Abstract
Small laboratory animals have provided significant information about melatonin regulation, yet most of these organisms are nocturnal and regulate melatonin synthesis by mechanisms that diverge from those of humans. For example, in all rodents examined, melatonin secretion occurs with a time lag of several hours after the onset of darkness; in addition, arylalkylamine N-acetyltransferase (AANAT), the key enzyme in melatonin synthesis, displays dynamic transcriptional activation specifically at night in all rodents studied to date. In ungulates and primates including humans, on the other hand, melatonin secretion occurs immediately during the early night and is controlled by circadian posttranscriptional regulation of AANAT. We hypothesize that the diurnal Octodon degus (an Hystricognath rodent) could serve as an improved experimental model for studies of human melatonin regulation. To test this, we monitored melatonin production in degus using pineal microdialysis and characterized the regulation of melatonin synthesis by analyzing degu Aanat. Degu pineal melatonin rises with little latency at night, as in ungulates and primates. In addition, degu Aanat mRNA expression displays no detectable diurnal variation, suggesting that, like ungulates and primates, melatonin in this species is regulated by a posttranscriptional mechanism. Compared with AANAT from all rodents examined to date, the predicted amino acid sequence of degu AANAT is phylogenetically more closely related to ungulate and primate AANAT. These data suggest that Octodon degus may provide an ideal model system for laboratory investigation of mechanisms of melatonin synthesis and secretion in diurnal mammals.
Keywords: arylalkylamine N-acetyltransferase (AANAT), degus, in vivo microdialysis, melatonin, pineal gland, posttranscriptional control
Introduction
Melatonin, an indolamine synthesized in the pineal gland at nighttime from serotonin, plays an important role in a multitude of physiological functions [1-4] and may be involved in human disorders including autism [5], cancer [6], and diabetes [7]. Arylalkylamine N-acetyltransferase (AANAT or serotonin N-acetyltransferase) is the key enzyme in the biosynthesis of melatonin. AANAT enzyme activity is high at night and low during the day as a result of clock-controlled adrenergic activation of the pineal gland at night [8]. At night, sympathetic nerve endings from cells located in the SCG release norepinephrine at nerve terminals in the pineal gland [9] that increases production of cAMP. The increased cAMP levels activate cAMP-dependent protein kinase A, which activates Aanat at night [10, 11].
Cyclic AMP triggering of melatonin production is conserved in all mammals. However, significant differences exist between the molecular regulation of Aanat in various species. In nocturnal rodents commonly used in laboratory investigations, melatonin production is rate limited by cAMP-dependent transcriptional activation of Aanat [11–13]. In diurnal mammals, however, posttranscriptional control of AANAT by PKA dominantly regulates melatonin production, since Aanat mRNA levels display very little diurnal variation [14–16]. The differential mechanisms of AANAT control result in marked differences in the dynamics of melatonin secretion at night. In nocturnal animals such as rats and hamsters, the onset of melatonin secretion is markedly delayed after dark onset [17]. In contrast, melatonin in humans rapidly surges following dark onset without latency [18]. The rapid release of melatonin following dark onset in non-rodent diurnal mammals and the expression of significant quantities of Aanat mRNA in the day demonstrate that AANAT is posttranscriptionally controlled during the diurnal cycle, likely by cAMP-mediated protection of AANAT from proteasome-dependent degradation [19]. A reasonable animal model to study mechanisms of melatonin regulation utilized in humans should (a) be diurnal, (b) exhibit rapid onset of melatonin synthesis at night; (c) demonstrate posttranscriptional activation of AANAT; and (d) be small enough to use in the laboratory setting while still permitting real-time, quantitative in vivo monitoring of melatonin secretion [20].
Not all small diurnal animals are appropriate for this purpose. For example, previous characterization of melatonin synthesis has been performed in the diurnal rodent Avicanthis ansorgei (Grass rats), which is classified along with rats and hamsters in the suborder Myomorpha. These rodents, which are small and breed with reasonable effciency for laboratory studies, regulate melatonin secretion with delayed kinetics similar to nocturnal rats [21]. Furthermore, grass rat AANAT is controlled by transcriptional regulation [21], demonstrating that diurnal mammals do not always utilize posttranscriptional control of melatonin synthesis. While these characteristics confirm the association between the timing of melatonin secretion (late versus early onset) and the regulation of melatonin synthesis (transcriptional versus posttranscriptional) seen in other animals, they also suggest that grass rat melatonin regulation is more similar to nocturnal rodents than to diurnal primates. To date, posttranscriptional control of AANAT and melatonin synthesis has not been reported in small laboratory mammals.
