Summary
We have generated a transgenic mouse line, Tg(Stra8-cre)1Reb (Stra8-cre), which expresses improved Cre recombinase under the control of a 1.4 Kb promoter region of the germ cell-specific stimulated by retinoic acid gene 8 (Stra8). cre is expressed only in males beginning at postnatal day (P)3 in early-stage spermatogonia, and is detected through pre-leptotene-stage spermatocytes. To further define when cre becomes active, we crossed Stra8-cre males with Tg(ACTB-Bgeo/GFP)21Lbe (Z/EG) reporter females and compared the expression of Enhanced Green Fluorescent Protein (EGFP) with the protein encoded by the zinc finger and BTB domain containing 16 (Zbtb16) gene, PLZF – a marker for undifferentiated spermatogonia. Co-expression of EGFP is observed in the majority of PLZF+ cells. We also tested recombination efficiency by mating Stra8-cre;Z/EG males and females with wild-type mice and examining EGFP expression in the offspring. Recombination is detected in >95% of Z/EG+ pups born to Stra8-cre;Z/EG fathers but in none of the offspring born to transgenic mothers, a verification that cre is not functional in females. The postnatal, premeiotic, male germ cell-specific activity of Stra8-cre makes this mouse line a unique resource to study testicular germ cell development.
Keywords: Cre recombinase, Stra8 promoter, spermatogonia, spermatocytes, Z/EG
These experiments were initiated to produce a transgenic mouse line that expresses cre in undifferentiated spermatogonia. To date, the use of cre-mediated recombination to inactivate genes in developing germ cells at specific stages has been limited by the restricted expression patterns of available cre drivers. Recombination in primordial germ cells is possible using the alkaline phosphatase, liver/bone/kidney Alpltm1(cre)Nagy mouse line (Lomeli et al., 2000). However, significant developmental events occur between the onset of cre at embryonic day (E)9.5 and the appearance of spermatogonia in the postnatal testis. Additional cre activity is detected in the placenta, intestine, and neural tube. Excision of floxed DNA in primary spermatocytes and elongating spermatids is achievable using synaptonemal complex protein 1 Tg(Sycp1-cre)4Min (Vidal et al., 1998) and protamine 1 Tg(Prm-cre)58Og (O'Gorman et al., 1997), respectively, but the temporal expression of these transgenes is too late to be of use in undifferentiated spermatogonia. Meanwhile, the expression of cre driven by the growth differentiation factor 9 (Gdf9) and zona pellucida 3 (Zp3) promoters is restricted to developing oocytes (Lan et al., 2004; Lewandoski et al., 1997).
We therefore selected the promoter of the premeiotic male and female germ cell-specific gene Stra8 to drive expression of improved Cre recombinase. Endogenous Stra8 is first expressed in ovarian germ cells at E12.5 and continues until E16.5, while in males it is first transcribed in early-stage spermatogonia in the postnatal testis and persists in premeiotic germ cells throughout adulthood (Menke et al., 2003; Oulad-Abdelghani et al., 1996). A 1.4 Kb Stra8 promoter fragment was recently fused to EGFP coding sequence, and the resulting transgene was shown to be a marker for spermatogonial stem cells (SSCs), a subpopulation of undifferentiated spermatogonia (Nayernia et al., 2004). We have used this same 1.4 Kb promoter region to generate Stra8-cre by inserting it upstream of the improved cre coding sequence, shown to exhibit improved expression over the conventional prokaryotic cre (Shimshek et al., 2002).
