Abstract
PspA is an important candidate for a vaccine with serotype-independent immunity against pneumococcal infections. Based on sequence relatedness, PspA has been classified into three families comprising six clades. We have previously addressed the cross-reactivity of antibodies against PspA fragments containing the N-terminal and proline-rich regions of PspA from clades 1 to 5 (PspA1, PspA2, PspA3, PspA4, and PspA5) by Western blot analysis and reported that anti-PspA4 and anti-PspA5 were able to recognize pneumococci expressing PspA proteins from all of the clades analyzed. We have now analyzed the functional capacity of these antibodies to bind and to mediate complement deposition on intact bacteria in vitro. Our results show that both PspA4 and PspA5 elicit antibodies that are able to bind and to mediate complement deposition efficiently on pneumococcal strains bearing PspA proteins from clades 1 to 5. Moreover, mice immunized with PspA4 and PspA5 were protected against an intranasal lethal challenge with strains expressing PspA proteins from the two major families. PspA4 and PspA5 are thus able to induce antibodies with a high degree of cross-reactivity in vitro, which is reflected in cross-protection of mice. We have also analyzed the contribution of the nonproline (NonPro) block within the conserved proline-rich region to the reactivity of anti-PspA antibodies, and the results indicate that N-terminal α-helical region, the blocks of proline repeats, and the NonPro region can influence the degree of cross-reactivity of antibodies to PspA.
Streptococcus pneumoniae is an important human pathogen, being responsible for millions of deaths worldwide every year. The pneumococcal disease burden could be greatly reduced by the use of the current seven-valent conjugate vaccine, but the high cost and restricted serotype coverage limit its widespread use, especially in developing countries. New-generation vaccines containing up to 13 serotypes are expected to increase vaccine coverage, but the serotype replacement in colonization and disease by nonvaccine serotypes observed with the use of the seven-valent conjugate vaccine (8-9, 11) further emphasizes the importance of the development of alternative vaccines. Protein antigens such as PspA (pneumococcal surface protein A) could be used to induce serotype-independent immunity at a low cost (24).
PspA is present in all isolated pneumococcal strains and was shown to be an important virulence factor, interfering with complement deposition (19, 21, 25), killing by apolactoferrin (23), and immune adherence to erythrocytes (12). It has been shown to induce protection in mice in carriage, pneumonia, and fatal systemic models (2, 4, 16). Mature PspA is composed of a mosaic structure with four domains: an α-helical N-terminal domain, a proline-rich region, a choline-binding domain, and a short hydrophobic tail (10, 27-28). PspA shows variability in the surface-exposed N-terminal region, and a classification was proposed based on sequence relatedness of the C-terminal portion of the α-helix, the clade-defining region. It has been classified into three families encompassing six clades. Family 1 (Fam1) is composed of clades 1 and 2, Fam2 includes clades 3, 4, and 5, and Fam3, which is rarely isolated, comprises clade 6 (10). Since the degree of similarity seems to be reflected in cross-reactivity, it has been proposed that a broad-coverage vaccine should contain at least one fragment from each of the two major families.
Immunization of healthy adults with a single recombinant fragment of PspA in a phase I clinical trial showed the induction of cross-reactive antibodies (14) that were able to induce passive protection in mice challenged intravenously (3). The natural exposure of adults to several pneumococcal strains might be responsible for the cross-reactivity detected, with the immunization with PspA acting as a booster dose.