The degu (Octodon degus) is a small diurnal mammal [22], which, unlike nocturnal rodents (such as laboratory rats, hamsters, mice) and diurnal rodents (such as grass rats and fat sand rats), is classified in the Hystricognath suborder [23]. The location of the degu pineal gland closely resembles that of other diurnal mammals and lies beneath the corpus callosum [24] adjacent to the posterior commissure and habenular commissure [25]. In contrast, in all other rodents, the pineal is superficially located at the confluence of the superior sagittal and transverse sinuses. Because of their manageable size and well-characterized behavior, degus have been used as diurnal animal models for circadian studies in several laboratories [26]. Melatonin production in degus is always high at night and low during the day, regardless of their behavior pattern [27]. These characteristics potentially make the degu an ideal laboratory animal model to study mechanisms of melatonin production in diurnal mammals.
In this study, we examine melatonin timing in degus using pineal microdialysis [20] and compare high-resolution pineal secretory profiles with those from rats [17]. In addition, we report the molecular cloning of the cDNA encoding degu AANAT and the diurnal patterns of degu Aanat mRNA expression. Our results demonstrate strong physiological and molecular similarities between degu and ungulate/primate regulation of melatonin.
Materials and methods
Animals
All animal protocols were reviewed and approved by the University of Michigan animal care and use committee. Adult degus (bred locally; 6–12 months) and rats (Sprague Dawley and Wistar strains from Harlan; 2–4 months) were used in this study. Animals were housed in temperature-controlled chambers with a light and dark cycle of 12:12 hr (lights on at 06:00 hr).
In vivo measurement of melatonin release using pineal microdialysis
Rats and degus were implanted with pineal microdialysis probes (see Ref. [20] for details). Following a recovery period of one day, animals were placed in microdialysis chambers. Microdialysis was performed with artificial CSF solution flowing continuously at 2 μL/min. The pineal dialysates were collected in a sample loop of an automated injector (Instech, Plymouth Meeting, USA), which delivers samples into an HPLC column (Sigma, St. Louis, USA) at 10 min intervals with the aid of an HPLC pump (Shimadzu, Tokyo, Japan). Fractionated pineal dialysates were analyzed online by a fluorescence detector. The automated control of the HPLC system including the handling and storage of the chromatograms were carried out with an external computer using Shimadzu chromatography software [20].
Aanat cloning
Degu pineal glands were collected at night, and total RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA, USA). One microgram of total RNA per pineal gland was reverse-transcribed by using SuperScript II RT (Invitrogen, Carlsbad, CA, USA). We first obtained a partial cDNA fragment of degu Aanat by PCR amplifying pineal cDNA using several degenerate primer pairs against conserved sequences of mammalian AANAT proteins. Three sets of forward-degenerate primer sequences included: 5’-GCYRYCRSCGGCGCCACAC-3’ and 5’-GCCARYGAGTTYCGYTGCCT-3’ and 5’-GYGYSTTTGAGATYGAGCG-3’. Three sets of reverse-degenerate primer sequences included: 5’-CCCGGYGCACNGYBGGCTG-3’ and 5’-AGBRYRTYCTCRCACATGAG-3’ and 5’-CYRARHYTCTSRTARAAGGG-3’. Next, the PCR products were sequenced to obtain the partial sequences of degu-specific Aanat cDNA. Based on the partial sequences of degu Aanat, degu-specific Aanat primers were designed to perform ‘Genome Walking’ (Clontech, Mountain View, CA, USA). Genomic DNA was prepared from degu brain using DNeasy kit (Qiagen). Four genomic libraries were generated by digesting individually with EcoRV, StuI, DraI, or PvuII. Each set of enzyme-digested genomic fragments was then ligated with an adaptor and subjected to primary amplification and the secondary amplification