Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that primarily affects immunocompromised individuals. Reverse genetics is commonly used to identify and characterize genes involved in a variety of cellular processes. In C. neoformans there is a limited set of positive selectable markers available to make gene deletions or other genetic manipulations. This has hampered the application of reverse genetics in this organism. We have adapted the Bacteriophage P1 Cre-loxP system for use in C. neoformans and successfully excised and reused the same drug marker, G418, to make two sequential gene deletions, lac1Δ and cap59Δ, in the same strain. This tool will allow investigators to make multiple sequential gene deletions in the same strain, which should facilitate the analysis of multigene families.
Keywords: Cre-recombinase, loxP, G418, Geneticin, NAT, nourseothricin, LAC1, CAP59
1. Introduction
Cryptococcus neoformans is an opportunistic fungal pathogen that primarily affects immunocompromised hosts. Initially infection can result in pneumonia and develop into a systemic infection that travels through the central nervous system, crossing the blood brain barrier resulting in cryptococcal meningitis. Infection with this organism is an AIDS defining illness (Casadevall and Perfect, 1998). C. neoformans grows well as haploid yeast cells, and has proved to be amenable to genetic manipulation. Additionally, with the sequencing of the cryptococcal genomes, JEC21, B3501 (Loftus et al., 2005), and H99 (jointly sequenced by Duke University, http://cgt.genetics.duke.edu, and the Broad Institute, http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans) the use of reverse genetics in determining a gene products’ involvement in cell integrity, cellular growth, known virulence factors, or virulence of C. neoformans is steadily increasing. Reverse genetics was used in the construction of a targeted gene deletion library that contains greater than 2000 gene deletions (Liu et al., 2008). These deleted candidate genes may prove to be involved in many of the previously mentioned processes.
Efficient selection of transformed cells requires a marker, and in C. neoformans both auxotrophic markers and positive selectable drug markers have been used successfully to generate deletion or mutant strains. The use of auxotrophic markers requires development of strains with the appropriate genetic background; therefore, they are not as suitable as positive drug markers for manipulation of clinical isolates or otherwise wild type strains. Currently, four positive selectable drug markers are used: nourseothricin (NAT) (McDade and Cox, 2001), phleomycin, geneticin (G418), and hygromycin (Hua et al 1999). The product of the nourseothricin acetyltransferase gene provides resistance to the aminoglycoside antibiotic nourseothricin (NAT) that interrupts protein synthesis (Mcdade and Cox 2000). Phleomycin is a copper-containing antibiotic that acts by interfering with DNA synthesis (Reiter et al., 1972). G418 is an aminoglycoside antibiotic that binds to the “A” site of the small ribosomal subunit; this action causes the ribosome to incorrectly read the genetic code (Yoshizawa et al., 1998). Hygromycin (HYG) has a similar mode of action to that of G418 by attacking the 30S ribosomal unit and thus interrupting protein translation (Broderson et al., 2000).
Using these four markers, many genes have been analyzed for multiple phenotypes, including virulence in animal models. The markers are also used for selection of gene modifications, such as the addition of epitope tags, point mutations, and swapping of promoter sequences. Additionally, they have been very effective when manipulating combinations of up to four genes. Strains with multiple gene deletions or modifications have been generated by sequential gene deletion: for example, using hygromycin, nourseothricin, and geneticin, three peroxiredoxins a.k.a. the thiol peroxidases (TSA1, TSA3, TSA4) were sequentially deleted to produce a triple TSA deletion strain (Baker et al., 2007; Giles et al., 2006; Missall et al., 2004). These markers have also been used to create multiple deletions through a combination of sequential gene deletion and genetic crosses: for example, a quadruple deacetylase deletion strain lacking all three chitin deacetylases, CDA1, CDA2, and CDA3, and the fungal polysaccharide deacetylase, FPD1 was made using all four markers and mating (Baker et al., 2007). Whether choosing to make multiple gene deletions through sequential gene deletion, mating, or a combination of the two, distinct markers for each gene are required. Because of the limited number of positive selectable markers for use in C. neoformans it can be difficult to utilize these strategies in large gene families such as the chitin synthases, which has more than four members (Banks et al., 2005).
In this study we demonstrate the use of the Cre-loxP system, originally described in Bacteriophage P1 (Sauer and Henderson, 1988), in C. neoformans. The Cre-loxP recombinase system includes two short asymmetric DNA sequences termed loxP sites (approximately 34 base pairs including two inverted repeats) and the Cre recombinase protein (38 kilodaltons). The Cre protein covalently binds to two short loxP sites and catalyzes recombination within them; any intervening DNA sequence is looped out and excised. The Cre-recombinase is the only protein required for this action.
The Cre-loxP system has been usurped for use in several organisms. For instance, two teams of investigators have used the Cre-loxP system in mammalian systems. Araki et al (2002) successfully incorporated the CRE gene into the germ line of mice (Araki et al., 2002). This created mouse lines that could be used for future gene deletions through gene trapping and targeting. Smith et al (1995) used this system in embryonic stem cells of mice to mediate chromosomal rearrangement during the expression of Cre-recombinase (Smith et al., 1995). The Cre-loxP system has also been used in the construction of recombinant adenoviral vectors (Hardy et al., 1997). To create recombinant viral vectors in adenoviruses for gene delivery, the Cre-loxP system has been placed into strain ψ5 of the adenovirus, with two loxP sites flanking the packaging site. When Cre-recombinase is expressed, the packaging site is deleted (Hardy et al., 1997).
The Cre-loxP recombination system has also been used in many fungal systems. It was successfully introduced into the non-pathogenic yeast, Saccharomyces cerevisiae (Sauer, 1987). By flanking the LEU2 gene with loxP sites this gene was successfully deleted during the transient expression of Cre-recombinase (Sauer, 1987). Additionally, the Cre-loxP system has been used in several other fungal systems, a few being: Yarrowia lipolytica, Neotyphodium coenophialum, Neotyphodium uncinatum, Epichloë festucae, Aspergillus nidulans, Schizosaccharomyces pombe, and Kluyveromyces marxianus (Fickers et al., 2003; Florea et al., 2009; Iwaki and Takegawa, 2004; Ribeiro et al., 2007; Watson et al., 2008). Furthermore, the Cre-loxP system was optimized for the deletion of targeted genes and marker recycling in the human fungal pathogen Candida albicans (Dennison et al., 2005).
