Abstract
A novel diagnostic immunoassay testing procedure for hepatitis B virus core antibody (anti-HBc) using homogeneous purified full-length hepatitis B virus core antigen (HBcAg) capsids obtained from Escherichia coli was compared with Abbott Architect anti-HBc chemiluminescent microparticle immunoassay (CMIA; indirect method) against a library of specimens. A monoclonal anti-HBc neutralization confirmatory assay was then used to determine the degree of discordance between specimens. The new assay was found to be superior in both sensitivity and specificity.
Antibodies (anti-HBc) to hepatitis B virus (HBV) core antigen (HBcAg) are present in current and past HBV infections. Anti-HBc antibodies represent a long-term serological marker of HBV infection, initially appearing during the acute phase of the infection and generally persisting thereafter (16). Thus, as anti-HBc is a universal marker of HBV infection, routine blood donor screening for anti-HBc has been implemented in some countries with low endemicity. It is a cost-effective method of screening. Such screening procedures have resulted in a decrease in the risk of posttransfusion HBV infections (12). In some individuals, the only serological marker of HBV infection is the presence of anti-HBc antibodies (10, 24), and thus detection of “anti-HBcAg alone” could reflect unrecognized “occult” HBV infection and physicians should consider investigating such patients with HBV molecular tests (21). Additionally, isolated anti-HBc can be used as a marker to assess the risk of HBV reactivation in patients undergoing therapy that could result in immunosuppression or patients who are HIV positive (17) or hepatitis C virus (HCV) positive (25).
With these uses in mind, having a more efficient and reliable assay for anti-HBc is desirable. Most current commercially available anti-HBc assays have poor sensitivity or specificity (2, 19) and can be attributed to the inferior performance of the competitive immunoassay, especially for detecting low-titer anti-HBc-reactive samples. False-positive reactivity can partially be attributed to unspecific activation of premature B lymphocytes causing the production of IgM, IgA, or IgM-related molecules without previous exposure to HBV (18, 19). The specificity of competitive assays for anti-HBc can be significantly improved by addition of mild reducing agents, but such modified procedures often lead to the loss of sensitivity, particularly for IgM anti-HBc (23). In this study, a novel immunoassay for anti-HBc based on the double-antigen sandwich enzyme-linked immunosorbent assay (ELISA) method is compared with a commercial anti-HBc assay, the Architect chemiluminescent microparticle immunoassay (CMIA).
MATERIALS AND METHODS
Expression and purification of full-length rHBcAg in E. coli.
Two full-length HBcAg genes were obtained by chemosynthesis: AB090269 (genotype D, named CpD) and AB014368 (genotype C, named CpC). The rare arginine codes in Escherichia coli (AGA and AGG) located in the HBcAg gene were changed to CGT. The two fragments were inserted into the pTO-T7 vector by NdeI and HindIII restriction sites and transformed into Escherichia coli BL21 cells for HBcAg expression. The transformant was cultured in LB medium at 37°C for 5 h and then further incubated for 8 h in the presence of 1 mM IPTG (isopropyl β-d-thiogalactoside). Cells were harvested and then centrifuged at 10,000 × g for 10 min, after which they were lysed by sonication. HBcAg in the supernatant was precipitated with 23% ammonium sulfate and collected by centrifugation. The precipitated fraction was dissolved with 4 M urea buffer (20 mM Tris-HCl, pH 8.0, containing 4 M urea, and 20 mM dithiothreitol [DTT]) and purified in a Q FF column and phenyl high-performance (HP) column (Amersham GE Health, Uppsala, Sweden) with the AKTA system (GE Health, Uppsala, Sweden). The purity and identity of the protein were analyzed by SDS-PAGE and Western blotting.
In vitro HBc capsid assembly.
HBcAg expressed in E. coli accumulates in a particulate state referred to herein as HBcAg capsids. The precipitated fraction of the salting-out process was dissolved in a buffer containing urea and DTT to dissociate the particles which might entrap turbid proteins and nucleic acids and hence interfere with the immunoassay. The purified HBcAg was first dialyzed against 20 mM Tris-HCl (pH 8.0), containing 4 M urea and 20 mM DTT to remove NaCl; and then against 20 mM Tris-HCl (pH 8.0), containing 20 mM DTT to remove urea; and finally against 20 mM sodium phosphate (pH 6.0), containing 300 mM NaCl to remove DTT and trigger spontaneous assembly. These processes were performed using the Cross-Flow filtration systems (Amersham GE Health, Uppsala, Sweden). The homogeneity of the assembled recombinant HBc (rHBc) capsids was then identified by transmission electron microcopy (JEM 2100; JEOL, Tokyo, Japan).