Because of the diversity observed in PspA, it is extremely important to analyze whether each fragment selected to compose a vaccine is indeed able to induce cross-protection. We have previously addressed the degree of cross-reactivity of antibodies to recombinant fragments including the N-terminal and proline-rich regions of PspA proteins from clades 1 to 5 (PspA1, PspA2, PspA3, PspA4, and PspA5) by Western blot analysis of 35 strains isolated in Brazil. As expected, we have observed higher cross-reactivity within the same clade. Within Fam1, anti-PspA1 serum also showed cross-reaction with PspA2-expressing strains, while anti-PspA2 showed reaction restricted to the same clade. Within Fam2, anti-PspA3 serum also showed reactivity restricted to PspA3-expressing strains, while anti-PspA5 and, more strikingly, anti-PspA4 sera showed a broad recognition capacity, being able to react with strains expressing PspA proteins from clades 1 to 5 (7). The ability of sera to recognize a pneumococcal strain by Western blot analysis does not necessarily correlate with their capacity to induce protection in vivo though. In fact, the levels of antibodies to PspA detected by enzyme-linked immunosorbent assay (ELISA) or through surface staining of the bacteria failed to provide a useful correlate of protection (22). Based on the strong evidence supporting the importance of complement in protection against pneumococcal disease, it was proposed that in vitro complement deposition mediated by antibody may be used as a surrogate assay for the prediction of protection induced by surface antigens of pneumococci (15). This work aimed at further characterizing antibodies against the PspA1, PspA2, PspA3, PspA4, and PspA5 N-terminal fragments in terms of their capacity to mediate C3 deposition on the surface of pneumococci expressing PspA proteins from different clades. Moreover, protection of mice against a lethal intranasal challenge with strains expressing PspA from Fam1 or Fam2 was also analyzed. The basis for the broad reactivity observed in the anti-PspA4 serum by Western blot analysis was also further investigated. Of the five PspA fragments analyzed, PspA4 was the only one containing a nonproline (NonPro) block within the proline-rich region. Not all native PspA proteins include this region: of 24 PspA sequences analyzed by Hollingshead and collaborators (10), 14 were shown to have this NonPro block. We have thus examined whether this region would be responsible for increased cross-reactivity.
MATERIALS AND METHODS
Construction of PspA fragments.
All cloning procedures were performed with Escherichia coli DH5α grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml). The plasmids encoding the N-terminal regions of PspA proteins from clades 1 to 5 (PspA1, PspA2, PspA3, PspA4, and PspA5) were previously described (7). Two new plasmids encoding PspA4 fragments were constructed: while the original construct (PspA4) encoded the complete N-terminal region plus the proline-rich region (containing a NonPro block within this region), PspA4AB contains only the N-terminal α-helical region without the entire proline-rich region, and PspA4Pro contains the N-terminal α-helical region plus the first block of prolines only, lacking both the NonPro and second proline blocks (Fig. 1). Both fragments were amplified by PCR from the original PspA4 construct using primers 5′ TAGCTCGAGACCATGGTAAGAGCAGAAGAAGCC 3′ (forward) and 5′ GGTACCTTAAGTCTCTTCTTCATCTCCATC 3′ (PspA4AB-reverse) or 5′ GGTACCTTATGGTTTTGGTGCTGGAGCT 3′ (PspA4Pro-reverse). The gene products were cloned into the pGEMT-easy vector (Promega), and the sequences were confirmed by DNA sequencing. The pGEMT-easy-pspA constructs were digested with XhoI and KpnI, and the resulting fragments were subcloned into the pAE 6×His vector (18), generating pAE-pspA4AB and pAE-pspA4Pro. A plasmid encoding a fusion between PspA3 and the proline-rich region (containing the NonPro block) of PspA4 was constructed by amplification of the proline-rich region of PspA4 using primers 5′ TAGTCTAGACCAGCGCCAGCTCCTCAA 3′ (NonPro F) and 5′ TAGGGTACCTTATGGTTGTGGTGCTGAAGCT 3′ (NonPro R). The gene product was cloned into the pGEMT-easy vector (Promega), and the sequence was confirmed by DNA sequencing. The pGEMT-easy-NonPro construct was digested with XbaI and KpnI, and the resulting fragment was subcloned into the 3′ end of pspA3 in pTG-pspA3NS (13). The fragment encoding the PspA3-NonPro fusion (Fig. 1) was digested using XhoI and KpnI and cloned into pAE 6×His, generating pAE-pspA3-NonPro.
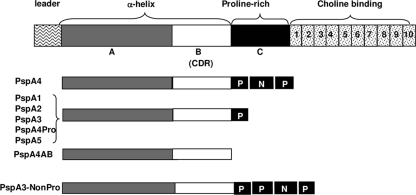
FIG. 1.
Schematic diagram of the PspA fragments. The whole PspA molecule, containing the N-terminal α-helical domain (A and B), the proline-rich region (C), and the choline-binding domain, is shown on the top. The recombinant fragments PspA1, PspA2, PspA3, PspA4, PspA4AB, PspA4Pro, PspA5, and Psp3-NonPro are represented with the distinct domains. P indicates the proline blocks, and NP indicates the NonPro block. CDR, clade-defining region.
PspA expression and purification.