The structure of the transgene is depicted in Figure 1a. After pronuclear injection of the transgenic construct and examination of resulting offspring (data not shown), one male was found to exhibit germline expression of Stra8-cre. This male was used to expand the line, and embryos were isolated from subsequent transgenic matings at two stages, E14.5 and E16.5, to assess transcriptional activity by RT-PCR (Fig. 1b). Surprisingly, none of the female embryos showed any cre mRNA expression during the window when endogenous Stra8 is active in the female. Male embryos, as expected, did not express the transgene. When testes were examined for cre transcript at E18, P3, and P7 (Fig. 1c), expression was first detected at P3 and became much stronger by P7. In P7 pups, various organs were isolated to assess the specificity of Stra8-cre expression (Fig. 1d). Of the eight organs examined, only testis showed the cre transcript. Postnatal ovaries, as with embryonic females, exhibited no cre expression, suggesting that the transgene is non-functional in females. To examine Stra8-cre protein expression in P7 males, anti-cre antibody was used on testis cross-sections (Fig. 1e). At this early stage of postnatal development, cre protein was detected in the majority of spermatogonia within the seminiferous tubules.
FIG. 1.
Testis-specific expression of Cre recombinase mRNA in postnatal Stra8-cre transgenic mice. (a) Schematic diagram of the transgene construct. A 1.4 Kb promoter fragment of Stra8 is inserted upstream of the 1.26 Kb cre coding sequence, separated by the SV40 splice donor/splice acceptor. -1400 and +7 denote the nucleotide positions of the Stra8 fragment, and restriction endonuclease recognition sites utilized in the construction of the 3.1 Kb transgene are listed. (b) cDNA from male and female embryos isolated at E14.5 and E16.5 was analyzed by PCR using cre and βActin primers. No cre band was detected in any of the males or females (denoted by symbols) at either time point. (c) cDNA from testes isolated at E18, P3, and P7 was analyzed by PCR for cre and βActin. A faint cre band was detected at P3, with a much stronger band at P7; – RT, no reverse transcription control. (d) cDNA from different organs isolated from P7 transgenic offspring was analyzed by PCR for cre and βActin: B, brain; H, heart; K, kidney; L, lung; S, spleen; T, testis; O, ovary; -RT, no reverse transcription on testis RNA. Only the testis shows a specific cre band. Faint, non-specific product was amplified from lung cDNA. (e) Cross-section of P7 transgenic seminiferous tubules. Cre protein is detected by anti-cre antibody (green); DNA is labeled with DAPI (blue).
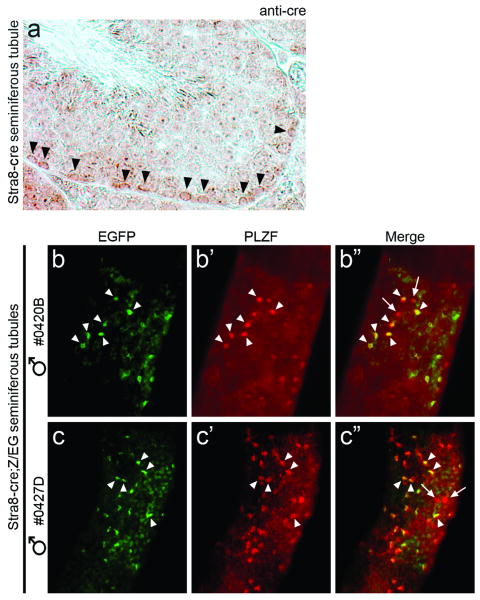
Immunohistochemistry was also performed on adult testis (Fig. 2a). Cre was observed in spermatogonia and pre-leptotene spermatocytes residing along the basement membrane. As positive staining was seen in different stages of the seminiferous epithelial cycle (Russell, 1990), we conclude that Stra8-cre expression begins in Type A spermatogonia and ends in pre-leptotene spermatocytes, the last developmental stage before the onset of meiosis. To determine whether Cre recombinase functions in undifferentiated spermatogonia, a subset of Type A, we mated Stra8-cre males to Z/EG reporter females that constitutively express a floxed Bgeo until cre-mediated excision, after which EGFP becomes active (Novak et al., 2000). Seminiferous tubules of Stra8-cre;Z/EG males were examined by whole-mount immunocytochemistry for the co-expression of EGFP and PLZF, a marker of undifferentiated spermatogonia (Buaas et al., 2004; Costoya et al., 2004). As shown in Figures 1b-c″, the majority of PLZF+ cells also expressed EGFP, indicating Stra8-cre activity. Additional, fainter EGFP+ cells that lack PLZF staining represent more differentiated germ cells (Figs. 1b″,c″). Interestingly, some PLZF+ spermatogonia did not exhibit EGFP, suggesting that cre becomes active in a subpopulation of undifferentiated spermatogonia.