The double-antigen sandwich immunoassay for anti-HBc.
The purified CpD protein was conjugated with horseradish peroxidase (HRP) by the NaIO4 oxidation method. The conjugate was purified by gel filtration chromatography on a Superdex 200HR column. The purified CpD-HRP conjugate was mixed with an equal volume of glycerin and stored at −20°C.
Microtiter wells (Yixinmei, Xiamen, China) were coated with 100 μl of a mixture of 4 μg/ml CpC solution (diluted in 50 mM Tris-HCl, pH 8.0) overnight. Thereafter, the wells were washed twice with phosphate-buffered saline (PBS; pH 7.4), followed by incubation with a blocking reagent (10% sucrose, 1% casein-Na, and 2% bovine serum albumin [BSA], in PBS, pH 7.4) at 37°C for 2 h. After removing the blocking solution, the wells were vacuum dried and stored at 4°C. A 50-μl specimen aliquot was mixed with 50 μl of assay buffer and then added to each well and incubated for 30 min at 37°C. After incubation, the wells were washed with a wash buffer (0.05% Tween 20 in PBS, pH 7.4). A 100-μl aliquot of CpD-HRP solution was added to each well and incubated for 30 min at 37°C. After rinsing 5 times with the wash buffer, 100 μl of tetramethylbenzidine (TMB) substrate solution (Wantai, Beijing, China) was added, and the mixture was then incubated for 15 min at 37°C. The absorbance was measured at 450 nm and 620 nm using a microplate reader (Tecan, Switzerland).
Determination of cutoff value.
A total of 2,656 serum samples from healthy blood donors collected from Xiamen Blood Centre were measured to characterize the optical density at 450 nm (OD450) with reference readings taken at 620 nm (data not shown), and the cutoff value for the sandwich ELISA was set as the mean + 3 standard deviations (SDs), equal to a negative control well + 0.12. The figure of 3 SDs was an arbitrary one which would cover 99.7% of the distribution.
Anti-HBc neutralization confirmatory assay.
A neutralization confirmatory assay based on an indirect chemiluminescent enzyme immunoassay (CLEIA) was used as the gold standard to determine the true status of a specimen for which a discrepancy from a different anti-HBc assay was found. In this assay, the CpC-coated wells were incubated with an anti-HBc monoclonal antibody (MAb) (16D5; 20 μg/well), anti-HB e antigen (anti-HBe) MAb (10D8; 20 μg/well), an unrelated antibody (anti-human papillomavirus type 18 [anti-HPV18] L1; 20 μg/well), and a blank assay buffer (20% fetal bovine serum in PBS) in parallel at 37°C for 30 min. The wells were then washed once, and 30-μl samples were added to each well filled with 70 μl of assay buffer and incubated at 37°C for 30 min. After five washes, a 100-μl aliquot of HRP-conjugated goat anti-human IgG solution was added to each well and incubated for 30 min at 37°C. The wells were then washed 5 more times, 100 μl of substrate solution (Luminol) was added for reaction, and the result was read by an Orin II microplate chemiluminescence reader (Berthod, DE). The neutralization rate was calculated as the percentage of the decreased relative light unit (RLU) value of the anti-HBc MAb-blocked well compared with the non-MAb-blocked well, and the neutralization rate of the anti-HBe and unrelated antibody was used to calculated specificity.
Monoclonal antibodies.
Monoclonal antibodies (MAbs) against HBV HBc or Hbe antigens were prepared using hybridoma technology. MAbs were purified using ammonium sulfate precipitation, followed by Mabselect Xtral affinity chromatography (Amersham GE Health, Uppsala, Sweden) from the ascites collected. Immunoglobulin concentrations were determined by spectrophotometry using a DU800 spectrometer (Beckman, CA). Commercial competitive anti-HBc and anti-HBe assays (Wantai, Beijing, China) were used to analyze the reactivity of all MAbs tested at a concentration of 50 μg/ml. Two MAbs (16D5 and 8D1) were specific anti-HBc antibodies that were reactive in the anti-HBc assay but nonreactive in the anti-HBe assay. Three MAbs, 20B11, 11E7, and 10D8, were found to be specific for anti-HBe and nonreactive in the anti-HBc assay. Five other MAbs, 11H4, 18B6, 19A6, 11H10, and 4B8, showed reactivity in both anti-HBe and anti-HBc assays.
Patient serum samples.