Here we report the use of the Cre-loxP system for marker recycling in C. neoformans. Specifically, we describe the reuse of the G418 drug marker to produce a strain with two unique gene deletions. Using a sequential deletion strategy, we successfully deleted the laccase gene, LAC1 (Wang et al., 1995), and the capsular gene, CAP59 (Chang and Kwon-Chung, 1994), using the same positive selectable marker.
2. Materials and Methods
2.1 Fungal strains and media
KN99α strain of C. neoformans serotype A was used as the wild type strain (Nielsen et al., 2003), and all deletions were made in this background. Strains were grown on rich medium, YPD (1% yeast extract, 2% bacto-peptone, and 2% dextrose) or YPG (1% yeast extract, 2% bacto-peptone, and 2% galactose). Selective YPD or YPG media contained 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) or 200 μg/ml geneticin (G418) (Invitrogen, Carlsbad, CA).
2.2 PCR and cloning conditions for G418-loxP deletion construct
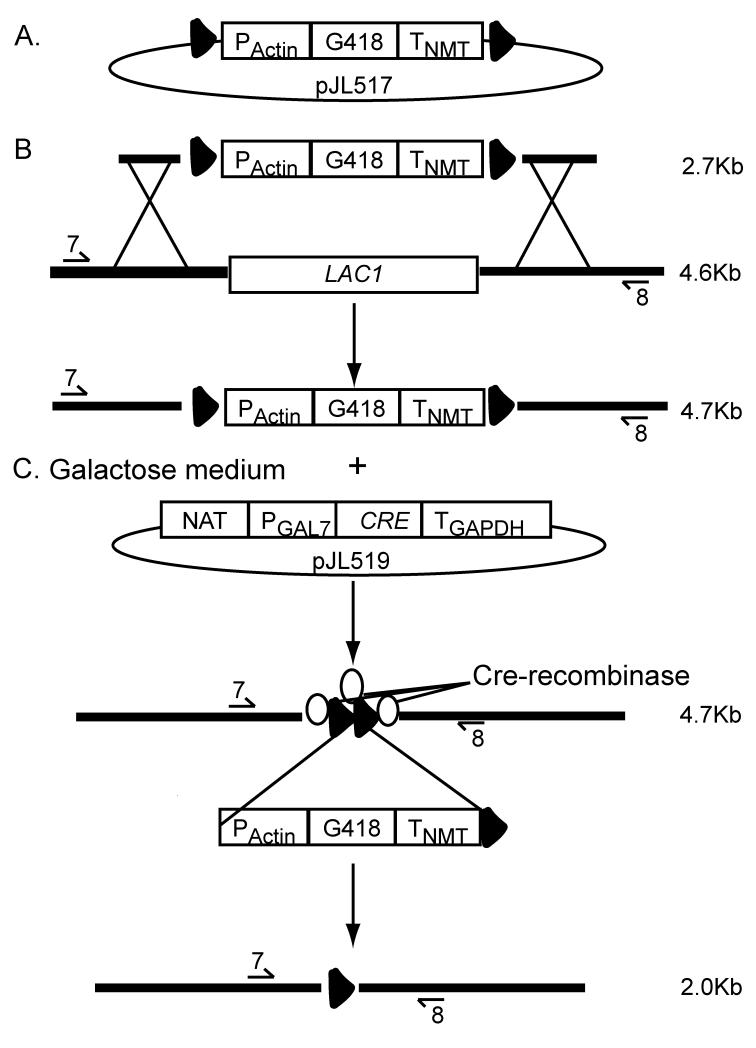
To add loxP sites to the geneticin drug marker we designed two primers that included the loxP sequence: the first primer consisted of 24 nt of the 5′ portion of the Actin promoter (PActin) that was added to the 3′ end of primer #319 reported by Guldener et al (Guldener et al., 1996), #319-Actin 5′-TCGACAACCCTTAATATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCTAGAaggatgtgagctggagagcggggc-3′. The second primer consisted of 24 nt of the 3′ end of the NMT1 terminator (TNMT) that was added to the 5′ end of primer #322 (Guldener et al., 1996), #322-NMT ACTACCTAATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTCGAGAGCTgggggacagatgatatccgacaag-3′. In both primer sequences the loxP sites are underlined. PCR reactions consisted of 50 pmols of each primer and 30 ng of pMH12-T vector template (G418 marker). Conditions were 94°C for 20 s, 60°C for 20 s, and 72°C for 4 m, cycled 35 times with a final extension of 72°C for 10 m. The PCR amplicon of 2.7 kb was gel purified using QIAquick gel extraction kit per manufactures instructions (QIAGEN, Valencia, CA). The fragments were cloned into the pCR2.1 vector and One Shot® TOP10 Chemically competent E. coli using the TOPO TA Cloning® Kit per manufactures instructions (Invitrogen, Carlsbad, CA). Inclusion of loxP sites flanking the G418 drug marker was confirmed by sequencing and the resulting vector was designated, pJL517 (Fig. 1).
Figure 1.
G418 marker recycling strategy. A. Flanking loxP sites (triangles) were added to the 2.7 kb G418 drug marker and ligated into a plasmid to create vector pJL517. B. Vector pJL517 and genomic DNA were used as templates in an overlap PCR that resulted in loxP-G418-loxP cassette flanked by the 5′ and 3′ UTR of LAC1. The native LAC1 (4.6 kb between primers #7 and #8) was replaced through homologous recombination with this construct. C. The strain created in B. was transformed with the uncut plasmid, pJL519 (CRE-recombinase); CRE was transiently expressed by growth on galactose induction medium. Translated Cre-recombinase (ovals) binds to the two-loxP sites. The 2.7 kb G418 construct is looped out, resulting in a PCR product that is 2.0 kb.