The pAE 6×His vectors containing the pspA constructs were used to transform BL21(DE3) SI E. coli competent cells (Invitrogen). Protein expression was induced in mid-log-phase cultures by the addition of 300 mM NaCl. The recombinant proteins, bearing an N-terminal histidine tag, were purified from the soluble fraction through affinity chromatography using Ni2+-charged resin (His Trap HP; GE HealthCare) in an Äkta Prime apparatus (GE HealthCare). Elution was carried out with 250 mM imidazole. The purified fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, dialyzed against 10 mM Tris-HCl (pH 8)-20 mM NaCl-0.1% glycine, and stored at −20°C.
Pneumococcal strains.
All of the strains used in this study were maintained as frozen (−80°C) stocks in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) in 20% glycerol. The serotypes, PspA clades, and sources of the strains are given in Table 1.
TABLE 1.
Pneumococcal strains used in this study
| Strain | Serotype | PspA clade | Source |
|---|---|---|---|
| St245/00 | 14 | 1 | Instituto Adolfo Lutz, São Paulo, Brazila |
| D39 | 2 | 2 | University of Alabama, Birminghamb |
| M10 | 11A | 3 | Universidade Federal de Goiás, Goiânia, Brazil |
| 3JYP2670 | 3 | 4 | University of Alabama, Birminghamb |
| ATCC 6303 | 3 | 5 | Instituto Adolfo Lutz, São Paulo, Brazila |
| A66.1 | 3 | 2 | University of Alabama, Birminghamb |
Kindly provided by Maria Cristina C. Brandileone.
Kindly provided by David E. Briles.
Animal immunization and challenge.
Animal experimental protocols were approved by the Ethics Committee of the Instituto Butantan (São Paulo, Brazil). Five- to 7-week-old female BALB/c mice from the Instituto Butantan (São Paulo, Brazil) were immunized intraperitoneally with 5 μg of PspA1, PspA2, PspA3, PspA4, or PspA5 adjuvanted with Al(OH)3 (50 μg Al3+). Animals were given three doses of protein at 14-day intervals. For the experiments with PspA4AB, PspA4Pro, and PspA4 or PspA3 and PspA3-NonPro, animals were immunized subcutaneously with three doses of protein (5 μg) adjuvanted with Al(OH)3 at 14-day intervals. The adjuvant alone was used as a control. Sera were collected from mice by retro-orbital bleeding 1 day before each dose and 14 days after the final dose. Mice were challenged 15 days after the last immunization. Animals were anesthetized through the intraperitoneal route with 200 μl of a 0.2% xylazine-1.0% ketamine mixture and then challenged through the inoculation of 50 μl of a suspension of strain A66.1 (4 × 106 CFU/animal) or ATCC 6303 (4 × 105 CFU/animal) into one nostril. Animals were observed for 10 days, and differences in survival were analyzed using the Fisher exact test.
Analysis of serum reactivity by ELISA.
Recognition of the different PspA proteins by antibodies to the recombinant proteins was analyzed in individual sera of four mice by ELISA. Polysorp 96-well plates (Nunc) were coated with the different PspA fragments (1 μg/ml), washed with phosphate-buffered saline (PBS) containing 0.1% Tween 20, and blocked with 10% nonfat dry milk in PBS. The plates were then incubated with serial dilutions of individual sera in PBS-1% bovine serum albumin at 37°C for 1 h. The plates were washed again and incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG, 1:15,000; Sigma) in PBS-1% bovine serum albumin at 37°C for 1 h. Following washes, antibodies were detected by adding OPD substrate (0.04% o-phenylenediamine in citrate-phosphate buffer [pH 5] containing 0.01% H2O2). After color development (10 min), the reaction was interrupted with H2SO4 and the A492 was determined. The reciprocal titer was considered the inverse of the last dilution of serum that registered an optical density above 0.1. Differences between groups were analyzed by Student's t test.
Antibody binding and complement deposition assays.
Antibody binding and complement deposition were analyzed as previously described (20). S. pneumoniae strains were grown in THY to a concentration of ∼108 CFU/ml (optical density at 600 nm, 0.4 to 0.5) and harvested by centrifugation at 2,000 × g for 2 min. The pellets were washed with PBS, resuspended in the same buffer, and incubated in the presence of heat-inactivated pooled sera from the third bleed of immunized mice for 30 min at 37°C. For antibody binding assays, samples were then washed with PBS and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG Fc (1:1,000; MP Biomedicals) on ice for 30 min. For complement deposition, bacteria were then washed with PBS, resuspended in gelatin Veronal buffer (Sigma), and incubated in the presence of freshly frozen normal mouse serum (from BALB/c mice as a source of complement) at 37°C for 30 min. After another wash with PBS, the samples were incubated with FITC-conjugated goat antiserum to mouse complement C3 (1:1,000; MP Biomedicals) on ice for 30 min. In both assays, bacteria were then washed twice with PBS, resuspended in 2% formaldehyde in PBS, and analyzed using a FACScalibur (BD Biosciences). Ten thousand gated events were acquired and analyzed in fluorescence intensity histograms.
