Abstract
The critical tumor suppressor p53 is mutated in over half of all human cancers. The majority of p53 cancer mutations are missense mutations, which can be classified into contact mutations that directly disrupt the DNA-binding motif of p53 but have modest impact on p53 conformation and structural mutations that greatly disrupt p53 conformation. Many p53 cancer mutants, including the hotspot mutations (R175H, R248W and R273H), not only lose p53-dependent tumor suppressor activities, but also acquire new oncogenic activities to promote cancer. Therefore, it is critical to elucidate the gain of oncogenic function of p53 cancer mutants. Employing humanized p53 mutant knock-in mouse models, we have identified a gain of oncogenic function shared by the most common p53 contact mutants (R273H and R248W) and structural mutant (R175H). This gain of function inactivates Mre11/ATM-dependent DNA damage responses, leading to chromosomal translocation and defective G2/M checkpoint. Considering the critical roles of ATM in maintaining genetic responses and therapeutic responses to many cancer treatments, the identification of this common gain of function of p53 cancer mutants will have important implication on the drug resistance of a significant portion of human cancers that express both the contact and structural p53 cancer mutants.
Keywords: p53, tumorigenesis, gain of function, genetic instability
Introduction
p53 is a critical tumor suppressor that maintains the genetic stability in mammals by playing multiple roles in cell cycle arrest, apoptosis, senescence and differentiation. All these functions of p53 could prevent the passage of DNA damage to the daughter cells. As a transcription factor, p53 functions by directly regulating the expression of hundreds of genes, products of which are responsible for mediating the p53-dependent functions (Wei et al., 2006). p53 is consisted of two N terminal transactivation domains, a core DNA-Binding domain and a C terminal oligomerization domain (Ko and Prives, 1996). Loss of p53 function is required for the progression of most cancers. In this context, p53 is somatically mutated in over 50% of all human cancers, and most of these mutations were missense mutations within the DNA-binding core domain (Hainaut and Hollstein, 2000). Based on their impact on p53 structure and function, p53 mutation can fall into two general classes: DNA contact mutations, which change the residues directly involved in contact with DNA but have modest impact on p53 conformation; structural mutations that dramatically alter the p53 conformation. Among all p53 mutations, there are four hotspot mutations at residues 175, 248, 249 and 273 (Hainaut and Hollstein, 2000). While both R248W and R273H mutations are contact mutation, R175H mutation is a structural mutation (Sigal and Rotter, 2000). All hotspot mutations abolish the wild type tumor suppression function of p53.
Expression of mutant p53 is correlated with the poor prognosis of the patients. Accumulating evidence has shown that p53 mutants not only lose their wild type p53 activity, but also acquire new oncogenic activities to promote cancer and drug resistance (Xu, 2006). For example, p53 mutants can help to transform cells, increase resistance of cells to chemotherapy, apoptosis (Sigal and Rotter, 2000). Downregulation of p53 mutants in cancer cells can reduce cell proliferation, tumorigeneic potential and chemotherapeutic resistance (Sigal and Rotter, 2000). Therefore, in order to improve the efficacy of therapy for human cancers expressing p53 cancer mutants, it is critical to elucidate the oncogenic gain of function of p53 cancer mutants. Previous studies have suggested that one mechanism to achieve the gain of function of p53 cancer mutants is through protein-protein interaction between p53 mutants and cellular proteins, leading to the disruption of the function of the cellular proteins. For example, certain p53 cancer mutants can interact with p73 and suppress p73 function (Li and Prives, 2007).
To investigate the impact of the p53 cancer mutations on the structure and function of p53, several groups have introduced the common p53 cancer mutations into the corresponding residues of the endogenous mouse p53 genes (Lang et al., 2004; Olive et al., 2004). These p53 knock-in mutant mice develop tumors with a similar kinetics as the p53−/− mice but with a more complex tumor spectrum, suggesting that these p53 mutants gain new oncogenic function. In order to better recapitulate the impact of the common p53 cancer mutations on the structure and function of p53, we employed the humanized p53 knock-in (HUPKI) allele to develop mouse models that express the most common p53 cancer mutants in the physiological context (Luo et al., 2001). By studying the common p53 contact mutation (R248W and R273H) knock-in mice, we recently discovered a gain of function of these p53 contact mutants in inducing genetic instability by inactivating ATM function (Song et al., 2007). ATM is the master regulator of cellular responses to DNA double-stranded break (DSB) damage (Xu, 2006). Since genetic instability, a hallmark of cancer, plays critical roles in tumor progression and drug resistance, it is important to determine whether this gain of function is common to other p53 cancer mutants. By establishing and analyzing the most common p53 structural mutant (R175H) knock-in mice, we provide compelling evidence that it is a common gain of function of p53 cancer mutants to induce genetic stability by disrupting the Mre11/ATM pathways.