using two nested Aanat primers. Primers used for downstream genome walking included primary primer 5’-CCGGATGAGATTCGACACTTCCTGACT-3’ and nested primer 5’-GTGTCCCGAGCTGTCCCTGGGCTGGTT-3’. Upstream genome walking was performed with primary primer 5’-CAGGTGCTGCAAGTAACGCCACAGCAG-3’ and nested primer 5’-ACAGAGCCCTTGCCCTGCTGCCGGAAG-3’. After the first round of genome walking, a 3 kb fragment from the PvuII genomic library spanning three coding exons of degu Aanat was isolated and sequenced. Based on this novel genomic sequence another set of reverse primers was used to amplify further upstream sequences by genomic walking: primary primer 5’-CAAAAATCCCATGTCTGAGAGCCATGA-3’ and nested primer 5’-GTGTTACATACCAGGACTTGGAAGCTG-3’. The second round of genome walking yielded a 2 kb fragment from the StuI library, which contained 1 kb of overlap with the PvuII fragment described above. The predicted degu Aanat coding regions were identified by comparing Aanat genomic and cDNA sequences from rat, mouse and human. To confirm the final sequence of the degu Aanat open reading frame, a full-length cDNA of degu Aanat was amplified from pineal cDNA by PCR using specific 5’ and 3’ primers which spanned the start and stop codons identified by genomic analysis and sequence comparison. This PCR product was cloned into the SalI and XbaI sites of pCMV-Sport6 (Invitrogen) and unique clones were completely sequenced on both strands.
AANAT enzyme activity assay
For analysis of cells expressing AANAT, assays were perfomed by the addition of substrate into the media of tissue culture cells. HEK293 cells were transfected with empty vector (control), or degu or rat Aanat cDNA expression vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Each transfection was performed in triplicate in 24-well plates using a total of 1 μg of DNA (200 ng of cDNA of interest and 800 ng of vector DNA). On the following day, transfected cells were incubated with 5-methoxytryptamine (final concentration 1 μM) for 3.5 hr at 37°C. Culture media was harvested and melatonin content was measured using HPLC.
Aanat gene expression analysis
Degus were killed over the course of one day at 4-hour intervals, and their pineal glands were collected immediately and stored in RNA later solution at −20°C. Total RNA was extracted using an RNeasy kit (Qiagen) and reverse-transcribed using random primers as above. Quantitative RT-PCR was performed on first-strand cDNA using primers designed to detect both degu and rat Aanat mRNA and products were quantified using SYBR green. Primer sequences used were: sense primer 5’-CCACACGCTCCCTGCCAGTGA-3’ and antisense primer 5’-GTGCAGTGTCAGCGACTCCTG-3’. Primers against 18S rRNA were used in separate reactions to normalize for total cDNA content.
Results
Pineal melatonin secretion was measured in freely moving degus and rats using online microdialysis [20] (Fig. 1). Melatonin onset phase (defined as the interval between the dark onset and melatonin rise to 20% of the daily maximum) in rats ranged from 98–420 min (Fig. 1; purple and green dots; see Ref. [17]), which is consistent with the delayed onset observed in all rodents examined previously. In contrast, degus melatonin secretion began 20–46 min after dark onset (Fig. 1; red dots). The early increase in melatonin in degus resembled the timing of melatonin secretion seen in sheep and humans [18] and is consistent with a posttranscriptional mechanism of melatonin regulation.
Fig. 1.
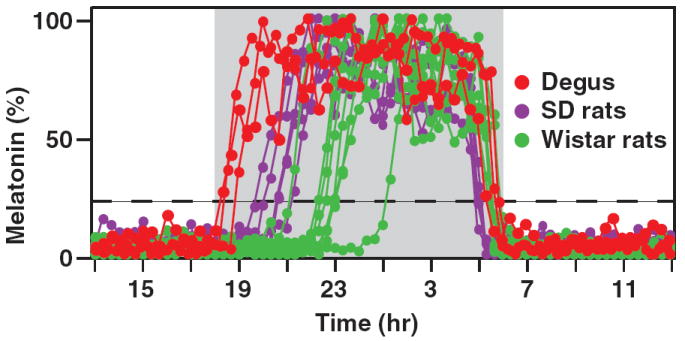
Onset of melatonin secretion is much earlier in degus than in rats. Melatonin secretion was monitored in degus (red dots, n = 3), Sprague Dawley (SD) rats (purple dots, n = 5), and Wistar rats (green dots, n = 8) using pineal microdialysis, and normalized to daily maximum levels. The dashed line represents 20% of the maximum levels. Animals were housed in LD12:12 hr with lights-off at 18:00 hr. Compared with rats, degus clearly display advanced onset of melatonin secretion.