One hundred twenty symptomatic HBV carriers (HBsAg-positive individuals with persistent elevation of alanine aminotransferase (ALT) levels (>1.5 times the upper limit of normal for at least 3 months), 245 asymptomatic HBV carriers (HBsAg-positive individuals with a normal ALT level and without a history of hepatitis), and 783 healthy individuals' serum samples (control cohort) were used to evaluate the concordance between the sandwich ELISA method and the Architect anti-HBc CMIA. All samples were collected from Quanzhou 1st Hospital (Quanzhou, Fujian, China), Xiamen Blood Centre (Xiamen, Fujian, China), and Xiamen CDC (Xiamen, Fujian, China).
HBV infectious markers.
HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc assays were performed using Architect i2000SR CMIA (Abbott Laboratories, Abbott Park, IL). HBsAg low titer (<2 IU on Architect HBsAg) samples were confirmed by other three HBsAg assays: the Monolisa HBsAg Ultra (Bio-Rad, Marnes La Coquette, France), the Hepanostika HBsAg Ultra (bioMérieux, Marcy l'Etoile, France), and the Murex V3 ELISA (Abbott Murex, Dartford, United Kingdom).
RESULTS
Characterization of the rHBcAg.
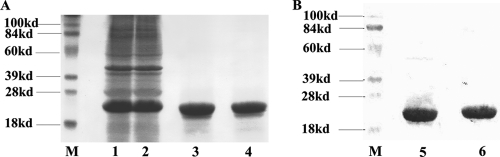
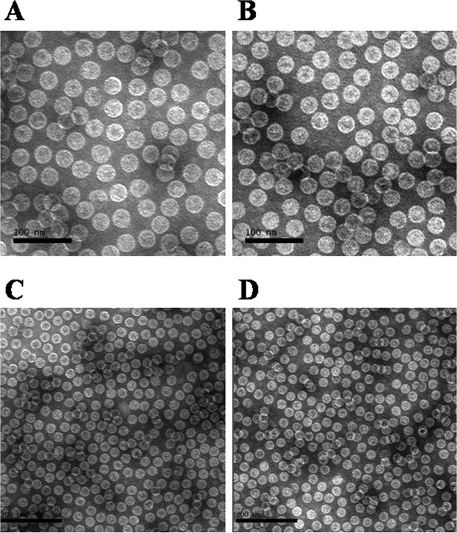
The CpC and CpD full-length rHBcAg were solubly expressed with high expression levels obtained (approximately 20% in total proteins) (Fig. 1A, lanes 1 and 2) in the BL21(DE3) cell line. The purified CpC and CpD proteins by ion-exchange chromatography and hydrophobic chromatography showed single bands on SDS-PAGE with an estimated molecular mass of about 22 kDa, respectively (Fig. 1A, lanes 3 and 4). The high purity of HBcAg proteins excluded the underlying interference against the immunoassay. Western blot analyses showed that both CpC and CpD reacted well with anti-HBc MAb (Fig. 1B), suggesting that the purified fractions were targets. After the in vitro capsid assembly, the rHBcAg formed as a homogeneous particulate structure similar in appearance to the regular 20-hedron structure of native HBcAg particle with a 27-nm diameter (Fig. 2A to D).
FIG. 1.
SDS-PAGE (A) and Western blot (B) analysis of CpC and CpD. (A) Purity of CpC and CpD. The concentration of separating gel was 13.5%, and proteins were visualized by Coomassie brilliant blue staining. Lanes 1 and 2 show the CpC- and CpD-positive fractions, respectively, after the sonication step. Lanes 3 and 4 show the purified CpC and CpD, respectively, by ion-exchange chromatography and hydrophobic chromatography. (B) Immunological characterization of purified CpC and CpD. Lanes 5 and 6 indicate the single bands of CpC and CpD, respectively (molecular mass, 22 kDa), identified by the monoclonal anti-HBc antibody. Lanes M, molecular mass marker lanes.
FIG. 2.
Transmission electron microscopy of the purified HBV core capsids: CpC (A and C) and CpD (B and D) particles. The scale is 100 nm for panels A and B and 200 nm for panels C and D.
Discrimination of the sandwich ELISA between anti-HBc and anti-HBe MAbs.
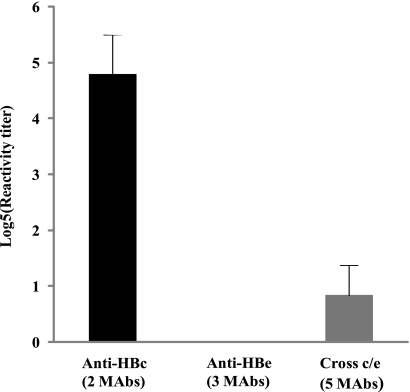
The MAbs were tested by the sandwich ELISA in serial dilution, and the reactivity titer that allowed for a reaction to occur for each MAb was calculated (Fig. 3). The 2 anti-HBc specific MAbs, 16D5 and 8D1, both at a concentration of 100 μg/ml had the highest reactivity (titer, ≥625), whereas the 3 anti-HBe-specific MAbs 20B11, 10D8, and 11E7 presented virtually no reactivity in the sandwich assay even for samples with a concentration higher than 100 μg/ml (titer, <1). A weak reactivity of the 5 cross-HBc/Hbe MAbs was also observed in the sandwich assay; however, this was observed only when antibodies at a concentration of 20 to ∼100 μg/ml were used (5≥ titer ≥1).