FIG. 2.
Initial Stra8-cre activity is detected in a subset of undifferentiated spermatogonia and its expression is observed through pre-leptotene stage spermatocytes. (a) Cross-section of a Stage VII seminiferous tubule isolated from a 6-week-old Stra8-cre mouse. Arrowheads point to pre-leptotene spermatocytes that express Stra8-cre, detected by anti-cre antibody. (b-c″) Whole-mount immunostaining of seminiferous tubules isolated from two different 2.5-week-old Stra8-cre;Z/EG mice, offspring resulting from a Stra8-cre male × Z/EG female cross. Arrowheads denote undifferentiated spermatogonia expressing the transcriptional repressor PLZF (red) that also express EGFP (green), a result of the excision of the Bgeo gene from the Z/EG allele by Stra8-cre (Merge, yellow). Arrows in b″ and c″ point to PLZF+ spermatogonia that do not express EGFP.
Finally, we tested recombination efficiency by mating Stra8-cre;Z/EG males with wild-type females, and Stra8-cre;Z/EG females with wild-type males. Offspring produced from these matings were examined for constitutive EGFP expression that would result from the transmission of the recombined Z/EG allele through transgenic sperm or eggs. Table 1 reveals that of the pups born to Stra8-cre;Z/EG fathers, >95% receiving the Z/EG allele exhibited recombination. In contrast, none of the pups born to Stra8-cre;Z/EG mothers receiving Z/EG showed recombination (Table 1). This finding verifies that cre is non-functional in females.
Table 1. Assessment of Stra8-cre recombination efficiency on the Z/EG target allele.
| breeding pair | # of offspring | % recombination in offspring | ||
|---|---|---|---|---|
| EGFP+ | lacZ+ | |||
| ♂ #2435 | ♀ | 66 | 2 | 97% |
| ♂ #2562 | ♀ | 17 | 0 | 100% |
| ♂ #2559 | ♀ | 25 | 1 | 96% |
| ♀ #3113 | ♂ | 0 | 16 | 0% |
| ♀ #2818 | ♂ | 0 | 12 | 0% |
♂ = male; ♀ = female; Z/EG = Tg(ACTB-BGeo/GFP)21Lbe
Exactly why Stra8-cre is male germ cell-specific is not clear. The promoter region used in this study contains both retinoic acid receptor-binding elements identified previously (Giuili et al., 2002); it is the same fragment that generated the Stra8-EGFP transgene, demonstrated to be a marker for SSCs (Nayernia et al., 2004). It is likely that the transgene lacks key enhancers important for expression in females. Nonetheless, these experiments have yielded a cre driver that will facilitate the assessment of gene function in postnatal, premeiotic male germ cells.
Materials and Methods
Generation of Transgenic Stra8-cre Mice
First, the -1400/+7 fragment of Stra8 genomic DNA (Nayernia et al., 2004), where +1 is the transcription start site, was isolated from FVB/NJ mouse DNA and amplified using PCR primers designed with an AvrII recognition site at the 5′ end of the forward primer, gStra8F1avrII (5′-TCT CCT AGG AAC TTG CCT CCA AGG GGG TAG G-3′), and an XhoI recognition site at the 5′ end of the reverse primer, gStra8R1xhoI (5′-TGC TCG AGA CGA CTG CCC GTC GCA GAA TAA G-3′). The resulting product was T/A cloned into pGEMTeasy vector, followed by excision using AvrII and XhoI. This fragment was then substituted for the GnRH promoter in the plasmid pL29mGnRH.iCre (Shimshek et al., 2002) via ligation following the excision of the GnRH fragment using AvrII and XhoI. Because pL29mGnRH.iCre contains two XhoI recognition sites, this ligation was performed in two steps. First, the Stra8 promoter was ligated to the plasmid vector backbone to produce a 4.39 Kb product. Second, the sequence between the two XhoI sites in the plasmid vector comprising the cre coding region (∼1.26 Kb) was amplified by PCR, T/A cloned into pGEMTeasy vector, digested with XhoI, and ligated to the XhoI-treated fragment that contained the Stra8 promoter. This ligation product, called pL29Stra8.iCre (∼5.65 Kb), was used to transform DH5α cells, and the plasmids from the resulting colonies were sequenced to verify proper insertion and orientation of fragments.