Results and Discussion
Generation of p53hkiR175H mice
To better recapitulate the impact of cancer mutations on the structure and function of human p53, we employed the humanized p53 knock-in (HUPKI) allele that is consisted primarily of human p53 sequence (amino acids 33-332) flanked by the conserved N- and C-terminus of mouse p53 (Luo et al., 2001). The strategy to create the p53hkiR175H (denoted R175H) mice was the same as that to generate p53hkiR248W and p53hkiR273H (denoted R248W and R273H) mice (Song et al., 2007) and described in Supplementary Figure 1. The entire cDNA of p53 in R175H MEFs was sequenced to verify that only R175H mutation but no other mutation was introduced into the HUPKI allele of R175H mice. We found that only spleen and thymus of R175H knick-in mice had strong expression of p53, possibly as a result of the physiologically occurring DNA damage response during V(D)J recombination.
Abolished p53 responses to DNA damage in R175H MEFs
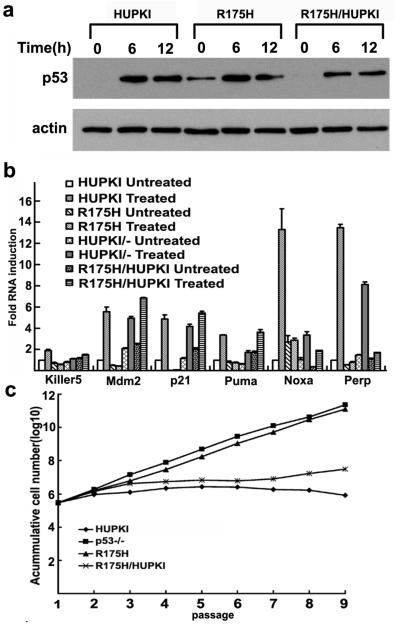
To determine the impact of R175H mutation on the WT p53-dependent functions, the p53 stability and activity in MEFs were analyzed post DNA damage. As expected, the protein levels of p53 were undetectable in HUPKI MEFs before DNA damage and significantly increased after Doxorubicin treatment that induces DNA DSB damage (Figure 1a). Similarly to R248W MEFs, the protein levels of p53 were constitutively high in R175H MEFs (Figure 1a), consistent with the notion that negative autoregulatory loop of p53 is abolished in p53 mutant MEFs.
Figure 1. p53 stability and activity after DNA damage in MEFs.
(a) Expression of p53 in HUPKI, R175H mutant and HUPKI/R175H heterozygous MEFs after treatment with Doxorubicin (0.5μM) for 6 and 12 hours. Actin was used as the loading control. (b) Induction of p53 target genes Keller 5, Mdm2, p21, Puma, Noxa and Perp in HUPKI, R175H, R175H/HUPKI and HUPKI/- MEFs. Total RNA was harvested 1 hour after 2Gy irradiation. The mRNA levels of each gene were determined by real time PCR and normalized with the mRNA levels of GAPDH. The mRNA levels in the untreated HUPKI MEFs were set to 1. (c) 3T3 assay to examine the replicative senescence of HUPKI, R175H, R175H/HUPKI and p53−/− MEFs. The assay was started with passage 5 MEFs of various genotypes.
To determine the impact of R175H mutation on WT p53-dependent activity, we analyzed the p53-dependent transcription in R175H MEFs after DNA damage. When analyzed by quantitative PCR, the expression of a number of p53 target genes, including p21, Bax and Mdm2, was significantly increased in the control HUPKI MEFs after DNA damage. However, the mRNA levels of these p53 targets were not increased in R175H MEFs after DNA damage (Figure 1b). Therefore, WT p53-dependent function is abolished in R175H MEFs.