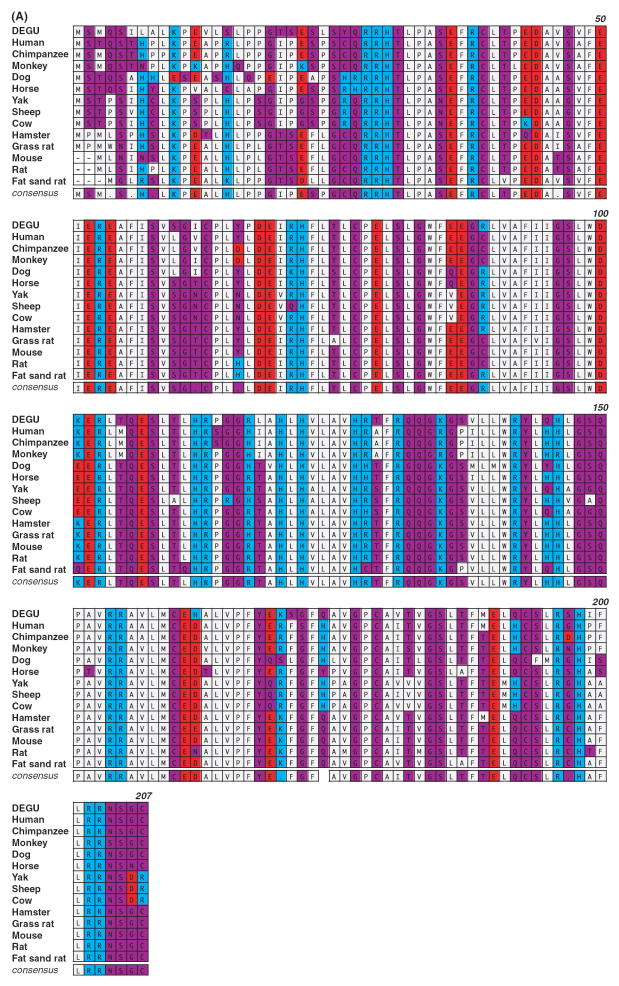
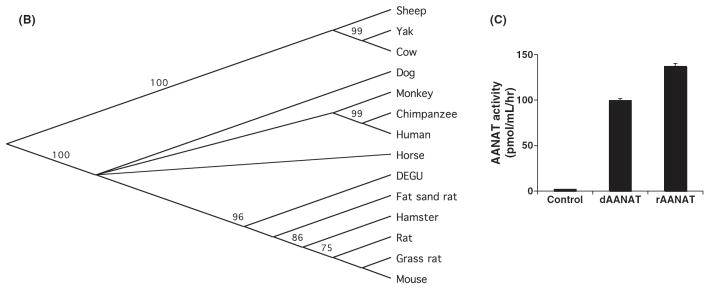
To generate molecular reagents to study degu AANAT that could be used for gene expression and phylogenetic analysis, we cloned degu Aanat cDNA through a multi-step process. Degenerate oligonucleotide probes against highly conserved regions of mammalian AANAT proteins were used to amplify a partial Aanat cDNA from nighttime degupineal glands. Specific degu Aanat primers, based on the partial clone, were used to amplify flanking upstream and downstream genomic sequences spanning the entire Aanat coding region as determined by comparison with rat, mouse and human coding segments. Unique 5’ and 3’ primers were then designed to amplify the full-length degu Aanat cDNA using nighttime pineal mRNA. The resulting degu Aanat cDNA precisely matched the genomic sequence and predicted an amino acid sequence of 207 amino acids (sequence accession number: FJ809749). The degu AANAT amino acid sequence shares a high degree of identity with rodents including hamsters (89%), mice (88%), grass rats (86%), laboratory rats (87%), and sand rats (89%). A slightly lower degree of identity is shared with primates including humans (83%), chimpanzees (83%) and monkeys (80%), and with ungulates including sheep (76%), cows (78%), and yaks (79%) (Fig. 2A). The degu AANAT sequence contains all key features of AANAT, including the catalytic core of the enzyme and N- and C-terminal regulatory regions [28]. Interestingly, phylogenetic analysis demonstrates that degu AANAT is positioned between ungulates/primates and rodents. Thus, degu AANAT is more closely related to the ungulates and primates proteins than are AANAT from any other diurnal or nocturnal rodent (Fig. 2B).
