain tissue and CSF from Tg2576 mice.
Methods
Immunoprecipitation using the anti-Aβ antibodies 6E10 and 4G8 was combined with either matrix-assisted laser desorption/ionization time-of-flight mass spectrometry or nanoflow liquid chromatography and tandem mass spectrometry.
Results
All fragments longer than and including Aβ1–17 displayed a tendency towards decreased levels upon γ-secretase inhibition, whereas Aβ1–15 and Aβ1–16 indicated slightly elevated levels during treatment.
Conclusion
These data suggest that Aβ1–15 and Aβ1–16 may be generated through a third metabolic pathway independent of γ-secretase, and that these Aβ isoforms may serve as biomarkers for secretase inhibitor treatment.
Key Words: Alzheimer's, Amyloid β, Mass spectrometry, γ-Secretase, Immunoprecipitation, Transgenic mouse
Introduction
Alzheimer's disease (AD) is the most common form of dementia, with characteristic pathological hallmarks that include intracellular neurofibrillary tangles and extracellular senile plaques [1]. The plaques consist mainly of β-amyloid (Aβ) [2,3,4], which is generated by cleavage of amyloid precursor protein (APP) by 2 different enzymes, β-and γ-secretase known as the amyloidogenic pathway [5]. In the alternative ‘non-amyloidogenic pathway’, α-secretase cleaves within the Aβ sequence precluding the formation of Aβ [6,7,8]. The β-site APP-cleaving enzyme 1 gene (BACE1) encodes the β-secretase activity [9,10,11,12], while γ-secretase is a protease complex consisting of at least 4 essential components: the homologous presenilins 1 and 2 (PS1 and 2), nicastrin, Aph-1 and Pen-2 [13]. The α-secretase candidates so far identified all belong to the ‘A Disintegrin and Metalloprotease’ (ADAM) family (ADAM9, ADAM10 and ADAM17) [14].
We and others have previously shown that the combination of immunoprecipitation (IP) and mass spectrometry (MS) is a useful analytical method in targeted Aβ proteomics for simultaneous identification and quantification of Aβ isoforms with high mass accuracy in a single analysis [15]. Using IP-MS, it has been shown that Aβ is manifested in several isoforms in the human brain and cerebrospinal fluid (CSF), having both C- and N-terminal truncations [16,17]. Further, IP-MS has also been used for analyzing the Aβ isoform pattern in the brains of transgenic (Tg) mice and Aβ isoforms in cell media [18,19].
We have recently shown that the levels of shorter Aβ isoforms (Aβ1–14, Aβ1–15 and Aβ1–16) are elevated in media from cells which have been treated with γ-secretase inhibitors [20]. Here, we analyze the effects of γ-secretase inhibition on the Aβ isoform pattern in CSF and the brain in a Tg mouse model of AD, Tg2576 [21]. We show that all Aβ isoforms longer than and including Aβ1–17 are γ-secretase dependent, whereas the shorter isoforms Aβ1–15 and Aβ1–16 are processed through a novel APP metabolic pathway.
Material and Methods
Experimental Animals
In this study, we used Tg2576 mice overexpressing APP with Swedish mutations (hAPP695.SWE) on a C57B6/SJL background [21]. Male Tg2576 mice were mated with female C57B6/SJL wild-type mice. Offspring were genotyped as described previously [22], and were used as hemizygotes (+/–) of APP and wild-type mice.
Treatment Protocol
The protocols for mouse treatment and sample collection were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania. Six-month-old Tg2576 mice were subcutaneously injected with either N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester (DAPT, 100 mg/kg, Calbiochem, Calif., USA), a γ-secretase inhibitor, or corn oil in 5% ethanol as control (n = 5/group, all females). Mouse CSF and brain tissues were collected 6 h after treatment.
CSF Collections
To collect CSF, mice were anesthetized by an intraperitoneal injection of ketamine hydrochloride (100 mg/kg) and xylazine (10 mg/kg). The skin and muscle above the cisterna magna were exposed and the meninges were punctured using a 30-gauge needle. CSF samples were aspirated from just below the surface of the punctured meninges using a P20 micropipette. The samples were stored at −80°C as described earlier [23,24].