FIG. 3.
Reactivity of different MAbs in the anti-HBc sandwich ELISA. The primary concentrations of all MAbs were adjusted to 100 μg/ml, which was then followed by 5-fold serial dilution. The y axis shows the log5 reactivity titer. The black bar to the left highlights the two anti-HBc-specific MAbs (16D5 and 8D1). Results for the 3 anti-HBe specific MAbs (11E7, 10D8, and 8D1) would be represented in the center; however, the actual reactivities cannot be seen because the levels are too low. The gray box to the right represents the five cross-anti-HBc/anti-HBe MAbs (11H4, 18B6, 19A6, 11H10, and 4B8).
Comparison of the analytical sensitivity for detecting anti-HBc between the sandwich ELISA and Architect CMIA.
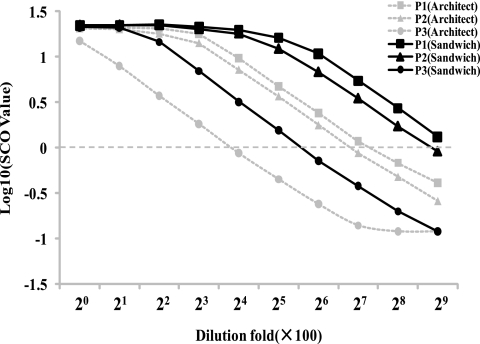
Three anti-HBc-positive sera were serially diluted in anti-HBc/anti-HBs/HBsAg-negative serum and were detected by the sandwich ELISA and Architect anti-HBc CMIA in parallel (Fig. 4). The analytical sensitivity for detecting anti-HBc of the sandwich ELISA is between 2- and 4-fold higher than that of the Architect anti-HBc CMIA. While a 2- to 4-fold difference is not considered significant in serological titers, the difference was consistently unidirectional.
FIG. 4.
Comparison between the sandwich ELISA and Architect anti-HBc CMIA. The broken horizontal line indicates the cutoff value.
Concordance between the sandwich ELISA and Architect CMIA in detecting samples in different clinical status.
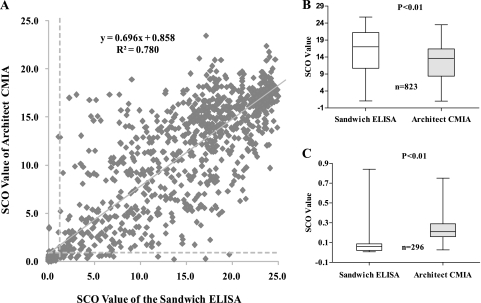
The anti-HBc status of serum samples was measured by the sandwich ELISA and Architect anti-HBc CMIA in parallel, and the concordance between the sandwich ELISA and Architect anti-HBc CMIA was calculated and is shown in Table 1. Of the 120 samples of HBV patients, all were positive in both the sandwich ELISA and Architect anti-HBc CMIA. Among 245 asymptomatic HBV carriers, 241 specimens were positive in the sandwich ELISA and Architect anti-HBc CMIA. The discrepancy lay with 1 positive sample (a1) in the sandwich ELISA, and the other three samples tested negative by both assays. Thus, the concordance between the 2 tests was 99.6% (95% confidence interval [CI], 97.8 to ∼99.9%). For the 783 samples from the control cohort, there were 462 positive cases and 293 negative cases with 28 discordant results, which include 24 sandwich-positive but Architect anti-HBc-negative and 4 Architect anti-HBc-positive but sandwich ELISA-negative cases. In reviewing the results for the total of 1,148 specimens tested, the concordance between sandwich ELISA and Architect anti-HBc CMIA was 97.8% (95% CI, 96.4 to ∼98.3) and there were a total of 29 discordant results. The signal/cutoff (SCO) value correlation for all samples in the two assays is shown in Fig. 5A. The correlation coefficient was 0.88, and the adjusted R2 = 0.78. The SCO level of the sandwich ELISA in 823 concordant positive samples was statistically significantly higher than that of the Architect anti-HBc CMIA and statistically significantly lower than that of the Architect anti-HBc CMIA in the 296 concordant negative samples (Fig. 5B and C).
TABLE 1.