RESULTS
PspA4 and PspA5 elicit antibodies with a high degree of cross-reactivity.
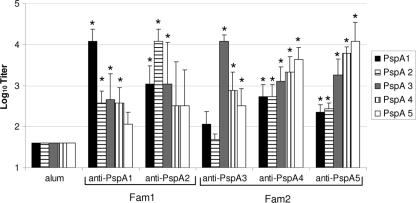
Sera collected from individual mice immunized with PspA1, PspA2, PspA3, PspA4, or PspA5 were analyzed by ELISA for reactivity against each recombinant PspA fragment. Data from sera collected after the second immunization are shown in Fig. 2. The pattern of cross-reactivity followed the classification of the different clades into the families, with the induction of significantly higher titers of anti-PspA antibodies against PspA proteins from the same family in all of the fragments tested compared to sera from animals inoculated with alum only. Reactivity with fragments from the other family was not always observed: anti-PspA1 (Fam1) did not elicit higher titers of antibodies reacting with PspA5 (Fam2), anti-PspA2 (Fam1) did not show reactivity with either PspA4 or PspA5 (Fam2), and anti-PspA3 (Fam2) did not react with either PspA 1 or PspA2 (Fam1). Both anti-PspA4 and anti-PspA5 showed increased reactivity with all five PspA fragments examined compared to sera from animals injected with alum.
FIG. 2.
Cross-reactivity of sera against PspA. Sera from four BALB/c mice immunized with PspA1, PspA2, PspA3, PspA4, or PspA5 were collected after the second immunization and analyzed individually by ELISA against each PspA fragment. The results are shown as the log10 of the titer. An asterisk indicates a statistically significant difference from animals immunized with alum (Student's t test, P ≤ 0.05). Results are representative of two experiments using sera from independent immunizations.
PspA4 and PspA5 elicit antibodies that bind and mediate C3 deposition on strains expressing different PspA proteins.
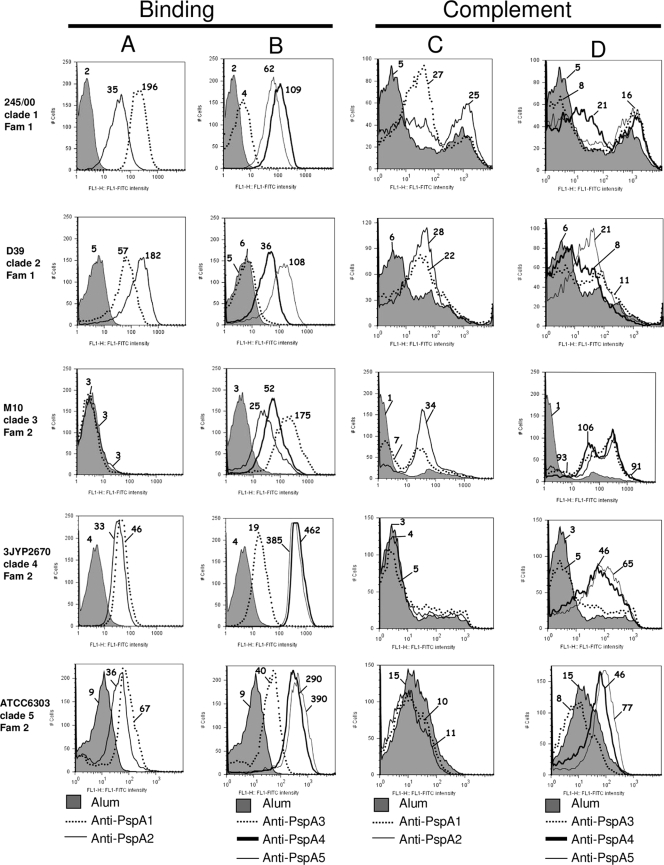
We further tested the ability of the anti-PspA antibodies to bind to the surface of pneumococci bearing different PspA proteins, and again the pattern of increased reactivity between clades of the same family was generally observed. Anti-PspA1 and anti-PspA 2 (Fam1) sera showed increased binding to St245/00 (clade 1, Fam1) and D39 (clade 2, Fam1), while binding to 3JYP2670 (clade 4, Fam2) and ATCC 6303 (clade 5, Fam2) was only slightly enhanced and no binding to M10 (clade 3, Fam2) at all was seen (Fig. 3A). Anti-PspA3 (Fam2) showed the narrowest cross-reactivity, being able to bind efficiently only to M10 (clade 3, Fam2). Restricted binding was seen for anti-PspA3 even to Fam2 strains 3JYP2670 (clade 4) and ATCC 6303 (clade 5), and no binding at all to Fam1 strains St245/00 (clade 1) and D39 (clade 2) was observed (Fig. 3B). Both anti-PspA4 and anti-PspA5 (Fam2) showed efficient binding to all of the strains tested (Fig. 3B). It is important to stress that the degree of binding varies with each individual pneumococcal isolate, so that we have only compared the median fluorescence values of the same strain with different sera.