2.2 PCR and Cloning conditions for the CRE construct for use in C. neoformans
We designed a Cre-recombinase construct driven by an endogenous galactose inducible promoter, PGAL7 (Fig. 1). The galactose promoters work by an inducible (galactose) and repressible (glucose) system (Ruff et al., 2009). The inclusion of the GAL7 promoter allowed for the transient expression of the Cre-recombinase on medium containing galactose, YPG (Fig. 1). Because it has been shown that using endogenous terminators greatly increases the stability of a transcript in C. neoformans (Hua et al., 2000) an endogenous terminator, TGAPDH, was included in this construct. An overlap PCR method (Davidson et al., 2002) was used to create the CRE-recombinase construct. Vector pSDM3155, kindly provided by J. J. Hooykaas, supplied the CRE template (Schrammeijer et al., 2003). Plasmid pSDM3155 does not contain a start or stop codon, therefore both were included in the design of the overlap primers 5-, 6-, 7-, and 8-GAL7CRE (Table 1). PCR amplifications were carried out as described above. Primers 6- and 7-GAL7CRE were used to amplify a 1kb fragment containing CRE with a start and stop codon. Primers 3- and 8-GAL7CRE and 5- and 10-GAL7CRE were used to amplify the PGAL7 and TGAPDH terminator fragments, respectively. The template for these reactions was 100 ng genomic DNA from the C. neoformans strain, JLCN587. This strain contains a NAT drug resistance construct attached upstream of a GAL7 promoter. The CRE coding sequence was fused downstream of the GAL7 promoter and upstream of the GAPDH terminator using primers 3- and 10-GAL7CRE. This construct, NAT:PGAL7:CRE:TGAPDH, was cloned into a vector that we designated, pJL519 (Fig. 1). The pJL519 vector containing the CRE recombinase construct was verified by sequencing (Retrogen, San Diego, CA).
Table 1.
Primers used for constructs and screening
| Primer Name | Primer Sequence*^ | |
|---|---|---|
| lac1Δ construct | LAC1-1 | CAGTATGTTCCTGATTC |
| LAC1-2 | GCTTGTTCAGATCGTCTCTCAGATAGCC | |
| LAC1-3 | GGTGTCCATAATACTCTATGCcaggaaacagctatgaccatg | |
| LAC1-4 | catggtcatagctgtttcctgGCATAGAGTATTATGGACACC | |
| LAC1-5 | catggtcatagctgtttcctgCCTCTAAGACATCCACTTTCC | |
| LAC1-6 | GGAAAGTGGATGTCTTAGAGGgttgtaaaacgacggccagtg | |
| LAC1-7 | TGCCTTATGCGCGAACTTCAGGTTCACC | |
| LAC1-8 | TTGATTTCGTTCGCTTGGTGTTTCTGCC | |
| LAC1-9 | TGTGAGTGTCGGTATAGCTAC | |
| LAC1-10 | GATCCCAATGCATTTGGACCC | |
| cap59Δ construct | CAP59-1 | TGTCGCCCAAGTCGACGG |
| CAP59-2 | CCCGTCCAACGTCTATACCATC | |
| CAP59-3 | GAACATTTCTTTCCCCGCCcaggaaacagctatgaccatg | |
| CAP59-4 | catggtcatagctgtttcctgGGCGGGGAAAGAAATGTTC | |
| CAP59-5 | cactggccgtcgttttacaacGCCTGGATGGTAGATTTCT | |
| CAP59-6 | AGAAATCTACCATCCAGGCgttgtaaaacgacggccagtg | |
| CAP59-7 | AACTTGTTATAGGCGGATACGG | |
| CAP59-8 | TGAGATGCTAAGATGATCTAGCAAG | |
| CAP59-9 | GGAGACATACTGAAGCAACG | |
| CAP59-10 | TGCTGGTGATCTTGATGGAC | |
| CRE construct | 3-GAL7CRE | GTGCGGGTTCAACGATTTTGcaggaaacagctatgaccatgattac |
| 5-GAL7CRE | cgcctgctggaagatggcgatTAGAGGGGGTTACAGTAGCAGAATAG | |
| 6-GAL7CRE | ctattctgctactgtaaccccctCTAATCGCCATCTTCCAGCAGGCG | |
| 7-GAL7CRE | gcactcaattctctcctgagaATGTCCAATTTACTGACCGTACAC | |
| 8-GAL7CRE | GTGTACGGTCAGTAAATTGGACATtctcaggagagaattgagtgc | |
| 10-GAL7CRE | GAAGACCTTGAATCTTAGACCCGTCATCCTTGTCTGGCTGAT | |
| CRE Screening | CRE Screening-5 | GTTTCACTGGTTATGCGGCGG |
| CRE Screening-6 | ATCGCCATCTTCCAGCAGGCGCAC | |
| CRE qPCR | CRE Screening-5 | GTTTCACTGGTTATGCGGCGG |
| CRE 3′ qPCR | ATCGCTATTTTCCATGAGTGAACGAAC | |
| Actin RT-1 | ACCATTGGTAACGAGCGATTCC | |
| Actin RT-2 | AAGGATAGAACCACCGATCCAG | |
| Excision Site Sequencing | ||
| LAC1-F-CREseq | GATTGACCATACTTGTGC | |
| LAC1-R-CREseq | TGATGAAGTCCGTTAAAG | |
| CAP59-F-CREseq | AAGTTCCGCGGTTGTTCGG | |
| CAP59-R-CREseq | ACACATGGGACTGGGTCG | |
Lowercase for lac1Δ and cap59Δ=drug marker cassette sequence; for CRE construct= marker sequence, PGAL7, and TGAPDH
Addition of start and stop codons to CRE construct primers underlined
2.4 Generation of deletion constructs
Overlap PCR gene deletion technology was used to generate gene-specific deletion cassettes of LAC1 or CAP59. Each reaction included a G418 cassette flanked by loxP sites (see above). Primers used in their construction are shown in Table 1. The amount of coding sequence deleted for LAC1 and CAP59, was 2.6 kb and 2.0 kb, respectively.