To study the potential dominant negative effects of R175H on the co-expressed WT p53, we analyzed p53-dependent DNA damage responses in R175H/HUPKI heterozygous mutant MEFs. Similar to that in HUPKI MEFs but in contrast to that in R175H MEFs, the p53 protein level is not detectable in R175H/HUPKI MEFs before DNA damage, suggesting no chronic activation of DNA damage pathways in the heterozygous mutant MEFs that can stabilize R175H (Figure 1a). Most of the analyzed p53-dependent gene expression in R175H/HUPKI MEFs after DNA damage is similar to that in HUPKI/- MEFs, suggesting that R175H is not efficient in dominantly suppressing the co-expressed WT p53 for some p53 target genes (Figure 1b). In contrast to R175H MEFs that undergo no replicative senescence, 3T3 assay indicates that R175H/HUPKI MEFs retain the p53-dependent replicative senescence (Fig. 1c). Therefore, p53-dependent functions are at least partially retained in the heterozygous mutant MEFs.
p53-dependent apoptosis in R175H thymocytes
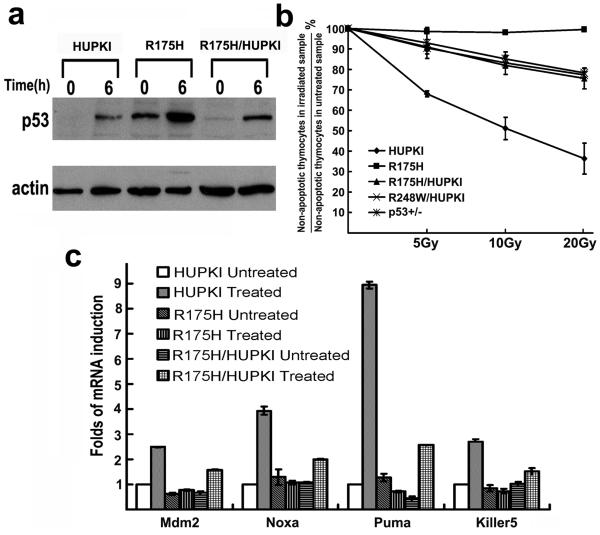
As expected for normal thymocytes, the protein levels of p53 were undetectable in HUPKI thymocytes before DNA damage but significantly increased after IR treatment (Figure 2a). Unlike the undetectable levels of p53 mutants in R248W thymocytes before DNA damage (Song et al., 2007), p53 protein levels were already high in R175H thymocytes before IR and further increased after IR (Figure 2a). Therefore, there exists a difference in the regulation of the stability of the p53 contact and structural mutants in thymocytes.
Figure 2. p53 stability and activity after DNA damage in thymocytes.
(a) Expression of p53 in HUPKI, R175H and HUPKI/R175H heterozygous thymocytes after 5Gy irradiation. Actin was used as the loading control. (b) p53-dependent apoptosis in HUPKI, R175H, HUPKI/R175H, HUPKI/R248W and p53+/− thymocytes 10 hours after treating with 5, 10 and 20Gy of IR. Mean values from three independent experiments are shown with standard deviation. (c) Expression of p53 target genes in HUPKI, R175H/HUPKI and R175H mutant thymocytes 6 hours after 5Gy of IR. mRNA levels of each gene were determined by real time PCR and normalized with the expression of GAPDH. The mRNA levels in the untreated HUPKI MEFs were set to 1.
While R175H is constitutively stabilized in thymocytes, these thymocytes were essentially resistant to p53-dependent apoptosis after IR treatment (Figure 2b). In addition, p53-dependent induction of target genes was abolished in R175H mutant thymocytes after IR treatment (Figure 2c). Therefore, R175H mutation abolishes the WT p53 functions in both MEFs and thymocytes. However, p53-dependent apoptosis in R175H/HUPKI thymocytes after IR is partially retained and at the same level as that in R248W/HUPKI and p53+/− thymocytes (Figure 2b).