FIG. 3.
Binding of antibodies and complement deposition in the presence of the different anti-PspA sera. Sera from mice immunized with PspA from Fam1 (PspA1 or PspA2) (A and C) or from Fam2 (PspA3, PspA4, or PspA5) (B and D) were tested for the ability to bind (1% serum, A and B) and to mediate deposition of C3 (10% serum, C and D) on S. pneumoniae strains bearing PspA proteins from clades 1 to 5. Serum from mice immunized with alum was used as a control for each strain and is represented by the gray area in each graph. Results are shown as fluorescence intensity histograms, and the median fluorescence intensity is indicated for each sample. Results are representative of two experiments using sera from independent immunizations.
As for complement deposition, anti-PspA1 and anti-PspA2 (Fam1) sera were able to efficiently mediate the deposition of C3 only on Fam1-bearing strains (St245/00 and D39) (Fig. 3C). Anti-PspA3 (Fam2) showed efficient deposition only on M10 (clade 3, Fam2) (Fig. 3D), while anti-PspA4 and anti-PspA5 were capable of efficient mediation of C3 deposition on all Fam2 strains (Fig. 3D). As for Fam1 strains, anti-PspA4 was able to mediate C3 deposition on St245/00 (clade 1) and anti-PspA5 on D39 (clade 2) at levels similar to those observed for the homologous Fam1 antibodies (Fig. 3D). Again, only median fluorescence values of the same strain with the different sera were compared.
PspA4 and PspA5 induce protection against challenges with Fam1- and Fam2-bearing strains.
Immunized mice were then submitted to a lethal intranasal challenge with pneumococci expressing PspA from Fam1 or Fam2. As shown in Table 2, animals immunized with PspA2 (Fam1) showed significant protection against a challenge with A66.1 (clade 2, Fam1) (P = 0.008). Moreover, PspA4 and PspA5 (Fam2) were also able to induce protection against A66.1, even though the strain expresses a PspA protein from a different family (P = 0.001 for PspA4 and P = 0.03 for PspA5). A66.1 was first reported to express only PspA2 (1), but subsequent work has shown it to express both PspA1 and PspA2 (3). We have amplified a single pspA band from this strain using primers LSM12 and SKH2 (17), and sequencing of cloned fragments confirmed only pspA2 clones. The expression of only PspA2 by A66.1 would be in accordance with the data showing protection after immunization with PspA2, but not with PspA1. Challenge with ATCC 6303 (clade 5, Fam2) was also performed, and as shown in Table 3, only protection within the same family was seen for PspA4 (P = 0.005) and PspA5 (P = 0.0007).
TABLE 2.
Survival after intranasal challenge with A66.1 (clade 2, Fam1)
| Immunizing antigen | No. alive/total | % Survival | P value |
|---|---|---|---|
| Alum | 0/6 | 0 | |
| Fam1 | |||
| PspA1 | 3/6 | 50 | 0.09 |
| PspA2 | 5/6 | 83a | 0.008 |
| Fam2 | |||
| PspA3 | 2/6 | 33 | 0.23 |
| PspA4 | 6/6 | 100a | 0.001 |
| PspA5 | 4/6 | 67a | 0.03 |
Statistically significant difference from animals inoculated with alum (Fisher exact test).
TABLE 3.
Survival after intranasal challenge with ATCC 6303 (clade 5, Fam2)
| Immunizing antigen | No. alive/total | % Survival | P value |
|---|---|---|---|
| Alum | 0/12 | 0 | |
| Fam1 | |||
| PspA1 | 3/12 | 25 | 0.11 |
| PspA2 | 2/12 | 17 | 0.24 |
| Fam2 | |||
| PspA3 | 3/11 | 27 | 0.09 |
| PspA4 | 6/11 | 55a | 0.005 |
| PspA5 | 8/12 | 67a | 0.0007 |
Statistically significant difference from animals inoculated with alum (Fisher exact test).