Concordance of Architect CMIA and sandwich ELISA anti-HBc levels in HBV detection in serum samples related to different clinical statuses
| Group and status (n) | No. of samples with result by Architect CMIA |
% Concordance (95% CI) | |
|---|---|---|---|
| + | − | ||
| Hepatitis B patients (120) | 100 (97.0∼100) | ||
| Sandwich anti-HBc+ | 120 | 0 | |
| Sandwich anti-HBc− | 0 | 0 | |
| Asymptomatic carriers (245) | 99.6 (97.8∼99.9) | ||
| Sandwich anti-HBc+ | 241 | 1a | |
| Sandwich anti-HBc− | 0 | 3 | |
| Common populations (783) | 96.4 (94.9∼97.6) | ||
| Sandwich anti-HBc+ | 462 | 24a | |
| Sandwich anti-HBc− | 4a | 293 | |
| Total (1,148) | 97.8 (96.4∼98.3) | ||
| Sandwich anti-HBc+ | 823 | 25 | |
| Sandwich anti-HBc− | 4 | 296 | |
A total of 29 discordant samples were further analyzed by a neutralization confirmatory assay. Finally, 28 samples (a1, b1 to b24, c1 and c2, and c4) were found to be anti-HBc positive.
FIG. 5.
(A) Correlation between the Architect CMIA and sandwich ELISA in detection of anti-HBc from 1,148 samples. The y axis represents the SCO values measured by the Architect CMIA, and the x axis represents those measured by the sandwich ELISA. The broken lines indicate the lowest detection limit for each assay. (B) Comparison of SCO values between the sandwich ELISA and Architect anti-HBc CMIA in 823 concordant positive specimens; (C) comparison of SCO values between the Architect CMIA and the sandwich ELISA in detecting 296 concordant negative specimens.
Analysis for the discordant samples between the sandwich ELISA and Architect CMIA.
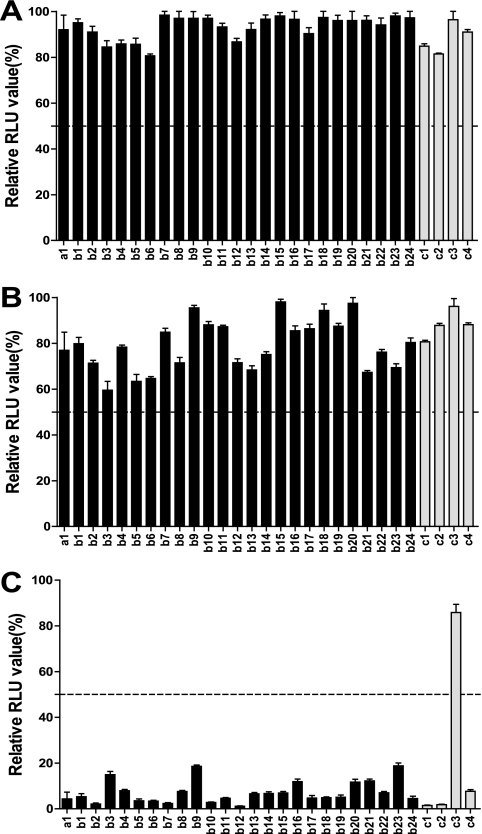
One asymptomatic HBV carrier (a1) and 28 (b1 to b24 and c1 to c4) in the control cohort had discordant anti-HBc result in the two assays. All samples with discordant anti-HBc results were tested by neutralization assay to determine their specificity. As the graph in Fig. 6 shows, all 25 sandwich+ and Architect− samples and 3 Architect+ and sandwich− samples were positive in the confirmatory assay, except 1 (c3) Architect+ and sandwich− sample. Otherwise, the a1 sample was HBsAg and HBV DNA positive and with undetectable anti-HBs, anti-HBeAg, and anti-HBe antibodies, suggesting that the individual is at the early stage of HBV infection. Among the other 24 (b1 to b24) sandwich+ and Architect− and 4 (c1 to c4) Architect+ and sandwich− samples from common populations, 22 samples and 1 sample, respectively, were anti-HBs positive. Thus, among the total of 1,148 samples, there were 851 anti-HBc-positive and 297 anti-HBc-negative cases. The sensitivity and specificity for the Sandwich ELISA were 99.7% (95% CI, 99.0 to ∼99.9%) and 100% (95% CI, 98.8 to ∼100%); those for the Architect CMIA were 97.1% (95% CI, 95.7 to ∼98.1%) and 99.7% (95% CI, 98.1 to ∼99.9%), respectively.
FIG. 6.