Finally, a 3.1 Kb fragment excised by AvrII and BsiWI from pL29Stra8.iCre was purified and microinjected into the pronuclei of fertilized eggs of FVB mice. The resulting transgenic mouse line was named FVB/N-Tg(Stra8-cre)1Reb (Stra8-cre). This line of Stra8-cre has been given to The Jackson Laboratory and JAX® Mice for distribution, with JAX Stock # 008208. For some experiments, Stra8-cre mice were crossed with STOCK Tg(ACTB-Bgeo/GFP)21Lbe/J (Z/EG) reporter mice obtained from The Jackson Laboratory. Wild-type FVB/NJ mice were also used in specific matings.
RT-PCR for cre mRNA Expression
Embryos were dissected from female mice at E14.5, E16.5, and E18 following timed matings of Stra8-cre intercrosses. The heads of the embryos were removed and the remaining caudal regions were prepared for total RNA isolation in dounce tissue homogenizers using TRIzol® Reagent (Invitrogen, Carlsbad, CA). Genotyping for the Sry gene was performed to confirm the sex of each embryo. Male and female pups were euthanized at P3 and P7, and the following organs were removed for total RNA isolation: brain (B), heart (H), kidney (K), lung (L), spleen (S) and testes (T) from males; ovaries (O) from females. Total RNA was reverse transcribed into cDNA using random hexamer primers (Invitrogen). To detect cre expression, a 179-bp fragment was amplified by PCR using primers ic202F (5′-GTG CAA GCT GAA CAA CAG GA-3′) and ic381R (5′-AGG GAC ACA GCA TTG GAG TC -3′) under the following conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec. and 60°C for 1 min., with a final extension at 72°C for 7 min. A 207-bp fragment of βActin was amplified as a control transcript using primers Actb-F70 (5′-CCA GTT CGC CAT GGA TGA CGA TAT-3′) and Actb-R277 (5′-GTC AGG ATA CCT CTC TTG CTC TG-3′) under the same thermocycling conditions. Products were separated on 3% (w/v) agarose gels.
Immunohistochemistry on Cross-Sections and Whole-Mount Tubules
For analysis of cross-sections, testes were isolated from postnatal Stra8-cre mice at various ages and fixed in neutral-buffered formalin [3.7% (v/v) formaldehyde in phosphate-buffered solution (PBS)] overnight at 4°C. Fixed testes were then dehydrated, embedded in paraffin wax, and cut into 5μm-thick sections. Following rehydration, antigen-retrieval was performed by boiling the cross-sections in 10 mM sodium citrate, pH 6.0, for 10 min. After rinsing in dH20, some cross-sections were incubated in 0.3% (v/v) hydrogen peroxide in MeOH for 20 min. to inhibit endogenous peroxidase activity. All samples were blocked in 3% (v/v) normal goat serum in PBS for 1 h at room temperature, followed by an overnight incubation at 4°C with anti-cre rabbit polyclonal antibody at 1:500 dilution (Novagen/EMD Biosciences, Madison, WI). After 2×10 min. washes in PBS, some cross-sections were incubated for 1h at room temperature with biotin-conjugated goat anti-rabbit antibody diluted 1:500 (Zymed/Invitrogen). For these samples, tertiary antibody incubation was performed by the addition of streptavidin-HRP antibody at 1:100 dilution for 20 min. Peroxidase activity was then visualized by using a DAB substrate kit following the manufacturer's instructions (Zymed/Invitrogen). Other cross-sections were incubated with AlexaFluor 488-conjugated goat anti-rabbit secondary antibody at 1:1000 dilution (Molecular Probes/Invitrogen). All samples were then rinsed in dH20. Coverslips were affixed with glycerol/vinyl alcohol medium and cross-sections were examined on a Nikon E600 microscope using brightfield optics.