Contribution of the NonPro region to the reactivity to PspA.
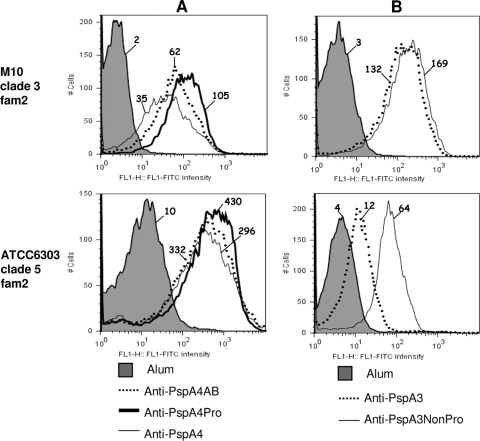
Of the five PspA fragments analyzed, PspA4 was the only one containing a NonPro region within the C-terminal proline-rich region. We have analyzed the contribution of both proline and NonPro regions to the cross-reactivity of PspA4. While the original PspA4 fragment contained the complete proline-rich region, PspA4AB contains only the α-helical region without the entire proline-rich region, and PspA4Pro contains the N-terminal α-helical region plus the first block of prolines only, lacking both the NonPro region and the second block of prolines (Fig. 1). We have analyzed the binding of anti-PspA4AB, anti-PspA4Pro, and anti-PspA4 antibodies to intact M10 (clade 3, lacks NonPro) and ATCC 6303 (clade 5, has NonPro). The capacity of antibodies to PspA4AB and PspA4Pro to bind to M10 was enhanced compared to that of antibodies to PspA4. Since M10 does not contain the NonPro region, reduced binding could be explained by a considerable amount of the antibodies induced after immunization with PspA4 being directed toward the NonPro region. Binding of antibodies to PspA4Pro was also slightly better than that of antibodies to PspA4AB, showing that the proline block is probably also immunogenic. Binding to ATCC 6303 followed a similar pattern, with higher binding for antibodies to PspA4Pro, followed by PspA4AB, but differences were not so evident (Fig. 4A). We next tested whether the addition of NonPro to PspA3 (which induced antibodies with very restricted cross-reactivity) would increase the recognition of different PspA proteins. Anti-PspA3 and anti-PspA3-NonPro showed similar binding to M10 (lacks NonPro), and the inclusion of the NonPro region clearly enhanced the cross-reactivity to ATCC 6303 (has NonPro) (Fig. 4B).
FIG. 4.
Binding of antibodies against the PspA4 and PspA3 constructs. Sera from mice immunized with PspA4AB, PspA4Pro, and PspA4 (A) or PspA3 and PspA3-NonPro (B) were tested for the ability to bind (1% serum) to S. pneumoniae strains bearing PspA proteins from clade 3 (M10, lacks NonPro) and clade 5 (ATCC 6303, has NonPro). Serum from mice immunized with alum was used as a control for each strain and is represented by the gray area in each graph. Results are shown as fluorescence intensity histograms, and the median fluorescence intensity is indicated for each sample. Results are representative of two experiments using sera from independent immunizations. FITC, fluorescein isothiocyanate.
DISCUSSION
Since PspA is a highly polymorphic antigen, the correct choice of fragments to be included in a vaccine formulation is crucial. We have tested the ability of sera raised against PspA fragments from clades 1 to 5 to bind to the surface of and to mediate complement deposition on pneumococci bearing different PspA proteins. Complement deposition assays have been proposed as a surrogate for prediction of protection (15). Previously, our group has also analyzed antibodies against PspA1, PspA3, and hybrid molecules, showing that binding and complement deposition mediated by anti-PspA1 and anti-PspA3 antibodies were restricted to the same family, whereas the hybrids were able to broaden this recognition to some extent (6). Subsequent work has shown that PspA4 and PspA5 were able to elicit antibodies with broad cross-reactivity, being able to recognize extracts from several strains expressing PspA proteins from clades 1 to 5 by Western blot analysis (7). We now show that anti-PspA4 and anti-PspA5 are also able to efficiently bind to the surface of pneumococci expressing PspA proteins from clades 1 to 5. As for C3 deposition, both sera were able to efficiently mediate complement deposition on pneumococci from the same family (clades 3, 4, and 5), whereas for Fam1 strains, anti-PspA4 was able to mediate C3 deposition on a clade 1-bearing strain and anti-PspA5 on a clade 2-bearing strain at levels similar to those observed for the homologous Fam1 antibodies. PspA3 elicited antibodies with the narrowest cross-reactivity, which is in accordance with our previous Western blot analysis results. These results show that the previously proposed clade-dependent immunity in Fam2 (6) may not be characteristic of all of the representatives of this family and is probably restricted to clade 3.