Fig. 2.
Degu Aanat cloning and analysis. (A) Amino acid sequence comparison of AANAT from degu and other mammalian species is shown. The sequence comparison was performed using MacVector ClustalW alignment program and was color-coded based on functionality of the amino acids. Acidic residues were marked in red, basic residues in blue, hydrophobic residues in gray, and hydrophilic residues in purple. The numbers indicated on the upper right lines denote the amino acid positions based on the degu AANAT sequence. (B) Phylogenetic analysis of degu AANAT. A diagram of the relationship between the degu AANAT amino acid sequences and AANATs from other mammalian species is shown. The phylogenic tree was constructed using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) method. The confidence index assigned to particular nodes in the tree (numbers shown) was estimated using the bootstrap re-sampling approach with Poisson-correction. Compared with the other rodents, the degus AANAT appears to be more closely related to those of ungulates and primates. (C) Degu AANAT expression in HEK293 cells. Cells transfected with empty vector (control), degu Aanat cDNA (dAANAT), or rat Aanat cDNA (rAANAT) were incubated with 5-methoxytryptamine (final concentration at 1 uM) for 3.5 hr at 37°C. Supernatants were harvested and measured for melatonin production using HPLC. The results were confirmed in at least three independent experiments. The activity of the degu enzyme was comparable to the activity of rat AANAT enzyme expressed in HEK293 cells.
The degu Aanat cDNA was cloned into a mammalian expression vector and expressed in cultured cells. HEK293 cells transfected with degu Aanat cDNA were exposed to exogenous substrate 5-methoxymelatonin and after an incubation period, the culture medium was assayed for melatonin (Fig. 2C). The melatonin synthetic activity of degu AANAT was comparable to the activity of rat AANAT expressed in HEK293 cells. Vector transfected cells did not express significant AANAT activity.
We examined the diurnal expression of degu Aanat mRNA to gain insight into the regulatory mechanism of melatonin synthesis in degus. Quantitative RT-PCR was performed on pineal mRNA samples from both degus and rats (Fig. 3). There was very little fluctuation in degu Aanat mRNA over the course of the day, with both day and night pineal glands exhibiting modest transcript levels. In contrast, rat pineal Aanat demonstrated dramatic increases during the night period and extremely low levels during the day, which is consistent with the known transcriptional regulation of Aanat in rats [11]. Thus, gene expression analysis combined with real-time measurements of degu pineal melatonin demonstrate that degu pineal melatonin synthesis in the night results from posttranscriptional regulation of AANAT.
Fig. 3.
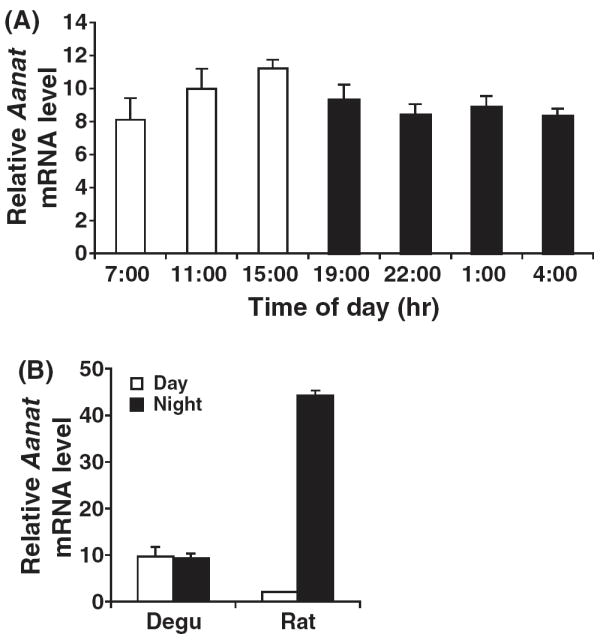
Aanat mRNA expression is constitutively expressed in the degu pineal gland. (A) Total RNA was extracted from degu pineal gland at 4-hr intervals over the course of a day. Quantitative RT-PCR was performed to measure degu Aanat mRNA levels, normalized to 18S RNA controls. Identical results were obtained in three experiments. (B) Total RNA was extracted from degu and rat pineal glands during the daytime (11:00–15:00 hr) or nighttime (1:00–4:00 hr). Aanat mRNA levels were measured as those in (A).