R175H mice are cancer prone
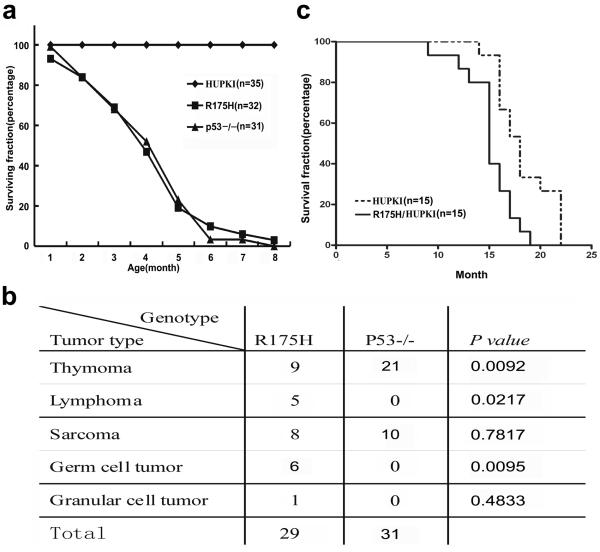
To test the impact of R175H mutation on p53-dependent tumor suppression, R175H mutant mice were monitored for the onset and spectrum of tumors and compared with p53−/− mice with similar genetic background. R175H mutant mice uniformly died of cancer with a similar kinetics as p53−/− mice (Figure 3a). However, the tumor spectrum observed in R175H mutant mice was more complex than that of p53−/− mice (Figure 3b). While thymic tumor and sarcomas were commonly observed in both R175H and p53−/− mice, other types of tumors including peripheral lymphomas (5 of 29) and germ-cell tumor (6 of 29) are only observed in R175H mice. The more complex tumor spectrum in R175H mutant mice supports the notion that R175H mutant gains new oncogenic function in promoting tumorigenesis. Consistent with the findings that p53-dependent apoptosis and senescence are partially retained in R175H heterozygous mutant mice, the onset of tumorigenesis in R175H/HUPKI mice is significantly delayed and suppressed in about half of the monitored mice when compared with that in R175H mice (Figure 3c).
Figure 3. R175H mice uniformly died of cancer.
(a) Survival curve of HUPKI, p53−/− and R175H mutant mice. N shows the number of mice monitored. Most of p53−/− and R175H mutant mice died within 6 months, while none of the HUPKI mice died within 8 months. (b) Tumor spectrum of p53−/− and R175H mutant mice. Two-tailed P-values are obtained from a Fisher's exact test using 2×2 contingency table. (c) Survival curve of R175H/HUPKI and HUPKI mice. Tumors were identified in 8 of the 15 R175H/HUPKI mice with one thymic lymphomas, one peripheral lymphomas and six sarcomas.
R175H mutant induces interchromosomal translocation
We have recently identified a gain of function of p53 contact mutants R248W and R273H in inducing interchromosomal translocation, the type of chromosomal translocation rarely seen in p53−/− cells (Bassing et al., 2003). Therefore, we tested whether this gain of function is also acquired by the common p53 structural mutant R175H. Interchromosomal rearrangement between Tcrβ loci on chromosome 6 and Tcrγ loci on chromosome 13 has been used as a predictor for the global interchromosomal translocation in mouse thymocytes (Kang et al., 2002; Lista et al., 1997). No Tcrβ-Tcrγ chromosomal translocations were detected in the thymocytes derived from p53−/− and HUPKI mice (Table 1). About 14% of pre-tumor thymus of R175 mice harbored the Tcrβ-γ translocation. In addition, similar frequency of translocation was detected in R175H heterozygous thymus, indicating that R175H can dominantly express the gain of function in the presence of co-expressed WT p53. Since the gain-of-function induced genetic instability is similar between the homozygous and heterozygous R175H mutant cells, the apparent delay and suppression of tumorigenesis in R175H heterozygous mutant mice indicate that the partially retained WT p53 function can effectively suppress tumorigenesis in R175H heterozygous mutant mice despite gain-of-function induced genetic instability.
Table 1.