2.5 Transformation of C. neoformans
KN99α was transformed using biolistic techniques. Cells were grown in YPD to late log-phase, concentrated, and plated onto YPD agar for transformation. The cells were bombarded with 0.6 μm gold beads (Bio-Rad, Richmond, CA) that were coated with DNA of the target construct according to the manufacturer’s recommendations. Following the transformation, the cells were incubated at 30°C for 4 h on nonselective medium to allow for recovery and then transferred with 0.8 ml sterile PBS to the appropriate selective medium. Transformants were observed in 3-5 days.
2.6 Analysis of transformants
To isolate stable transformants, all transformants were passaged five times on nonselective YPD medium and then tested for resistance to the appropriate selective marker. Only those transformants that grew equally well on selective and nonselective media were considered to be stable. A three-primer PCR screen was used to verify homologous integration at both the 5′ and 3′ ends of the deletion cassette. In this manner, homologous recombinants can be distinguished from wild type. A PCR screen with primers flanking the deletion construct was used to amplify the entire integration region and determined that a single copy of the transforming DNA had been inserted at the desired locus. Southern blots were performed to screen for single integration in the genome. Single bands were observed on all Southern blots when hybridized with a selectable marker-specific probe. All deletion strains generated for this work had a single deletion construct integrated by homologous recombination at the appropriate locus and no other insertions in the genome (data not shown). At least three independent isolates for each deletion were obtained.
2.7 Transformation and analysis of Cre-recombinase mediated drug maker loss
Confirmed lac1ΔG418R and lac1Δ G418Scap59ΔG418R strains were transformed using biolistic techniques as described. Cells were grown in YPG to late log-phase, concentrated, and plated onto YPG agar for transformation and subsequent expression of CRE by the PGAL7. The cells were bombarded with 0.6 μm gold beads (Bio-Rad, Richmond, CA) coated with 0.5, 1.5, or 5 μg plasmid DNA (pJL519) as a non-integrative construct that contains a nourseothricin-resistance cassette. Following transformation, the cells were incubated at 30°C for 4 h on nonselective YPG medium then transferred to YPG with 100 μg/ml nourseothricin. To isolate geneticin sensitive transformants, nourseothricin resistant transformants were picked and passaged 5X for two days on nonselective non-induction YPD medium. The last passage was onto YPD and YPD supplemented with 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) or 200 μg/ml G418 (Invitrogen, Carlsbad, CA). Transformants that grew only on YPD without drug were selected for further screening. Primers seven and eight (Table 1) for each deletion construct (lac1Δ or cap59Δ) were used to PCR amplify the locus of the original gene replacement for each positive transformant. To determine the loss of the plasmid containing the CRE gene, all positive transformants were screened with CRE-specific primers that amplify a 500 bp fragment of CRE (Table 1). Loss of the 2.7 kb G418 cassette was determined by gel electrophoresis. The genomic region surrounding the excision site was sequenced to verify genomic integrity and retention of one loxP site (Protein and Nucleic Acid Chemistry Laboratory, Washington University, Saint Louis, MO).
2.8 Quantitative PCR of CRE
Total RNA was extracted from lyophilized lac1ΔG418R + 5 μg pJ516 (CRE vector) or 5 μg pHL001 (NAT vector) cultures that had been grown 48 hr in either YPD or YPGlcNAc with 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) using Agilent total RNA isolation kit (Agilent Technologies, Wilmington, DE) per manufactures instructions for yeast cell cultures. First-strand cDNA was made using 1 μg total RNA using the iScript cDNA synthesis kit (BioRad, Hercules, CA). This cDNA was used as template in a real-time PCR using SsoFast EvaGreen Supermix (BioRad, Hercules, CA) according to the manufacturer’s recommendations. A Biorad CFX96 thermal cycler was programmed with the following two step PCR cycles: Initial denaturation for 30 s at 98°C then 10 s at 98°C, 35 s at 60°C, with a plate read repeated in the second step for a total of 35 cycles. A melting curve was performed at the end of the reaction to confirm a single product. Standard curves were performed for each primer set and efficiencies calculated. The data were normalized to Actin cDNA expression included with each experiment.
2.9 Genomic DNA preparation
Genomic DNA was prepared by a modification of the glass bead DNA extraction protocol described. Briefly, C. neoformans cells were suspended in a microfuge tube in 500 μl lysis buffer (50 mM Tris-HCl, pH 7.5, 20 mM EDTA, 1% SDS), with 400 mg glass beads (425-600 μm; Sigma G-9268, St Louis, MO). Cells were disrupted by vortexing 10 m, followed by 10 m incubation at 70°C. After brief vortexing, 200 μl 5M KOAc and 200 μl 5M NaCl were added. The tubes were placed on ice for 20 m and centrifuged at 14,000 rpm for 20 m. The supernatant was mixed with 500 μl phenol/chloroform and spun for 5 m at 14,000 rpm. The aqueous phase was then mixed with 500 μl chloroform and spun for 5 m at 14,000 rpm. The DNA was then precipitated by addition of 500 μl isopropanol, dried, and resuspended in 50 μl deionized water.
2.10 Southern hybridizations
Approximately 10 μg of genomic DNA from each strain was digested with various restriction endonucleases according to the manufacturer’s recommendations. Restriction fragments were separated on a 1% agarose gel and transferred to nylon membranes using a Turbo-Blot apparatus (Schleicher & Schuell) with 10X SSC as transfer buffer. Probes for Southern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P] dCTP (GE Bio-Sciences AA0005, Piscataway, NJ) according to the manufacturer’s instructions. The blots were incubated in 10 ml of buffer (1X phosphate buffer, 7% SDS) solution for 1 h at 65°C, then probe was added to this solution, and the blots were hybridized at 65°C overnight. The blots were washed twice in 2X SSC, 0.1% SDS at room temperature for 10 min and once for 10 min in 0.2X SSC, 0.1% SDS that had been prewarmed to 65°C.