Anti-HBc neutralization confirmatory assay for 29 discordant samples between the sandwich ELISA and the Architect anti-HBc CMIA. For each test, relative light unit (RLU) values were calculated with the well containing MAbs compared with the well not containing MAbs. Only values below 50% (indicated by a broken horizontal line) were determined as anti-HBc positive. (A) Unrelated antibody (anti-HPV 18 L1)-blocked wells; (B) anti-HBe MAb-blocked wells; (C) anti-HBc MAb-blocked wells.
DISCUSSION
In this technique, the production of homogenous and stable HBcAg particles is critical. Purified monomer HBcAg proteins are subunits for making capsids. Although numerous studies have demonstrated that the production of truncated version of HBcAg that mimic the sequence of HBeAg in E. coli can be accomplished easily in terms of yield and purification (27, 28), nevertheless, the interdimer disulfide bridges that are formed by the Cys-183 at the C terminus of between HBcAg dimers increased the stability of HBc capsids (14, 15). The expression of full-length HBcAg was difficult since it has an arginine-rich C terminus that was coded for by rare codons in E. coli. This problem was resolved by optimizing certain codons in this region. Since HBcAg expressed in E. coli spontaneously accumulates in a particulate state - HBcAg capsids, under such denaturing conditions, HBcAg capsids might dissolve into subunits, which were rinsed during purification and then assembled into homogenous and stable particles. In contrast with routine method, the presented procedure yielded proportionately more particles (more than 95%; data not shown) and better homogeneity. The novel immunoassay for anti-HBc measurement using a sandwich ELISA method was built around such particles. Compared with Abbott Architect CMIA anti-HBc assay, the new sandwich ELISA has improved sensitivity and specificity.
The clinical significance of using anti-HBc lies in several clinical situations. HBV reactivation has been reported during cancer chemotherapy, during HIV infection, and after kidney and bone marrow transplantation (5, 7, 20, 22). There are at least 2 reported cases of HBV reactivation with liver inflammation after rituximab administration, and one patient died from liver failure (3); the other reactivation occurred in a patient with preexisting anti-HBs and anti-HBc antibodies (4). When a patient is diagnosed to be reactive for “anti-HBc alone,” several explanations might apply to this phenomenon. First, a certain proportion of “anti-HBc-alone” individuals will be false positive, depending both on the anti-HBc test used and the HBV prevalence of the population investigated. Using the sandwich ELISA should reduce this concern significantly because of its sensitivity and specificity profile. Second, “anti-HBc-alone” individuals may be in the window phase of an acute HBV infection when HBsAg disappears followed by anti-HBs a few weeks later. The significance of “anti-HBc-alone” individuals in this window period is that such individuals are probably infectious. Third, the status “anti-HBc alone” can reflect an HBV infection which was resolved many years or decades earlier. Fourth, a large fraction of “anti-HBc-alone” patients can be assumed to have an unresolved chronic infection with low-grade, possibly intermittent virus production. Such individuals may have detectable serum HBV DNA (8.1%) and are potentially infectious (13). The negative HBsAg assay is unclear in these cases, but this result can be caused by very low concentrations of HBsAg, fixation of HBsAg in immune complexes, or by HBsAg mutations (11, 26). From a practical point of view, there seems to be no clear-cut distinction between “late immunity” and “unresolved infection.” Indeed, in a study of 5,511 blood donor samples from 3 hospitals showed that 203 (3.7%) of randomly selected blood donors were confirmed as “anti-HBc alone.” Of these, 11 (5.4%) were HBV-DNA positive, as detected by nested PCR. All samples had HBV-DNA levels below 400 copies/ml, and all were genotype D. Thus, HBV was present but below the detectable limit for the assay used. Hence, routine screening for anti-HBc may be required as an additional preventive measure for controlling transmission of HBV via blood transfusion (8). Fifth, “anti-HBc alone” may represent the suppression of HBV replication by HCV coinfection. Knoll et al. (13) showed that in 112/550 “anti-HBc alone” individuals (20.4%) were coinfected with HCV, and patients with HCV coinfection more often had an elevated ALT activity or chronic liver damage. The detrimental effect of HCV coinfection has previously been reported in HBsAg-positive patients with chronic HBV infection (6, 9). Finally, in a study of 379 samples with detectable anti-HBc, 155 (40.9%) were “anti-HBc only.” HBV DNA was detected in 6/151 (4%), all of which had a viral load of <400 copies per ml. Anti-HIV was found in 50/151 (33.1%) and anti-HCV in 14/151 (9.3%) patients. Only one of the HIV-infected patients had detectable HBV DNA. Phylogenetic analysis of the HBV surface gene from three patients showed a variety of genotypes (A, E, and G). One sequence had a mutation in codon 144, which has previously been reported to give false-negative HBsAg results. Thus, “anti-HBc only” is a common phenomenon in the clinical virology laboratory, but only a small proportion of samples had detectable HBV DNA. The presence of HBsAg mutants with a possible false-negative HBsAg test result is of concern. Samples with the result “anti-HBc only” could be used to monitor the emergence of these mutants (1). The assay presented here should assist with the resolution of diagnostic difficulties with HBV.