Discussion
Accumulating evidence that melatonin plays a role in human health and disease underscores the importance of studying the mechanisms of melatonin synthesis. Significant previous work has unraveled the cellular and biochemical mechanisms of melatonin synthesis in nocturnal rodent models. However, as more species have been studied, it has become clear that nocturnal rodent models do not recapitulate the kinetics and posttranscriptional regulation of AANAT and melatonin synthesis that characterize ungulates and primates, including humans. Our studies suggest that degus represent a more faithful model of human melatonin regulation. Importantly, as a small diurnal mammal, we demonstrate that degus can be monitored using real-time pineal microdialysis, which can be used for detailed mechanistic studies in vivo [12, 20, 29].
Our studies unequivocally show that degus are superior to rats as a model for ungulate/primate melatonin regulation, since (a) degu melatonin increases rapidly at night, and (b) degu Aanat mRNA does not vary from day to night. As an animal model, degus have a number of advantages compared with ungulates and primates, including ease of care and favorable costs. In addition, degus are relatively small and can thus be bred and caged in large numbers, making biochemical and molecular studies statistically feasible. Furthermore, the diurnality of degus is clearly an advantage, since correlations to human studies that relate circadian dysfunction to states such as sleep can be more intuitively interpreted (compared with nocturnal rodent studies).
Cloning and molecular analysis of degu Aanat in this study show that degu AANAT is regulated posttranscriptionally, since Aanat is expressed constitutively throughout the diurnal cycle in the degu pineal gland. This contrasts with the dynamic regulation of diurnal Aanat transcription in rats [11], mice [30], hamsters [31], and diurnal grass rats [21]. Comparative promotor analysis of rat and degu Aanat genes would help elucidate mechanisms of the diurnal transcription of Aanat in rodents. Interestingly, the N-terminal regulatory domain of degu AANAT is more closely related to sequences found in primates than other rodents. Thus, posttranslational control of AANAT activity may play a key role in the early rise of melatonin in degu pineal gland in vivo. Further work will be required to test whether molecular mechanisms of degu AANAT regulation are indeed more similar to primates and mediated by conserved N-terminal sequences.
Degus (Octodon degus) belong to the infraorder Hystricognathi and are distinct from the members of Myomorpha suborder that include nocturnal rodents such as laboratory rats, mice, hamsters, and diurnal rodents including grass rats and sand rats. Molecular analysis suggests that degus are more closely related to guinea pigs and rabbits than to members of Myomorpha (http://animal.nibio.go.jp/research/e_gonadotropin.html). In fact, it is debated whether Hystricognathi should be re-classified into the order Lagomorpha (which includes rabbits, hares, and pikas) rather than Rodentia, where they currently reside [32–34]. Phylogenetic analysis of degu AANAT in our studies demonstrates that compared with AANAT from any animal examined in the Myomorpha suborder, the degu AANAT protein is more closely related to ungulate and primate proteins. As mentioned above, diurnal grass rats exhibit transcriptional regulation of Aanat, while other diurnal animals exhibit posttranscriptional regulation. These data support the assertion that the phylogenetic position of an organism, not their diurnality or nocturnality of behavior, determines regulatory mechanisms of melatonin synthesis [21].
In conclusion, the early onset of melatonin secretion, the posttranscriptional control of AANAT, and the phylogenetic lineage of degus AANAT together strongly position the degu as a valuable laboratory animal model to understand melatonin synthesis in non-rodent diurnal mammals such as humans. Future application of in vivo microdialysis of degus and detailed molecular analysis of degus AANAT should yield more accurate insights into the precise mechanisms of regulation of melatonin synthesis in primates, including humans.