2.11 Analysis of melanin production
Cells of each strain were taken from solid YPD medium and spread onto glucose-free asparagine medium containing 2% bacto-agar (1g/liter L-asparagine, 0.5 g/liter MgSO4, 3 g/liter KH2PO4, 1 mg/liter thiamine) plus 1mM L-Dopa. Incubation was at 30°C for 3-5d in the dark. Melanin production was determined visually.
2.12 Analysis of capsule formation
Strains were streaked onto DME plates (13.4 g/L Dulbecco’s Modified Eagle’s Medium (Sigma, St. Louis, MO), 25 mM MOPS pH 7.0, 1.8% agar) and incubated for 3d at 30°C. Individual isolates were resuspended in 1:4 India Ink:H2O solution. Cells were observed through an Olympus AHBT3 microscope at 1000X magnification.
3. Results
3.1 Constructs generated for recycling of the selectable drug marker G418
We were interested in creating a drug marker for use in C. neoformans that could be excised after being used to screen for a genetic change, and then reused to produce multiple gene deletions in the same strain. To accomplish this, we took advantage of the Cre-loxP system that requires two main components for the excision of DNA: the first consists of two-loxP sites flanking either side of a given DNA sequence; the second is a functional Cre-recombinase protein. For the first component, we utilized PCR to engineer two loxP sites to flank the geneticin drug marker, and inserted the product into a plasmid (see Material and Methods), to create the vector designated, pJL517 (Fig. 1). To assemble the second component, we designed a Cre-recombinase construct driven by an endogenous galactose inducible promoter, PGAL7. The galactose promoters work by an inducible (galactose) and repressible (glucose) system (Ruff et al., 2009). Hence, inclusion of the GAL7 promoter allowed for the transient expression of the Cre-recombinase on medium containing galactose, YPG. The construct containing NAT:PGAL7:CRE:TGAPDH was made using overlap PCR (see material and methods) and cloned into the vector we designated pJL519 (Fig. 1).
3.2 Deletion of LAC1 with loxP-G418-loxP cassette
Cryptococcal cells can use L-3,4-dihydroxyphenylalanine (L-dopa) as a precursor to produce the pigment melanin. The primary enzyme that is responsible for this reaction is encoded by LAC1 (Missall et al., 2005; Williamson, 1994). Because the loss of melanin production is an easily scored phenotype, we selected LAC1 as our first gene deletion candidate using the loxP-G418-loxP cassette. We used overlap PCR (Davidson et al., 2002) to make a deletion cassette that contained the loxP-G418-loxP construct flanked by approximately 1 kb of the 5′ and 3′ UTR of LAC1. The deletion cassette was biolistically transformed into a wild type strain, KN99α. Transformants resistant to G418 were selected for further screening.
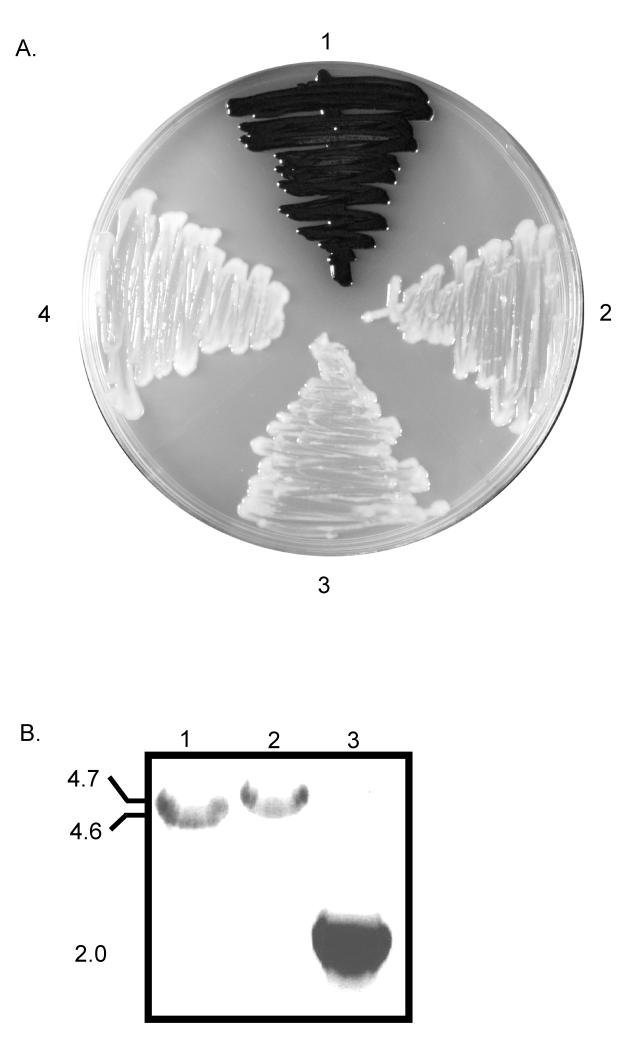
Transformation of KN99α with the construct resulted in the homologous replacement of genomic LAC1 (Fig. 1), which was confirmed by phenotype, PCR, and Southern blot (Fig. 2 and data not shown). After five days of growth on L-dopa medium, the wild type strain produced abundant melanin, which caused the colonies to appear black (Fig. 2). In contrast the lac1ΔG418R deletion strain appeared phenotypically similar to a previously generated negative control strain lac1Δ(Fig. 2). PCR analyses using LAC1 specific primers (7 and 8) that lie outside the region of homologous recombination were used to determine if the loxP-G418-loxP cassette replaced the endogenous LAC1 gene at its native locus. The PCR product of 4.7 kb indicated the loxP-G418-loxP cassette had replaced the wild type LAC1 (4.6 kb) in the lac1ΔG418R strain (Fig. 2). Southern blot analysis confirmed that only one integration event occurred in this strain during transformation (data not shown). These data indicated that LAC1 was deleted in the lac1ΔG418R strain and the inclusion of loxP sites flanking the G418 marker did not adversely affect the deletion cassette.