We have performed an intranasal lethal challenge of immunized mice, and the groups that received PspA4 or PspA5 showed statistically significantly higher survival after a challenge with A66.1, which expresses PspA2 (Fam1), and ATCC 6303, which expresses PspA5 (Fam2). These results thus show that both fragments would be able to confer broad protection against pneumococci expressing PspA proteins from the two major families. Moreover, these results are in accordance with the broad reactivity of antibodies to PspA4 and PspA5 with pneumococcal strains bearing different PspA proteins detected both in Western blot experiments and in functional assays of binding and complement deposition on intact bacteria. Fusion proteins composed of PspA fragments from Fam1 and Fam2 have been proposed as an alternative for the induction of broad protection using recombinant protein (6) or recombinant attenuated Salmonella (26). In the latter work, oral immunization of mice with Salmonella strains expressing PspA fusion proteins was shown to induce serum antibodies that were able to bind and to mediate complement deposition on strains expressing PspA proteins from clades 1 to 5 and also to provide protection against challenges with multiple pneumococcal strains. Our results show the potential of immunization with a single PspA fragment to induce cross-protection.
We have also analyzed the contribution of the NonPro region to the cross-reactivity of antibodies to PspA. The exclusion of the NonPro region of PspA4 led to the induction of antibodies with a higher capacity to bind to a strain that lacks this region, which indicates that a considerable amount of the antibodies induced by immunization with PspA4 might be directed to the NonPro region. The block of proline repeats also seemed to increase binding to the strains tested. Moreover, fusion of the entire proline-rich region containing NonPro to PspA3 (which was previously shown to induce antibodies with reduced cross-reactivity) was able to enhance the binding of antibodies to a strain that expresses PspA containing the NonPro region. Though it was not possible to define precisely the exact contribution of each region to the cross-reactivity of the antibodies, these results indicate that the N-terminal α-helical region, the blocks of proline repeats, and also the NonPro region can influence the degree of cross-reactivity, depending on whether the strain tested expresses a PspA protein containing the NonPro region or not. It has been previously suggested that the proline-rich region may protect mice against a pneumococcal challenge (5).
In conclusion, our results show that immunization with both PspA4 and PspA5 elicits antibodies with a functional capacity to recognize and to mediate complement deposition on a broad range of pneumococcal isolates. More importantly, PspA4 and PspA5 were able to induce protection against a challenge with one strain expressing PspA from Fam1 and one strain from Fam2 (the two major families), indicating that these antigens have potential to be used to as vaccine antigens to induce broad protection against different pneumococcal strains.
Acknowledgments
This work was supported by FAPESP, CNPq, Fundação Butantan, and the Millenium Institute-Gene Therapy Network (MCT-CNPq) (Brazil).