Acknowledgments
We thank Ms. Catherine Autin and Megan H. Hagenauer for help with animal care, Ms Yaxi Chen for laboratory assistance. This work was support by grants NS057583 (to JB), and NS054724 (to MMW).
References
- 1.Maldonado MD, Murillo-Cabezas F, Terron MP, Flores LJ, Tan DX, Manchester LC, Reiter RJ. The potential of melatonin in reducing morbidity-mortality after craniocerebral trauma. J Pineal Res. 2007;42:1–11. doi: 10.1111/j.1600-079X.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- 2.Simko F, Paulis L. Melatonin as a potential antihypertensive treatment. J Pineal Res. 2007;42:319–322. doi: 10.1111/j.1600-079X.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- 3.Reiter RJ. The melatonin rhythm: both a clock and a calendar. Experientia. 1993;49:654–664. doi: 10.1007/BF01923947. [DOI] [PubMed] [Google Scholar]
- 4.Skene DJ, Arendt J. Human circadian rhythms: physiological and therapeutic relevance of light and melatonin. Ann Clin Biochem. 2006;43:344–353. doi: 10.1258/000456306778520142. [DOI] [PubMed] [Google Scholar]
- 5.Melke J, Goubran Botros H, Chaste P, et al. Abnormal melatonin synthesis in autism spectrum disorders. Mol Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiu SY. Towards rational and evidence-based use of melatonin in prostate cancer prevention and treatment. J Pineal Res. 2007;43:1–9. doi: 10.1111/j.1600-079X.2007.00451.x. [DOI] [PubMed] [Google Scholar]
- 7.Peschke E. Melatonin, endocrine pancreas and diabetes. J Pineal Res. 2008;44:26–40. doi: 10.1111/j.1600-079X.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 8.Klein DC, Schaad NL, Namboordiri MA, et al. Regulation of pineal serotonin N-acetyltransferase activity. Biochem Soc Trans. 1992;20:299–304. doi: 10.1042/bst0200299. [DOI] [PubMed] [Google Scholar]
- 9.Drijfhout WJ, Van Der Linde AG, Kooi SE, et al. Norepinephrine release in the rat pineal gland: the input from the biological clock measured by in vivo microdialysis. J Neurochem. 1996;66:748–755. doi: 10.1046/j.1471-4159.1996.66020748.x. [DOI] [PubMed] [Google Scholar]
- 10.Borjigin J, Li X, Snyder SH. The pineal gland and melatonin: molecular and pharmacologic regulation. Annu Rev Pharmacol Toxicol. 1999;39:53–65. doi: 10.1146/annurev.pharmtox.39.1.53. [DOI] [PubMed] [Google Scholar]
- 11.Borjigin J, Wang MM, Snyder SH. Diurnal variation in mRNA encoding serotonin N-acetyltransferase in pineal gland. Nature. 1995;378:783–785. doi: 10.1038/378783a0. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Borjigin J. N-acetyltransferase is not the rate-limiting enzyme of melatonin synthesis at night. J Pineal Res. 2005;39:91–96. doi: 10.1111/j.1600-079X.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 13.Roseboom PH, Coon SL, Baler R, et al. Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology. 1996;137:3033–3045. doi: 10.1210/endo.137.7.8770929. [DOI] [PubMed] [Google Scholar]
- 14.Ackermann K, Bux R, Rub U, et al. Characterization of human melatonin synthesis using autoptic pineal tissue. Endocrinology. 2006;147:3235–3242. doi: 10.1210/en.2006-0043. [DOI] [PubMed] [Google Scholar]
- 15.Coon SL, Roseboom PH, Baler R, et al. Pineal serotonin N-acetyltransferase: expression cloning and molecular analysis. Science. 1995;270:1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- 16.Craft CM, Murage J, Brown B, et al. Bovine arylalkylamine N-acetyltransferase activity correlated with mRNA expression in pineal and retina. Brain Res Mol Brain Res. 1999;65:44–51. doi: 10.1016/s0169-328x(98)00336-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Borjigin J. Relationship between nocturnal serotonin surge and melatonin onset in rodent pineal gland. J Circadian Rhythms. 2006;4 doi: 10.1186/1740-3391-4-12. online journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wehr TA. The durations of human melatonin secretion and sleep respond to changes in daylength (photoperiod) J Clin Endocrinol Metab. 1991;73:1276–1280. doi: 10.1210/jcem-73-6-1276. [DOI] [PubMed] [Google Scholar]