Figure 2.
Evaluation of the deletion of LAC1 and removal of G418 marker. Strains were grown five days on L-dopa medium at 30°C in dark. A. 1. KN99α, production of abundant black melanin pigment. 2. lac1Δ (negative control strain), showing no discernible pigment production causing white appearance of colonies. 3. lac1ΔG418R, white appearance of colonies indicates that LAC1 has been successfully deleted with the loxP-G418-loxP construct. 4. lac1ΔG418S, transient expression of Cre-recombinase, G418 construct was excised and the strain remained unable to produce melanin. B. PCR products using LAC1 specific primers #7 and #8 for 1. wild type LAC1, 2. lac1ΔG418R, and 3. lac1ΔG418S. Molecular weights listed to the left side of gel image.
3.3 Transient expression of Cre-recombinase and G418 removal
In order to remove the loxP-G418-loxP construct from the lac1ΔG418R strain, we transiently expressed the Cre-recombinase protein using the galactose inducible CRE construct described in Fig. 1. We anticipated that the Cre-recombinase protein known to target the two-loxP sites would be translated and shuttled to the nucleus where it would loop out the G418 selectable marker (as described in Fig. 1). The culmination of these events should result in a strain deleted for LAC1 and the G418 construct. We transformed the lac1ΔG418R with 1.5 μg of plasmid DNA containing the CRE gene. After three to five days on inducible YPG-NAT selection, 19 colonies were recovered and moved to non-induction YPD medium. These putative transformants were passaged as described in Materials and Methods. Passaging aids in the loss of vector that might be maintained extra-chromosomally. For the last selection, the 19 colonies were replicated on non-induction YPD, YPD-G418, and YPD-NAT. All 19 strains were sensitive to NAT; however, only one of the 19 displayed sensitivity to both NAT and G418. This data indicated that the G418 marker had been looped out and lost, and that the NAT marker was also no longer present in this strain. PCR analysis of this transformant, lac1ΔG418S, using primers LAC1-7 and LAC1-8 indicated that 2.7 kb of DNA corresponding to the length of the G418 marker had been lost at the LAC1 locus. PCR analysis using CRE-screening primers −5 and −6 that amplify a 500 bp region of the CRE gene confirmed that the CRE vector was absent from the this strain (data not shown). This indicated that after transformation onto galactose medium the Cre-recombinase was expressed and caused the loss of the 2.7 kb G418 fragment. This produced a G418 sensitive LAC1 deletion strain, and growth of this strain on L-dopa medium confirmed the LAC1 gene was still deleted (Fig. 2.).
3.4 Cre-recombinase specific excision
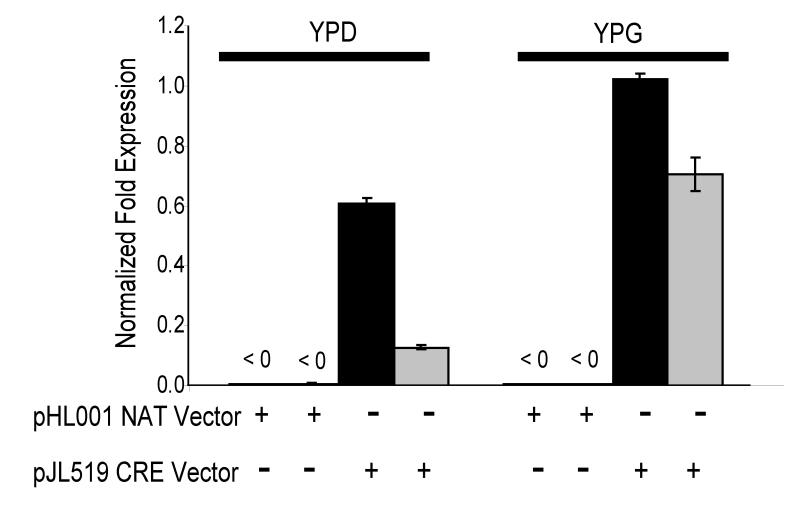
To determine if the excision of G418 from the lac1ΔG418S strain was due to Cre-recombinase we used quantitative PCR to measure CRE expression under inducing (galactose) and non-inducing (glucose) conditions in the lac1ΔG418R strain transformed with the pJL519 vector. In two independent isolates the data indicated that CRE transcript levels were present even during non-inducing conditions, but expression levels increased during inducing conditions (Fig 3). Of note, both of these strains were also determined to be sensitive to G418 (data not shown). We further sequenced this locus and confirmed the expected presence of 262 nucleotides corresponding to one loxP site and vector DNA sequence that was amplified during the construction of the deletion cassette (Table 1). Additionally, sequencing confirmed that the 5′ and 3′ untranslated region of the LAC1 gene remained intact (data not shown). To determine the rate of background excision (i.e. CRE independent excision) three independent transformations of the lac1ΔG418R strain were done with an empty vector. No G418S colonies were recovered from these transformations (data not shown). Taken together the data suggest that the excision of the loxP-G418-loxP drug marker was due to Cre-recombinase.
Figure 3.
CRE quantitative PCR of transformed lac1ΔG418R strains. lac1ΔG418R was transformed with either 5 μg of pJL519 or an empty vector (pHL001), passaged four times on selective medium, then grown 48 h in YPD or YPG containing 100 μg/ml nourseothricin. Two independent isolates from transformations are shown. (+) Indicates the transformation vector used on the strains. Bars at top of graph indicate growth medium YPD (uninduced) and YPG (induced). CRE expression levels were normalized to Actin expression level for each strain in each condition. Data is from three replicates, error bars SEM (+/−).