Mouse Brain Tissue Collection
After CSF collection, the mice were decapitated after lethal anesthesia. Different regions of the exposed brains were dissected, i.e. the cortex, hippocampus, cerebellum and remaining regions. All brain samples were stored at −80°C until further study, as described earlier [25].
Aβ Extraction
The brain samples (30–120 mg) were homogenized (Pellet Pestle®, Sigma-Aldrich, St. Louis, Mo., USA) on ice in Tris-buffered saline (20 mM Tris, 137 mM NaCl, pH 7.6) with complete protease inhibitor tablets (Roche, Basel, Switzerland). The extraction ratio (brain tissue: Tris-buffered saline) was 1:5 (w/v). Formic acid (FA) was added to the sample (final concentration 70%) followed by sonication (power: 15 amplitude microns; tune: ‘middle’) and centrifugation at 30,000 g for 1 h at 4°C. The FA-soluble Aβ extract was dried and dissolved in FA, and finally neutralized using 0.5 M Tris.
Immunoprecipitation and Mass Spectrometry
IP using the KingFisher magnetic particle processor (Thermo Scientific, Waltman, Mass., USA) and mass spectrometric analysis using MALDI-TOF MS were performed as described earlier [17]. Briefly, an aliquot (8 μg) of the anti-Aβ antibody 6E10 (Signet Laboratories, Dedham, Mass., USA), which is reactive to amino acids 1–17, was added to 50 μl magnetic Dynabeads M-280 Sheep Anti-Mouse IgG (Invitrogen, Carlsbad, Calif., USA). The antibody-coated beads were added to 5–10 μl CSF, and diluted to 1 ml in 0.025% Tween 20 in phosphate-buffered saline (pH 7.4). After washing, using the KingFisher magnetic particle processor, the Aβ isoforms were eluted using 100 μl 0.5% FA. The neutralized Aβ extract from brain tissue was immunoprecipitated by adding 50 μl 6E10-coated beads (8 μg) and 50 μl 4G8-coated beads (8 μg, Signet, reactive to amino acid 17–24).
MALDI-TOF MS measurements were performed using an Autoflex instrument (Bruker Daltonics, Bremen, Germany) operating in linear mode at 19 kV acceleration voltage. Each spectrum represents an average of 1,500 shots acquired 75 at a time. The MALDI samples were prepared with the seed layer method using α-cyano-4-hydroxycinnamic acid as the matrix.
LC-MS/MS was conducted by nanoflow liquid chromatography coupled to electrospray ionization Fourier transform ion cyclotron resonance tandem mass spectrometry (LC-ESI-FTICR-MS/MS) with an Ettan MDLC (GE Healthcare, Uppsala, Sweden) coupled to an LTQ-FT (ThermoFisher Scientific, Bremen, Germany), a hybrid linear quadrupole ion trap-Fourier transform ion cyclotron resonance mass spectrometer equipped with a 7-T magnet, as described previously [26].
Results
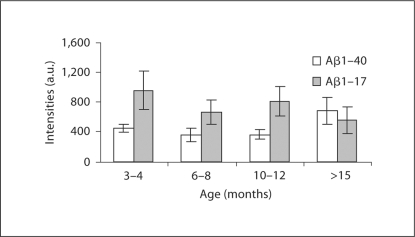
To test whether the IP-MS method using MALDI-TOFMS was sufficiently sensitive for analyzing such small volumes as 5–10 μl CSF, a pilot study was conducted using untreated Tg2576 mice ranging in age from 3–4 months up to older than 15 months. We analyzed 4 age groups with 3 mice in each group. Using IP-MS, 6 different peaks at different mass-to-charge ratios (m/z) were reproducibly detected, which by mass were assigned to be 6 isoforms of Aβ (Aβ1–15, Aβ1–16, Aβ1–17, Aβ1–19, Aβ1–38 and Aβ1–40). The data revealed an age-dependent shift in the Aβ pattern; the older Tg2576 mice produced more Aβ1–40 and less Aβ1–17 than the younger animals (fig. 1). Mice aged 6–7 months were used for the γ-secretase-inhibitor treatment study.