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Arulanandam, B. P., J. M. Lynch, D. E. Briles, S. Hollingshead, and D. W. Metzger. 2001. Intranasal vaccination with pneumococcal surface protein A and interleukin-12 augments antibody-mediated opsonization and protective immunity against Streptococcus pneumoniae infection. Infect. Immun. 69:6718-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briles, D. E., E. Ades, J. C. Paton, J. S. Sampson, G. M. Carlone, R. C. Huebner, A. Virolainen, E. Swiatlo, and S. K. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., S. K. Hollingshead, J. C. Paton, E. W. Ades, L. Novak, F. W. van Ginkel, and W. H. Benjamin, Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339-348. [DOI] [PubMed] [Google Scholar]
- 5.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darrieux, M., E. N. Miyaji, D. M. Ferreira, L. M. Lopes, A. P. Lopes, B. Ren, D. E. Briles, S. K. Hollingshead, and L. C. Leite. 2007. Fusion proteins containing family 1 and family 2 PspA fragments elicit protection against Streptococcus pneumoniae that correlates with antibody-mediated enhancement of complement deposition. Infect. Immun. 75:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darrieux, M., A. T. Moreno, D. M. Ferreira, F. C. Pimenta, A. L. de Andrade, A. P. Lopes, L. C. Leite, and E. N. Miyaji. 2008. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J. Med. Microbiol. 57:273-278. [DOI] [PubMed] [Google Scholar]
- 8.Frazão, N., A. Brito-Avo, C. Simas, J. Saldanha, R. Mato, S. Nunes, N. G. Sousa, J. A. Carrico, J. S. Almeida, I. Santos-Sanches, and H. de Lencastre. 2005. Effect of the seven-valent conjugate pneumococcal vaccine on carriage and drug resistance of Streptococcus pneumoniae in healthy children attending day-care centers in Lisbon. Pediatr. Infect. Dis. J. 24:243-252. [DOI] [PubMed] [Google Scholar]
- 9.Hicks, L. A., L. H. Harrison, B. Flannery, J. L. Hadler, W. Schaffner, A. S. Craig, D. Jackson, A. Thomas, B. Beall, R. Lynfield, A. Reingold, M. M. Farley, and C. G. Whitney. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 196:1346-1354. [DOI] [PubMed] [Google Scholar]
- 10.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, S. S., R. Platt, S. L. Rifas-Shiman, S. I. Pelton, D. Goldmann, and J. A. Finkelstein. 2005. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116:e408-413. [DOI] [PubMed] [Google Scholar]
- 12.Li, J., D. T. Glover, A. J. Szalai, S. K. Hollingshead, and D. E. Briles. 2007. PspA and PspC minimize immune adherence and transfer of pneumococci from erythrocytes to macrophages through their effects on complement activation. Infect. Immun. 75:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyaji, E. N., W. O. Dias, M. M. Tanizaki, and L. C. Leite. 2003. Protective efficacy of PspA (pneumococcal surface protein A)-based DNA vaccines: contribution of both humoral and cellular immune responses. FEMS Immunol. Med. Microbiol. 37:53-57. [DOI] [PubMed] [Google Scholar]
- 14.Nabors, G. S., P. A. Braun, D. J. Herrmann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 15.Ochs, M. M., W. Bartlett, D. E. Briles, B. Hicks, A. Jurkuvenas, P. Lau, B. Ren, and A. Millar. 2008. Vaccine-induced human antibodies to PspA augment complement C3 deposition on Streptococcus pneumoniae. Microb. Pathog. 44:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogunniyi, A. D., R. L. Folland, D. E. Briles, S. K. Hollingshead, and J. C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne, D. B., A. Sun, J. C. Butler, S. P. Singh, S. K. Hollingshead, and D. E. Briles. 2005. PspA family typing and PCR-based DNA fingerprinting with BOX A1R primer of pneumococci from the blood of patients in the USA with and without sickle cell disease. Epidemiol. Infect. 133:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos, C. R., P. A. Abreu, A. L. Nascimento, and P. L. Ho. 2004. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz. J. Med. Biol. Res. 37:1103-1109. [DOI] [PubMed] [Google Scholar]
- 19.Ren, B., M. A. McCrory, C. Pass, D. C. Bullard, C. M. Ballantyne, Y. Xu, D. E. Briles, and A. J. Szalai. 2004. The virulence function of Streptococcus pneumoniae surface protein A involves inhibition of complement activation and impairment of complement receptor-mediated protection. J. Immunol. 173:7506-7512. [DOI] [PubMed] [Google Scholar]
- 20.Ren, B., A. J. Szalai, S. K. Hollingshead, and D. E. Briles. 2004. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect. Immun. 72:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren, B., A. J. Szalai, O. Thomas, S. K. Hollingshead, and D. E. Briles. 2003. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect. Immun. 71:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche, H., A. Hakansson, S. K. Hollingshead, and D. E. Briles. 2003. Regions of PspA/EF3296 best able to elicit protection against Streptococcus pneumoniae in a murine infection model. Infect. Immun. 71:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaper, M., S. K. Hollingshead, W. H. Benjamin, Jr., and D. E. Briles. 2004. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect. Immun. 72:5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai, S. S. 2006. Streptococcus pneumoniae protein vaccine candidates: properties, activities and animal studies. Crit. Rev. Microbiol. 32:139-153. [DOI] [PubMed] [Google Scholar]
- 25.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xin, W., Y. Li, H. Mo, K. L. Roland, and R. Curtiss III. 2009. PspA family fusion proteins delivered by attenuated Salmonella enterica serovar Typhimurium extend and enhance protection against Streptococcus pneumoniae. Infect. Immun. 77:4518-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]