3.5 Recycling of G418 marker and subsequent removal
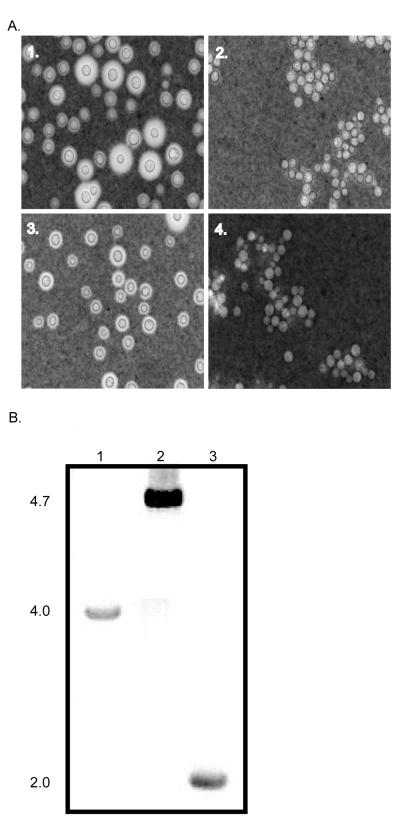
To establish that the G418 marker could be reused, we deleted the CAP59 gene from the lac1ΔG418S strain. Cap59 is involved in the production of the polysaccharide capsule that surrounds the cell wall of C. neoformans and the deletion of this gene has an easily recognizable phenotype. The capsule can be induced in low iron (Dulbecco’s Modified Eagle’s medium; DME) and/or in the presence of CO2. Strains deleted for the CAP59 gene produce little to no capsule upon induction (Granger et al., 1985). Transformation of lac1ΔG418S strain with the deletion construct containing loxP-G418-loxP flanked by 5′ and 3′ UTR of the CAP59 gene resulted in the homologous replacement of genomic CAP59 as confirmed by PCR and Southern blot analysis (data not shown). After three days of growth on DME medium, cells were stained with India ink for the visualization of capsule (see Materials and Methods). The lac1ΔG418Scap59ΔG418R deletion strain displayed the typical acapsular phenotype with its induced capsule being markedly smaller than both the wild type KN99α and parental lac1ΔG418S strains (Fig. 4.). Additionally, the lac1ΔG418Scap59ΔG418R cells displayed a clumping phenotype typical of the acapsular control strain, cap59Δ (Fig. 3). These data suggested that CAP59 was deleted with the loxP-G418-loxP construct and indicated the successful reuse of the G418 marker.
Figure 4.
Evaluation of the deletion of CAP59 and removal of G418 marker. A. To induce capsule formation the strains were grown three days on low iron medium, DME, at 30°C in dark. Strains were stained with 1:1 India Ink:H2O and observed through an Olympus AHBT3 microscope at 1000X magnification. 1. KN99α, production of capsule surrounding the cell wall. 2. cap59Δ (negative control strain), upon induction no discernible capsule is produced and cells clump. 3. lac1ΔG418S, production of capsule surrounding the cell wall is unaffected by the deletion of LAC1. 4. lac1ΔG418Scap59ΔG418R, no discernible capsule is produced and cells clump similar to negative control. B. PCR products using CAP59 specific primers #7 and #8 for 1. wild type CAP59, 2. lac1ΔG418Scap59ΔG418R, and 3. lac1ΔG418Scap59ΔG418S. Molecular weights listed to the left side of gel image.
Once the deletion of CAP59 was confirmed, we again transiently expressed the Cre-recombinase protein using the construct described above. To boost the number of putative transformants recovered, we changed the amount of transforming plasmid from 1.5 μg to 0.5 μg or 5.0 μg. As no putative transformants were recovered from the transformation utilizing 0.5 μg of transforming vector decreasing the amount of transforming DNA had the opposite affect that we desired. In contrast, 32 putative transformants from the 5.0 μg transformation were recovered, selected, and screened as described in material and methods. Of these, 14 were found to be sensitive to both G418 and NAT. This suggested that the G418 marker was excised and the CRE-recombinase vector had been lost. However, PCR analysis with the CRE-screening primers −5 and −6 indicated that three of the 14 maintained the CRE gene. This data suggested that either the CRE gene had inserted ectopically or it was still being maintained extrachromosomally in the genome of these three strains. The other 11 strains were negative for the CRE sequence, which indicated that the CRE vector was not maintained (data not shown). Therefore, by increasing the amount of the Cre-recombinase vector in the transformation from 1.5 μg (amount used in the initial transformation of the lac1ΔG418R strain) to 5.0 μg the number of positive transformants having Cre-mediated marker excision increased. Seeking to further increase the number of transformants having an excised marker event we added a native nuclear localization signal to the CRE-recombinase construct. However, we found this inclusion did not increase the number of recovered transformants (data not shown). PCR analysis of the eleven-lac1ΔG418Scap59ΔG418S strains using primers CAP59#7 and CAP59#8 indicated that the 2.7 kb G418 fragment had been excised at the CAP59 locus (Fig. 4). Three independent isolates were sequenced and as previously noted for the excision of the G418 drug marker from the LAC1 locus, we found 262 nucleotides corresponding to one loxP site and expected vector DNA sequence. Again, like LAC1, the 5′ and 3′ untranslated region of the CAP59 gene remained intact (data not shown). These data indicated that the Cre-loxP system could be used successfully in C. neoformans for the removal of DNA flanked by two-loxP sites, and that this strategy can be used repeatedly.