Fig. 1.
MALDI-TOFMS intensities for Aβ117 and Aβ140. The mass spectrometric signals for Aβ117 were divided by 3.
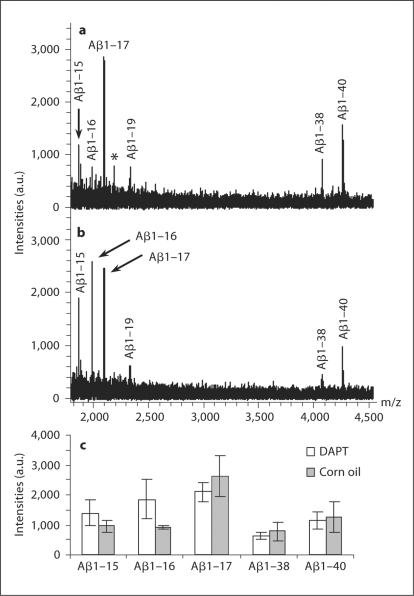
Using IP-MS, a study including 5 mice treated with γ-secretase inhibitor (DAPT) and 5 treated with corn oil (vehicle) was performed. All 6 isoforms were reproducibly detected in the CSF of all mice. As seen in figure 2a,b, treatment with DAPT greatly increased the mass spectrometric signal for Aβ1–16 (p < 0.01, Mann-Whitney U exact test). The signal corresponding to Aβ1–15 showed a tendency towards increased levels, while the signals for the longer isoforms, including Aβ1–17, indicated slightly decreased levels (fig. 2c).
Fig. 2.
MALDI-TOF mass spectra showing the Aβ isoform pattern in CSF from vehicle (a) and γ-secretase-inhibitor treated (DAPT; b) mice. * Unidentified peak. The average MALDI-TOF mass spectrometric signals for the detected Aβ isoforms are displayed from the 2 different treatment groups (c).
From the same mice included in the CSF study, the hippocampus, cortex and cerebellum were analyzed using IP and MALDI-TOFMS to study changes in the Aβ isoform pattern in response to γ-secretase inhibition. The detected Aβ isoforms were also confirmed using LC-MS/MS (data not shown). Figure 3a–b displays MALDI-TOF mass spectra from FA-extracted Aβ isoforms from the hippocampus, where Aβ1–15, Aβ1–17 Aβ1–19, Aβ1–38, Aβ1–40 and Aβ1–42 were reproducibly detected in all mice. Aβ1–16 was not detected in all samples, and was omitted from the evaluation. The same trend as for CSF was noted; the short Aβ isoforms indicated slightly increased levels, while isoforms longer than and including Aβ1–17 showed a tendency towards decreased levels (fig. 3c). Similar data were obtained in the cerebellum (fig. 3d) and cortex (fig. 3e), although the number of reproducibly detected Aβ isoforms differed between the different brain regions.
Fig. 3.
MALDI-TOF mass spectra showing the Aβ isoform pattern in the hippocampus from γ-secretase inhibitor- (DAPT; a) and vehicle- (b) treated mice. * Unidentified peaks. The average MALDI-TOF mass spectrometric signals for the detected Aβ isoforms are displayed from the 2 different treatment groups in the hippocampus (c), cerebellum (d) and cortex (e).
Discussion
Using MALDI-TOFMS measurement on 6E10 immunoprecipitated Aβ from CSF, a distinct pattern of 6 different peaks at different m/z was detected in both treated and untreated mice. They were, by mass, assigned to be 6 isoforms of Aβ(Aβ1–16, Aβ1–17, Aβ1–18, Aβ1–19, Aβ1–38 and Aβ1–40). In human CSF, more than 20 different Aβ isoforms have been detected [15], while in this study on mouse CSF, 6 isoforms were reproducibly detected. The fewer Aβ isoforms detected in mouse CSF probably reflects the small volume of CSF available for analysis (5–10 μl, compared with 1 ml in human CSF studies).