Acknowledgments
We thank James Wai-Kuo Shih and Mun H. Ng for helpful discussions and comments on the manuscript.
This work was supported by Key Special Subjects of Infectious Diseases grants 2008ZX10002-012 and 2009ZX10004-704 and Outstanding Young Researchers of Fujian Province grant 2009J06020.
We declare there are no conflicts of interest.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Alhababi, F., T. A. Sallam, and C. Y. Tong. 2003. The significance of ‘anti-HBc only’ in the clinical virology laboratory. J. Clin. Virol. 27:162-169. [DOI] [PubMed] [Google Scholar]
- 2.Caspari, G., H. J. Beyer, G. Elbert, K. Koerner, P. Muss, F. W. Schunter, A. Uy, W. Gerlich, R. Thomssen, and H. Schmitt. 1989. Unsatisfactory specificities and sensitivities of six enzyme immunoassays for antibodies to hepatitis B core antigen. J. Clin. Microbiol. 27:2067-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czuczman, M. S., A. J. Grillo-Lopez, C. A. White, M. Saleh, L. Gordon, A. F. LoBuglio, C. Jonas, D. Klippenstein, B. Dallaire, and C. Varns. 1999. Treatment of patients with low-grade B-cell lymphoma with the combination of chimeric anti-CD20 monoclonal antibody and CHOP chemotherapy. J. Clin. Oncol. 17:268-276. [DOI] [PubMed] [Google Scholar]
- 4.Dervite, I., D. Hober, and P. Morel. 2001. Acute hepatitis B in a patient with antibodies to hepatitis B surface antigen who was receiving rituximab. N. Engl. J Med. 344:68-69. [DOI] [PubMed] [Google Scholar]
- 5.Dhedin, N., C. Douvin, M. Kuentz, M. F. Saint Marc, O. Reman, C. Rieux, F. Bernaudin, F. Norol, C. Cordonnier, D. Bobin, J. M. Metreau, and J. P. Vernant. 1998. Reverse seroconversion of hepatitis B after allogeneic bone marrow transplantation: a retrospective study of 37 patients with pretransplant anti-HBs and anti-HBc. Transplantation 66:616-619. [DOI] [PubMed] [Google Scholar]
- 6.Di Marco, V., O. Lo Iacono, C. Camma, A. Vaccaro, M. Giunta, G. Martorana, P. Fuschi, P. L. Almasio, and A. Craxi. 1999. The long-term course of chronic hepatitis B. Hepatology 30:257-264. [DOI] [PubMed] [Google Scholar]
- 7.Dusheiko, G., E. Song, S. Bowyer, M. Whitcutt, G. Maier, A. Meyers, and M. C. Kew. 1983. Natural history of hepatitis B virus infection in renal transplant recipients—a fifteen-year follow-up. Hepatology 3:330-336. [DOI] [PubMed] [Google Scholar]
- 8.El-Zaatari, M., H. Kazma, M. Naboulsi-Majzoub, M. Haidar, F. Ramlawi, Z. Mahfoud, and S. Ramia. 2007. Hepatitis B virus DNA in serum of ‘anti-HBc only’-positive healthy Lebanese blood donors: significance and possible implications. J. Hosp. Infect. 66:278-282. [DOI] [PubMed] [Google Scholar]
- 9.Gaeta, G. B., G. Stornaiuolo, D. F. Precone, S. Lobello, M. Chiaramonte, T. Stroffolini, G. Colucci, and M. Rizzetto. 2003. Epidemiological and clinical burden of chronic hepatitis B virus/hepatitis C virus infection. A multicenter Italian study. J. Hepatol. 39:1036-1041. [DOI] [PubMed] [Google Scholar]
- 10.Grob, P., W. Jilg, H. Bornhak, G. Gerken, W. Gerlich, S. Gunther, G. Hess, H. Hudig, A. Kitchen, H. Margolis, G. Michel, C. Trepo, H. Will, A. Zanetti, and I. Mushahwar. 2000. Serological pattern “anti-HBc alone”: report on a workshop. J. Med. Virol. 62:450-455. [DOI] [PubMed] [Google Scholar]
- 11.Joller-Jemelka, H. I., A. N. Wicki, and P. J. Grob. 1994. Detection of HBs antigen in “anti-HBc alone” positive sera. J. Hepatol. 21:269-272. [DOI] [PubMed] [Google Scholar]
- 12.Kleinman, S. H., M. C. Kuhns, D. S. Todd, S. A. Glynn, A. McNamara, A. DiMarco, and M. P. Busch. 2003. Frequency of HBV DNA detection in US blood donors testing positive for the presence of anti-HBc: implications for transfusion transmission and donor screening. Transfusion 43:696-704. [DOI] [PubMed] [Google Scholar]