4. Discussion
We produced a recyclable drug marker and Cre-recombinase vector for use in C. neoformans and demonstrated their successful use for the sequential deletion of LAC1 and CAP59 genes in the same strain. This entire process utilized only two drug markers, one for the loxP-G418-loxP construct and one for the transient expression of Cre-recombinase (NAT), and created a strain that can be transformed again with both drug markers. The excision of the loxP flanked G418 drug marker was not highly efficient in C. neoformans. However, depending upon the system, others have reported a varying range of efficiencies from as low as 0.5% (Neotyphodium uncinatum and Epichloë festucae (Florea et al., 2009)) to nearly 100% (Candida albicans and Yarrowia lipolytica (Dennison et al., 2005; Fickers et al., 2003)). Even with this intermediate efficiency, a major advantage of using the Cre-loxP system in C. neoformans is that investigators can exploit just two positive selectable markers for the generation of multiple sequential gene deletions.
It is interesting that we observed expression of the episomal CRE-recombinase during uninduced growth conditions, as well as the level of induced expression being only two-four fold above that of uninduced (Fig. 3). When the same galactose promoter, PGAL7, was incorporated into the chromosomal locus of ADE2, a greater than 20-fold induction was observed compared to the complete absence of gene expression in uninduced cultures (Ruff et al., 2009). These differences between episomal and chromosomal expression may indicate that the control of gene expression on episomes with the galactose promoters is not as efficient as that of control on chromosomes or that glucose-repression/galactose-induction is location dependant. Even though expression was observed during uninduced conditions, G418 sensitive colonies were never recovered without induction (data not shown). This indicated that a critical level of CRE expression might be necessary for the removal of the loxP-G418-loxP drug marker from the strains.
Because many gene families in C. neoformans contain multiple members, the Cre-loxP system can be especially useful. For example, redundancy in the antioxidant defense system is due to the layers of components that contribute to the viability of the cell (Giles et al., 2006). The antioxidant gene family includes four catalases (Giles et al., 2006), three thiol peroxidases (Missall et al., 2004), two glutathione peroxidases (Missall and Lodge, 2005), superoxide dismutase (Giles et al., 2005), and an alternative oxidase (Akhter et al., 2003). The Cre-loxP system could be useful when dealing with complex gene families, which require multiple gene deletions for functional dissection.
This system could prove to be a major advantage when trying to establish synergism between deletions that cause severe cellular defects. For instance, many single gene deletions result in a mutant strain that does not transform or mate, which makes establishing phenotypic synergism between two or more gene deletions problematic. The loxP sites could be engineered around both genes and then the plasmid containing Cre-recombinase introduced. It is likely that simultaneous excision of both genes would occur, thus allowing phenotypic analysis of the double mutant. Potentially, this system could even be expanded to encompass several genes being deleted simultaneously.
In summary, we have adapted the Bacteriophage P1 Cre-loxP system for use in C. neoformans. The possibilities and usefulness of this tool for manipulating the C. neoformans genome are numerous. We anticipate that this tool will be of widespread use and will hopefully become indispensable in the Cryptococcal community.
Acknowledgements
This work was supported by NIH-NIAID grants R01AI072185 and R01AI050184 to JKL. The authors would like to thank Kim Gerik, Leona Campbell and Jack Ruff for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhter S, et al. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 2003;71:5794–5802. doi: 10.1128/IAI.71.10.5794-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, et al. Site-directed integration of the CRE gene mediated by Cre recombinase using a combination of mutant lox sites. Nucl. Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LG, et al. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–67. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks IR, et al. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderson DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell Press. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. ASM; Washington, D.C.: 1998. [Google Scholar]
- Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;138:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Dennison PMJ, et al. Gene disruption in Candida albicans using a synthetic, codon-optimized Cre-loxP system. Fungal Genet Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Fickers P, et al. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Meth. 2003;55:727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Florea S, et al. Elimination of marker genes from transformed filamentous fungi by unselected transient transfection with a Cre-expressing plasmid. F Fungal Genet Biol. 2009;46:721–730. doi: 10.1016/j.fgb.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Giles SS, et al. Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot Cell. 2005;4:46–54. doi: 10.1128/EC.4.1.46-54.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, et al. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell. 2006;5:1447–1459. doi: 10.1128/EC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger DL, et al. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;72:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, et al. A new efficient gene disruption cassette for repeated use in budding yeast. Nucl. Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S, et al. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, et al. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin Diagn Lab Immun. 2000;7:125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Takegawa K. A set of loxP marker cassettes for Cre-mediated multiple gene disruption in Schizosaccharomyces pombe. Biosci Biotech Bioch. 2004;68:545–550. doi: 10.1271/bbb.68.545. [DOI] [PubMed] [Google Scholar]
- Liu ZH, et al. Cloning and characterization of a novel chitinase gene (chi46) from Chaetomium globosum and identification of its biological activity. Appl Microbiol Biot. 2008;80:241–252. doi: 10.1007/s00253-008-1543-x. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade H, Cox G. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Missall TA, Lodge JK. Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:487–489. doi: 10.1128/EC.4.2.487-489.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missall TA, et al. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot Cell. 2005;4:202–208. doi: 10.1128/EC.4.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missall TA, et al. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol Micro. 2004;51:1447–1458. doi: 10.1111/j.1365-2958.2004.03921.x. [DOI] [PubMed] [Google Scholar]
- Nielsen K, et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter H, et al. Mode of action of phleomycin on Bacillus subtilis. J. Bacteriol. 1972;111:586–592. doi: 10.1128/jb.111.2.586-592.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro O, et al. Application of the Cre-loxP system for multiple gene disruption in the yeast Kluyveromyces marxianus. J Biotechnol. 2007;131:20–26. doi: 10.1016/j.jbiotec.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Ruff JA, et al. Three galactose inducible promoters for use in C. neoformans var. grubii. Fungal Genet Biol. 2009;46:9–16. doi: 10.1016/j.fgb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. PNAS USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, et al. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucl. Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJH, et al. A site–directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination. Nature Genetics. 1995;9:376–385. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Cryptococcus neoformans melanin and virulence: Mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AT, et al. Gene tagging and gene replacement using recombinase-mediated cassette exchange in Schizosaccharomyces pombe. Gene. 2008;407:63–74. doi: 10.1016/j.gene.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: Identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, et al. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]