Analysis of the Aβ isoform pattern in CSF from mice of varying ages indicated an age-dependent increase in the longer and more aggregation-prone Aβ peptides. If verified in human studies, this may provide an explanation for the strong age-dependency of amyloid pathology in the brain.
The isoform patterns varied in different brain regions. In the IP extract from approximately 30 mg hippocampus, 6 different isoforms were detected compared with only 3 in the cortex (93–115 mg). This may be due to either the method used not being sufficiently sensitive for detecting low abundant isoforms or that the shorter isoforms (Aβ1–15, Aβ1–17 and Aβ1–19) are absent in the cortex. Further, by normalizing the peaks to the sum of the intensities of the 3 most abundantly detected isoforms in all brain regions (Aβ1–38, Aβ1–40 and Aβ1–42), the cortex displayed an approximate 2-fold increase in the Aβ1–40/Aβ1–42 ratio (in both DAPT and untreated). Aβ1–42 also had the highest mass spectrometric signal in the cortex. It should be noted that the ratio between the Aβ1–42 and Aβ1–40 peaks in the mass spectrum cannot be interpreted as a direct reflection of their relative abundance since the ionization efficiency might be different for the 2 peptides and that Aβ1–42 is more hydrophobic and less soluble than Aβ1–40. However, these data suggest that Aβ1–42 is present at higher levels in the cortex relative to the other brain areas studied.
In the two APP processing pathways described in the literature, the ‘amyloidogenic’ and ‘non-amyloidogenic’, either β- or α-secretase cleaves APP followed by γ-secretase-mediated cleavage of the remaining C-terminal APP fragment [27]. We show here that CSF Aβ1–15 and Aβ1–16 indicated slightly increased levels, whereas the levels of all fragments longer than and including Aβ1–17 were slightly decreased in response to γ-secretase inhibitor treatment. These data are in agreement with previous results on γ-secretase-inhibitor-treated cells [20]. Similar results were obtained using IP-MS on formic-acid-extracted Aβ isoforms from the hippocampus, cerebellum and cortex. This suggests that all fragments longer than and including Aβ1–17 depend on γ-secretase, directly or indirectly, while Aβ1–15 and Aβ1–16 do not. These shorter Aβ isoforms may be processed along a third APP-processing pathway involving concerted β- and α-secretase-mediated cleavages of APP. Additional knowledge on this putative third APP processing pathway may be gained from ongoing secretase inhibitor studies in humans.
Acknowledgements
This work was supported by grants from the Swedish Research Council (projects 2006-6227, 2006-2740 and 14002), Alzheimer's Association (award number NIRG-08-90356), cNEUPRO, the Sahlgrenska University Hospital, the Inga-Britt and Arne Lundberg Research Foundation, the Gothenburg Medical Society, Swedish Brain Power, Stiftelsen Gamla Tjänarinnor, the Åke Wiberg Foundation, Gun och Bertil Stohnes Stiftelse, Alzheimer Foundation, Sweden, NIH (AG17586, AG17586, AG10124).