- 19.Schomerus C, Korf HW, Laedtke E, et al. Selective adrenergic/cyclic AMP-dependent switch-off of proteasomal proteolysis alone switches on neural signal transduction: an example from the pineal gland. J Neurochem. 2000;75:2123–2132. doi: 10.1046/j.1471-4159.2000.0752123.x. [DOI] [PubMed] [Google Scholar]
- 20.Borjigin J, Liu T. Application of long-term microdialysis in circadian rhythm research. Pharmacol Biochem Behav. 2008;90:148–155. doi: 10.1016/j.pbb.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garidou ML, Gauer F, Vivien-Roels B, et al. Pineal arylalkylamine N-acetyltransferase gene expression is highly stimulated at night in the diurnal rodent, Arvicanthis ansorgei. Eur J Neurosci. 2002;15:1632–1640. doi: 10.1046/j.1460-9568.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 22.Refinetti R. Variability of diurnality in laboratory rodents. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2006;192:701–714. doi: 10.1007/s00359-006-0093-x. [DOI] [PubMed] [Google Scholar]
- 23.Opazo JC. A molecular timescale for caviomorph rodents (Mammalia, Hystricognathi) Mol Phylogenet Evol. 2005;37:932–937. doi: 10.1016/j.ympev.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Wright JW, Kern MD. Stereotaxic atlas of the brain of Octodon degus. J Morphol. 1992;214:299–320. doi: 10.1002/jmor.1052140306. [DOI] [PubMed] [Google Scholar]
- 25.Uria H, Antolin I, Tolivia D, et al. The pineal gland of the trumpet-tailed rat (Octodon degus) J Pineal Res. 1992;13:174–183. doi: 10.1111/j.1600-079x.1992.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee TM. Octodon degus: a diurnal, social, and long-lived rodent. Ilar J. 2004;45:14–24. doi: 10.1093/ilar.45.1.14. [DOI] [PubMed] [Google Scholar]
- 27.Vivanco P, Ortiz V, Rol MA, et al. Looking for the keys to diurnality downstream from the circadian clock: role of melatonin in a dual-phasing rodent, Octodon degus. J Pineal Res. 2007;42:280–290. doi: 10.1111/j.1600-079X.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 28.Klein DC. Arylalkylamine N-acetyltransferase: ‘the Timezyme’. J Biol Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- 29.Borjigin J, Deng J, Sun X, et al. Diurnal pineal 3-O-sulpho-transferase 2 expression controlled by beta-adrenergic repression. J Biol Chem. 2003;278:16315–16319. doi: 10.1074/jbc.M300828200. [DOI] [PubMed] [Google Scholar]
- 30.Roseboom PH, Namboodiri MA, Zimonjic DB, et al. Natural melatonin ‘knockdown’ in C57BL/6J mice: rare mechanism truncates serotonin N-acetyltransferase. Brain Res Mol Brain Res. 1998;63:189–197. doi: 10.1016/s0169-328x(98)00273-3. [DOI] [PubMed] [Google Scholar]
- 31.Gauer F, Poirel VJ, Garidou ML, et al. Molecular cloning of the arylalkylamine-N-acetyltransferase and daily variations of its mRNA expression in the Syrian hamster pineal gland. Brain Res Mol Brain Res. 1999;71:87–95. doi: 10.1016/s0169-328x(99)00175-8. [DOI] [PubMed] [Google Scholar]
- 32.D’erchia AM, Gissi C, Pesole G, et al. The guinea-pig is not a rodent. Nature. 1996;381:597–600. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- 33.Graur D, Hide WA, Li WH. Is the guinea-pig a rodent? Nature. 1991;351:649–652. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- 34.Reyes A, Gissi C, Pesole G, et al. Where do rodents fit? Evidence from the complete mitochondrial genome of Sciurus vulgaris. Mol Biol Evol. 2000;17:979–983. doi: 10.1093/oxfordjournals.molbev.a026379. [DOI] [PubMed] [Google Scholar]