- 13.Knoll, A., A. Hartmann, H. Hamoshi, K. Weislmaier, and W. Jilg. 2006. Serological pattern “anti-HBc alone”: characterization of 552 individuals and clinical significance. World J. Gastroenterol. 12:1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nath, N., K. Hickman, S. Nowlan, D. Shah, J. Phillips, and S. Babler. 1992. Stability of the recombinant hepatitis B core antigen. J. Clin. Microbiol. 30:1617-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman, M., F. M. Suk, M. Cajimat, P. K. Chua, and C. Shih. 2003. Stability and morphology comparisons of self-assembled virus-like particles from wild-type and mutant human hepatitis B virus capsid proteins. J. Virol. 77:12950-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ollier, L., C. Laffont, A. Kechkekian, A. Doglio, and V. Giordanengo. 2008. Detection of antibodies to hepatitis B core antigen using the Abbott ARCHITECT anti-HBc assay: analysis of borderline reactive sera. J. Virol. Methods 154:206-209. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Rodriguez, M. T., B. Sopena, M. Crespo, A. Rivera, T. Gonzalez del Blanco, A. Ocampo, and C. Martinez-Vazquez. 2009. Clinical significance of “anti-HBc alone” in human immunodeficiency virus-positive patients. World J. Gastroenterol. 15:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, E. F., J. A. Weare, R. Randell, P. V. Holland, G. Madsen, and R. H. Decker. 1991. Characterization of a reduction-sensitive factor from human plasma responsible for apparent false activity in competitive assays for antibody to hepatitis B core antigen. J. Clin. Microbiol. 29:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallberg, M., and L. O. Magnius. 1989. Enzyme immunoassay for anti-hepatitis B core (HBc) immunoglobulin G1 and significance of low-level results in competitive assays for anti-HBc. J. Clin. Microbiol. 27:849-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vento, S., G. di Perri, R. Luzzati, M. Cruciani, T. Garofano, C. Mengoli, E. Concia, and D. Bassetti. 1989. Clinical reactivation of hepatitis B in anti-HBs-positive patients with AIDS. Lancet i:332-333. [DOI] [PubMed] [Google Scholar]
- 21.Vitale, F., F. Tramuto, A. Orlando, G. Vizzini, V. Meli, G. Cerame, W. Mazzucco, R. Virdone, U. Palazzo, M. R. Villafrate, A. Tagger, and N. Romano. 2008. Can the serological status of anti-HBc alone be considered a sentinel marker for detection of occult HBV infection? J. Med. Virol. 80:577-582. [DOI] [PubMed] [Google Scholar]
- 22.Wands, J. R., C. M. Chura, F. J. Roll, and W. C. Maddrey. 1975. Serial studies of hepatitis-associated antigen and antibody in patients receiving antitumor chemotherapy for myeloproliferative and lymphoproliferative disorders. Gastroenterology 68:105-112. [PubMed] [Google Scholar]
- 23.Weare, J. A., E. F. Robertson, G. Madsen, R. Hu, and R. H. Decker. 1991. Improvement in the specificity of assays for detection of antibody to hepatitis B core antigen. J. Clin. Microbiol. 29:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber, B., W. Melchior, R. Gehrke, H. W. Doerr, A. Berger, and H. Rabenau. 2001. Hepatitis B virus markers in anti-HBc only positive individuals. J. Med. Virol. 64:312-319. [DOI] [PubMed] [Google Scholar]
- 25.Wedemeyer, H., M. Cornberg, B. Tegtmeyer, H. Frank, H. L. Tillmann, and M. P. Manns. 2004. Isolated anti-HBV core phenotype in anti-HCV-positive patients is associated with hepatitis C virus replication. Clin. Microbiol. Infect. 10:70-72. [DOI] [PubMed] [Google Scholar]
- 26.Weinberger, K. M., T. Bauer, S. Bohm, and W. Jilg. 2000. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J. Gen. Virol. 81:1165-1174. [DOI] [PubMed] [Google Scholar]
- 27.Wingfield, P. T., S. J. Stahl, R. W. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, J., F. Schodel, and D. L. Peterson. 1992. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J. Biol. Chem. 267:9422-9429. [PubMed] [Google Scholar]