For whole-mount immunocytochemistry, seminiferous tubules were dissected from testes isolated from postnatal Stra8-cre;Z/EG mice at various ages and processed as previously described (Payne and Braun, 2006). Anti-EGFP mouse monoclonal antibody (JL-8; Clontech, Mountain View, CA) and anti-PLZF rabbit polyclonal antibody (H-300, Santa Cruz Biotechnology) were used in these studies at 1:50 dilution. AlexaFluor-conjugated secondary antibodies were diluted 1:1000 (Molecular Probes/Invitrogen). Tubules were examined by epifluorescence microscopy with FITC and TRITC filter sets.
Assessment of Stra8-cre Recombination Efficiency
Matings were established between Stra8-cre;Z/EG males and wild-type FVB/NJ females, and between Stra8-cre;Z/EG females and wild-type FVB/NJ males. Offspring produced from these matings were euthanized at 3-5 d of age and genotyped for the presence of EGFP. Heads were removed from the euthanized pups, and the remaining torsos were rinsed in PBS and sectioned in half. One half was examined for EGFP expression by visualizing tissues using epifluorescence microscopy with an FITC filter. The other half was fixed in 2% (v/v) paraformaldehyde in PBS on ice for 1.5 h, then washed 3×15 min. in PBS containing 2mM MgCl2, 0.01% (w/v) sodium deoxycholate, and 0.02% (v/v) NP-40. Torsos were stained overnight at room temperature with 0.5 mg/ml X-Gal, 5mM potassium ferrocyanide, and 5mM potassium ferricyanide. Following 3×10 min. washes in PBS, torsos were examined for lacZ staining using brightfield microscopy. Mice positive for EGFP expression and negative for lacZ staining were scored as successfully exhibiting recombination at the Z/EG allele, transmitted through either sperm or egg from the transgenic parent.
Acknowledgments
The authors thank Dr. Rolf Sprengel for the kind gift of plasmid pL29mGnRH.iCre. Correspondence and requests for materials should be addressed to R.E. Braun at bob.braun@jax.org.
Literature Cited
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Giuili G, Tomljenovic A, Labrecque N, Oulad-Abdelghani M, Rassoulzadegan M, Cuzin F. Murine spermatogonial stem cells: targeted transgene expression and purification in an active state. EMBO Rep. 2002;3:753–759. doi: 10.1093/embo-reports/kvf149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan ZJ, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod. 2004;71:1469–1474. doi: 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262:303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Li M, Jaroszynski L, Khusainov R, Wulf G, Schwandt I, Korabiowska M, Michelmann HW, Meinhardt A, Engel W. Stem cell based therapeutical approach of male infertility by teratocarcinoma derived germ cells. Hum Mol Genet. 2004;13:1451–1460. doi: 10.1093/hmg/ddh166. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulad-Abdelghani M, Bouillet P, Decimo D, Gansmuller A, Heyberger S, Dolle P, Bronner S, Lutz Y, Chambon P. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol. 1996;135:469–477. doi: 10.1083/jcb.135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C, Braun RE. Histone lysine trimethylation exhibits a distinct perinuclear distribution in Plzf-expressing spermatogonia. Dev Biol. 2006;293:461–472. doi: 10.1016/j.ydbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. Histological and Histopathological Evaluation of the Testis. St. Louis: Cache River Press; 1990. p. 286. [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Vidal F, Sage J, Cuzin F, Rassoulzadegan M. Cre expression in primary spermatocytes: a tool for genetic engineering of the germ line. Mol Reprod Dev. 1998;51:274–280. doi: 10.1002/(SICI)1098-2795(199811)51:3<274::AID-MRD6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]