Percentage of t(6, 13) chromosomal translocation in pre-tumor mouse thymocytes of HUPKI, p53−/−, R175H and R175H/HUPKI mice.
| Genotype | HUPKI | P53−/− | R175H | R175H/HUPKI |
|---|---|---|---|---|
| Number of mice analyzed |
20 | 19 | 35 | 36 |
| Number of translocation (Percentage) |
0 | 0 | 5 (14%) |
5 (13.8) |
Impaired ATM function in R175H cells
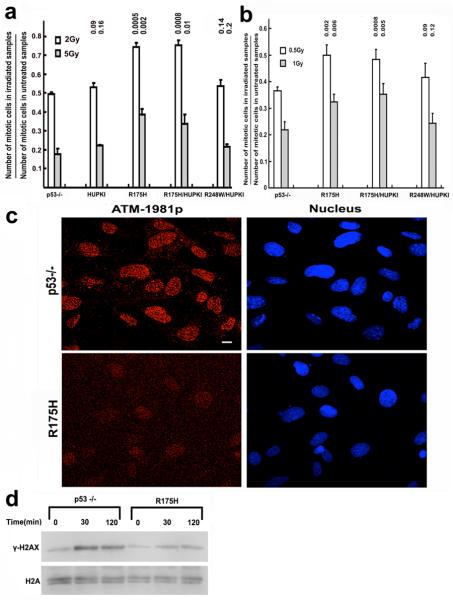
ATM kinase is critical to suppress chromosomal translocations and activate cell cycle checkpoints after IR (Xu, 2006). Therefore, we analyzed the G2/M checkpoint in R175H cells after IR. While the G2/M checkpoint was normal in p53−/− MEF cells, it was significantly impaired in both R175H homozygous and heterozygous MEFs (Figure 4a). In contrast, while G2/M checkpoint is impaired in R248W MEFs after IR (Song et al., 2007), the G2/M checkpoint was normal in R248W heterozygous MEFs after IR. Consistent with this finding, the G2/M checkpoint was similarly impaired in the proliferating R175H homozygous and heterozygous thymocytes (Figure 4b). Therefore, p53 structural mutant R175H has a dominant effect on the co-expressed WT p53 in expressing this gain of function.
Figure 4. Impaired G2-M checkpoint and ATM activation in R175H mutant cells after irradiation.
G2-M checkpoint in MEFs (a) and proliferating thymocytes (b) of various genotypes. Mean values from three independent experiments are shown with standard deviation. P values between p53−/− and other samples are indicated. (c) IRIF of ATM-1981p in p53−/− and R175H MEFs 30 mins after 2Gy irradiation. Nucleus is countered stained with DEPI. (d) Phosphorylation of H2AX (γ-H2AX) in p53−/− and R175H mutant MEFs after 2Gy of IR.
Upon DNA DSB damage, activated ATM, which is autophosphorylated at ser1981 (Bakkenist and Kastan, 2003), was recruited to the site of DNA damage and form irradiation induced foci. When compared with that in p53−/− MEFs, the IRIF of phosphorylated ATM was significantly impaired in R175H MEFs, indicating that ATM activation is impaired in R175H MEFs after IR (Figure 4c). In support of this notion, the ATM-dependent phosphorylation of H2AX (γ-H2AX), was reduced in R175H MEFs after IR when compared with that in p53−/− MEFs (Figure 4d). Therefore, p53 R175H mutant inactivates ATM after DNA DSB damage.
R175H impairs the recruitment of MRN/ATM to DSBs
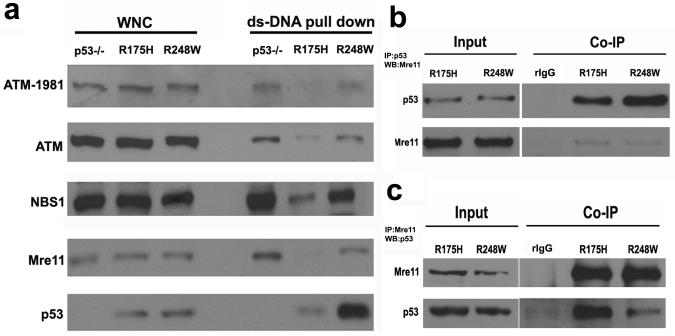
As MRN complex played a very important role in activating ATM (Lavin, 2007), we tested the hypothesis whether R175H mutant can influence the MRN-mediated recruitment of ATM to the DNA DSB damage sites. Employing a DSB DNA pull-down assay to show the association of MRN complex and ATM with DNA DSBs, we showed that the association of Mre11 complex with DSBs was significantly reduced in the nucleus p53 R175H MEFs when compared with that in p53−/− MEF nucleus (Figure 5a). This suggests that the recruitment of MRN complex to DSBs was impaired in R175H cells. Considering the important role of MRN in recruiting ATM to the site of DNA damage, the binding of phosphorylated ATM to DSBs was also reduced in the R175H nucleus when compared with p53−/− cells (Figure 5a). Therefore, both p53 contact and structural mutants impair the recruitment of MRN/ATM to DNA DSBs.