References
- 1.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 3.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong CW, Quaranta V, Glenner GG. Neuritic plaques and cerebrovascular amyloid in Alzheimer disease are antigenically related. Proc Natl Acad Sci USA. 1985;82:8729–8732. doi: 10.1073/pnas.82.24.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreasson U, Portelius E, Andersson ME, Blennow K, Zetterberg H. Aspects of beta-amyloid as a biomarker for Alzheimer's disease. Biomark Med. 2007;1:59–78. doi: 10.2217/17520363.1.1.59. [DOI] [PubMed] [Google Scholar]
- 6.Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ. Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science. 1990;248:1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 7.Haass C, Hung AY, Schlossmacher MG, Teplow DB, Selkoe DJ. Beta-amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J Biol Chem. 1993;268:3021–3024. [PubMed] [Google Scholar]
- 8.Haass C, Schlossmacher MG, Hung AY, Vigo-Pelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB, et al. Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature. 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 9.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G. Identification of a novel aspartic protease (ASP 2) as beta-secretase. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 10.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 11.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 12.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B. Aph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 14.Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMS family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 15.Portelius E, Zetterberg H, Gobom J, Andreasson U, Blennow K. Targeted proteomics in Alzheimer's disease: focus on amyloid-beta. Expert Rev Proteomics. 2008;5:225–237. doi: 10.1586/14789450.5.2.225. [DOI] [PubMed] [Google Scholar]
- 16.Piccini A, Russo C, Gliozzi A, Relini A, Vitali A, Borghi R, Giliberto L, Armirotti A, D'Arrigo C, Bachi A, Cattaneo A, Canale C, Torrassa S, Saido TC, Markesbery W, Gambetti P, Tabaton M. Beta-amyloid is different in normal aging and in Alzheimer disease. J Biol Chem. 2005;280:34186–34192. doi: 10.1074/jbc.M501694200. [DOI] [PubMed] [Google Scholar]
- 17.Portelius E, Tran AJ, Andreasson U, Persson R, Brinkmalm G, Zetterberg H, Blennow K, Westman-Brinkmalm A. Characterization of amyloid beta peptides in cerebrospinal fluid by an automated immunoprecipitation procedure followed by mass spectrometry. J Proteome Res. 2007;6:4433–4439. doi: 10.1021/pr0703627. [DOI] [PubMed] [Google Scholar]
- 18.Philipson O, Hammarstrom P, Nilsson KP, Portelius E, Olofsson T, Ingelsson M, Hyman BT, Blennow K, Lannfelt L, Kalimo H, Nilsson LN. A highly insoluble state of Abeta similar to that of Alzheimer's disease brain is found in Arctic APP transgenic mice. Neurobiol Aging. 2009;30:1393–1405. doi: 10.1016/j.neurobiolaging.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Sweeney D, Gandy SE, Sisodia SS. The profile of soluble amyloid beta protein in cultured cell media: detection and quantification of amyloid beta protein and variants by immunoprecipitation-mass spectrometry. J Biol Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- 20.Portelius E, Price E, Brinkmalm G, Stiteler M, Olsson M, Persson R, Westman-Brinkmalm A, Zetterberg H, Simon AJ, Blennow K: A novel pathway for amyloid precursor protein processing. Neurobiol Aging 2009, E-pub ahead of print. [DOI] [PubMed]
- 21.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 22.Uryu K, Laurer H, McIntosh T, Pratico D, Martinez D, Leight S, Lee VM, Trojanowski JQ. Repetitive mild brain trauma accelerates Abeta deposition, lipid peroxidation, and cognitive impairment in a transgenic mouse model of Alzheimer amyloidosis. J Neurosci. 2002;22:446–454. doi: 10.1523/JNEUROSCI.22-02-00446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cirrito JR, May PC, O'Dell MA, Taylor JW, Parsadanian M, Cramer JW, Audia JE, Nissen JS, Bales KR, Paul SM, DeMattos RB, Holtzman DM. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J Neurosci. 2003;23:8844–8853. doi: 10.1523/JNEUROSCI.23-26-08844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMattos RB, Bales KR, Parsadanian M, O'Dell MA, Foss EM, Paul SM, Holtzman DM. Plaque-associated disruption of CSF and plasma amyloid-beta (Abeta) equilibrium in a mouse model of Alzheimer's disease. J Neurochem. 2002;81:229–236. doi: 10.1046/j.1471-4159.2002.00889.x. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara T, Hong M, Zhang B, Nakagawa Y, Lee MK, Trojanowski JQ, Lee VM. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron. 1999;24:751–762. doi: 10.1016/s0896-6273(00)81127-7. [DOI] [PubMed] [Google Scholar]
- 26.Portelius E, Brinkmalm G, Tran A, Zetterberg H, Westman-Brinkmalm A, Blennow K. Identification of novel APP/Abeta isoforms in human cerebrospinal fluid. Neurodegener Dis. 2009;6:87–94. doi: 10.1159/000203774. [DOI] [PubMed] [Google Scholar]
- 27.Selkoe DJ, Schenk D. Alzheimer's disease: molecular understanding predicts amyloid-based therapeutics. Annu Rev Pharmacol Toxicol. 2003;43:545–584. doi: 10.1146/annurev.pharmtox.43.100901.140248. [DOI] [PubMed] [Google Scholar]