Figure 5. Impaired recruitment of MRN complex to the DSBs damage sites.
(a) Protein levels of ATM, ATM-1981p, Mre11, NBS1 and p53 in the dsDNA pull down using the nuclear extracts of p53−/−, R175H and R248W mutants. The whole nuclear extracts were used as the input control. (b, c) Co-immunoprecipitation to show the Interaction between p53 mutants and Mre11 in MEF. Whole cell lysates from R175H and R248W mutants were immunoprecipitated with anti-p53 antibody (b) or anti-Mre11 antibody (c) respectively, followed by western blot for Mre11 or p53. For both controls, the R175H mutant cell lysates were immunoprecipitated with rabbit IgG.
As a mechanism to understand how R248W impairs the recruitment of MRN complex to the site of DNA damage, we found that R248W mutant physically interacts with Mre11 (Song et al., 2007). To test whether p53 structural mutant also possesses this function, we tested the interaction between R175H and Mre11 with reciprocal co-immunoprecipitation. Our findings indicate that R175H mutant also interacts with Mre11 (Figures 5b, c). In summary, these findings support a common gain of oncogenic function of p53 cancer mutants in disrupting the recruitment of MRN/ATM to the site of DNA damage by physically interacting with Mre11, leading to ATM inactivation. These findings have important implications on the mechanisms underlying this gain of function of p53 cancer mutants. While all these mutations abolish the WT p53-dependent function, only R175H but not R248W and R273H mutations have significant impact on the p53 structure (Hainaut and Hollstein, 2000). The differential impact of the common p53 cancer mutations on p53 structure indicates that the gain of function in inactivating Mre11/ATM by complexing with Mre11 is not the result of certain structural changes induced by p53 mutations. In addition, when compared with R248W, R175H binds more Mre11 than R248W but is retained less at the DNA DSBs, suggesting that R248W could be recruited to the DNA DSBs in an Mre11-complex independent manner.
Based on the findings that Mre11 can interact with WT p53 after high levels of DNA damage in normal cells, it is possible that DNA damage induced posttranslational modifications of p53 promote the interaction between Mre11 and p53 cancer mutants (Carbone et al., 2002). Therefore, as a result of the lack of WT p53 function, the accumulation of DNA damage in the p53 mutant expressing cells leads to chronic activation of DNA damage responses and constitutive posttranslational modification of p53 mutants (Bartkova et al., 2005; Gorgoulis et al., 2005; Song et al., 2007). If this is the case, it could be predicted that this gain of function of p53 cancer mutants is shared by other p53 structural and contact mutants. In addition, the interaction between p53 cancer mutants and transcription factors such as NF-Y after DNA damage could contribute to the gain of function in disrupting the cell cycle checkpoints by modulating the expression of cell cycle genes (Di Agostino et al., 2006).
The identification of a common gain of function of p53 cancer mutants in inducing genetic instability by disrupting ATM has significant implication on current cancer therapy. Since R248W, R273H and R175H mutations contribute to about 15% of all p53 cancer mutations, our findings predict that this gain of function of p53 cancer mutants affects at least 8% of all human cancers. Many current cancer therapies such as radiotherapy or treatment with topoisomerase inhibitor eliminate the cancer cells by inducing DNA DSB damage into their genome. Considering the combination of the increased genetic instability due to impaired ATM activation and the lack of effective cell death pathways in p53 mutant cancer cells, these cancer therapies might promote tumorigenesis in the p53 mutant expressing cancer cells that survive the treatment. Our findings suggest that destabilization of p53 mutants and restoration of ATM function will improve the efficacy of current therapies in suppressing cancer cells expressing p53 cancer mutants.
Materials and Methods
Western blotting and co-immunoprecipitation analysis
Protein extracts from 4×105 MEFs or 5×105 thymocytes were resolved on 6–10% SDS–PAGE gel and transferred to nitrocellulose membrane, which was probed with a monoclonal antibody against p53 (pAb1801; Santa Cruz Biotechnology, Santa Cruz, CA), or β-actin (Santa Cruz Biotechnology), or γ-H2AX (Cell Signaling Technology). The membrane was subsequently probed with a horseradish peroxidase-conjugated secondary antibody and developed with ECL PLUS (Amersham, Piscataway, NJ). For co-immunoprecipitation experiment, 1–3 mg of whole cell protein extracts were immunoprecipitated with antibodies against Mre11 (Novus, NB100-142) or p53 (FL393, Santa Cruz Biotechology). The amount of p53, Mre11 in the immunoprecipitate was analyzed by western blotting.
Cell-cycle analysis
Cell cycle was analyzed as previously described (Song et al., 2007). Asynchronously growing MEFs or proliferating thymocytes stimulated for 48 hr by PMA (5ng/ml) and ionomycin (500ng/ml) (EMD Biosciences, SD, CA) were irradiated with various dosages and harvested 1 h after IR. Cells in mitosis were identified by staining with propidium iodide for DNA content and FITC-conjugated antibody recognizing histone H3 phosphorylated at Ser 10 (Upstate, Charlottsville, VA).
3T3 proliferation assay of MEFs
Passage 5 MEFs of different genotypes were serially passaged on 60mm dishes once every three days at a plating density of 3×105 cells/plate. At each passage, three plates were counted and the mean value presented. The accumulative cell number at each passage = (the increase fold of the cell number) × (the total cell number of the previous passage).
Histology analysis
Tumor samples were fixed in 10% buffered formalin, embedded in paraffin and sliced. All sections were stained with hematoxylin and eosin for histological assessment as previous described (Chao et al., 2006).
Double strand DNA pull-down assay
This assay was performed as described(Song et al., 2007). Briefly, biotinylated dsDNA (2 kb), which was generated by PCR amplification of pcDNA3.1 with biotinylated T7 primer and reverse primer, was immobilized on streptavidin paramagnetic beads (Dynabeads M-280; Invitrogen) with a density of 0.1μgDNA/10μl beads. MEFs were irradiated with 2Gy and harvested 30min later. One hundred μg of nuclear extracts from MEFs were used for each pulldown assay. Immobilized DNA (0.2 μg) was mixed with the nuclear extracts, incubated at room temperature for 30 min and incubated for another 2 h at 4C. Beads were washed with ice-cold buffer D (10 mM Tris at pH 7.6, 100 mM NaCl) and boiled for 5 min in SDS sample buffer. dsDNA-associated proteins were analysed by western blotting with specific antibodies.
Immunoflorescence microscopy
MEFs grown on coverslip were exposed to 2 Gy irradiation, and 30 min later, fixed with methanol:acetone (50:50). The fixed cells were blocked with 3% BSA in PBS for 30 min at room temperature and incubated with mouse monoclonal antibody recognizing phosphorylated ATM at 1987 (1:600 dilution) or a rabbit polyclonal antibody against H2AX-139p (1:200) in 1% BSA in PBS for overnight at 4□. After washing, the cells were incubated with secondary antibodies for 1 hr at room temperature. After mounting on glass slides with VECTASHIELD mounting media with DAPI (Vector Laboratories, Burlingame, CA), images were acquired by Olympus confocal microscope.
Supplementary Material
Acknowledgement
We thank Dr. M. Hollstein for the HUPKI construct. This work was supported by grants from NIH (R01 CA94254) and DOD Breast Cancer Research Program (W81XWH-08-1-0381) to Y.X. □
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–70. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633–40. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. Embo J. 2006;25:2615–22. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- Kang J, Bronson RT, Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. Embo J. 2002;21:1447–55. doi: 10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Lista F, Bertness V, Guidos CJ, Danska JS, Kirsch IR. The absolute number of trans-rearrangements between the TCRG and TCRB loci is predictive of lymphoma risk: a severe combined immune deficiency (SCID) murine model. Cancer Res. 1997;57:4408–13. [PubMed] [Google Scholar]
- Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–8. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–93. [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–80. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Xu Y. DNA damage: a trigger of innate immunity but a requirement for adaptive immune homeostasis. Nat Rev Immunol. 2006;6:261–270. doi: 10.1038/nri1804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.