Abstract
Genes that are essential for viability represent potential targets for the development of anti-infective agents. However, relatively few have been determined in the filamentous fungal pathogen Aspergillus fumigatus. A novel solution employing parasexual genetics coupled with transposon mutagenesis using the Fusarium oxysporum transposon impala had previously enabled the identification of 20 essential genes from A. fumigatus; however, further use of this system required a better understanding of the mode of action of the transposon itself. Examination of a range of conditions indicated that impala is activated by prolonged exposure to low temperatures. This newly identified property was then harnessed to identify 96 loci that are critical for viability in A. fumigatus, including genes required for RNA metabolism, organelle organization, protein transport, ribosome biogenesis, and transcription, as well as a number of noncoding RNAs. A number of these genes represent potential targets for much-needed novel antifungal drugs.
Fungal diseases, particularly those caused by the aspergilli, remain a substantial problem (12, 43). The clinical infections caused by the aspergilli range from immediately life-threatening invasive aspergillosis, a systemic infection which results in high mortality levels (65%) even when treated (41), to the array of more long-term infections termed chronic aspergillosis (7) which are particularly problematic due to the length of therapeutic intervention coupled with drug toxicity and the emergence of resistance (46). More recently, an association between Aspergillus fumigatus and other airborne fungi and severe allergic asthma (severe asthma with fungal sensitization [SAFS]) has been identified (8, 9). It is estimated that SAFS accounts for around 4% of the severe forms of asthma, with A. fumigatus the predominant causative agent. Treatment options for Aspergillus infection are, however, limited as only four classes of drugs are available to treat systemic infections and their use is not without problems, including toxicity, resistance, and limitations in their mode of use (48, 50, 51).
Despite the need for novel drugs to combat fungal infection, antifungal drug discovery is limited by the availability of suitable well-validated drug targets. Several functional genomic studies have been undertaken to identify essential genes that would provide possible new routes to antifungal therapy. Recent work with Saccharomyces cerevisiae has shown that >10% of its genes are required for survival on rich growth medium, suggesting almost 900 potential targets for drug development (15). However, S. cerevisiae is a limited model for filamentous fungi, which display radically different morphology and complex multicellular growth and developmental biology. This is reflected in the larger gene complement of filamentous fungi, with 10,000 to 14,000 genes compared to the ∼6,000 of yeast (37). Comparative studies with Candida albicans indicate that only 61% of the S. cerevisiae essential genes are present in the Candida essential gene set, emphasizing the importance of performing essential gene screens directly on the organism of interest (45).
Unfortunately, the identification of essential genes in A. fumigatus is not a straightforward task. Despite the recent improvements in strains and techniques for filamentous fungi (4, 38), directed mutagenesis is still a rather involved and time-consuming process. Large-scale genome-wide disruption studies of the aspergilli have been proposed but as yet have not been performed. Additionally, the difficulties in screening for genes whose loss renders the organism inviable make such studies impractical. A novel solution to this conundrum was developed almost simultaneously by two different research groups (6, 13, 14). The use of the parasexual cycle to construct diploid isolates combined with transposon-based mutagenesis enabled the identification of 20 essential genes (Fig. 1). However, attempts to replicate this study met several difficulties, particularly with relation to the use of the transposable element imp160::pyrG as a mutagen.
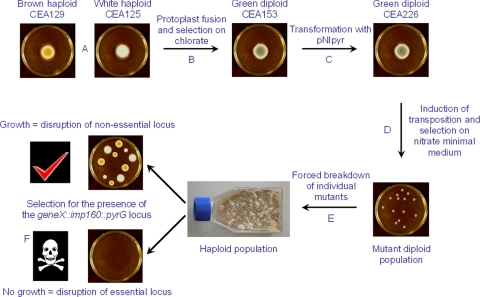
Fig. 1.
Schematic representation of the essential gene screen. A green diploid unable to grow in the absence of supplemental pyrimidines or to use nitrate as a sole nitrogen source (CEA153; w1/+ +/r7 cnx1/+ niaD4/niaD2 pyrG1/pyrG1) was generated from the parasexual fusion of brown (r7 niaD2 pyrG1) and white (w1 cnx1 pyrG1) haploid strains and subsequent selection on chlorate (A and B). The diploid was transformed with plasmid pNIpyr, which harbors the imp160 transposon integrated within the promoter of the A. nidulans niaD gene and tagged with an A. nidulans pyrG cassette (C). Strain CEA226 has a single copy of the NdeI-digested pNIpyr plasmid integrated at an undefined location. Large numbers of mutants are generated by activation of the imp160::pyrG transposon at 10°C. Mobilization of the transposon is detected by selecting for the restoration of the niaD gene by growth on nitrate-minimal medium; reintegration of the imp160::pyrG mutagen is ensured by performing the selection on medium lacking pyrimidines (D). Each individual mutant is picked and plated on medium containing the mitotic destabilizer benomyl, revealing a population of haploid progeny (E). This population is screened for the presence of the mutating imp160::pyrG element by plating on medium lacking pyrimidines (F). Absence of growing colonies is indicative of insertion of the transposon at loci critical for viability.
Here we present our novel finding that mobilization of the imp160::pyrG element is induced by exposure to low temperatures. Exploiting this, we demonstrate the first inducible transposable mutagenesis system for use in filamentous fungi and we present the results from a screen of 17,138 mutants in which we identify 96 essential loci, including two putative noncoding RNAs (ncRNAs).
MATERIALS AND METHODS
Strain construction.
Media and growth conditions are described elsewhere (14). A. fumigatus diploid strains CEA225 and CEA226 were generated from haploid spore color mutant isolates (white [w1] and brown [r7]), and their background has been described in detail elsewhere (14). Briefly, a diploid was formed by crossing white spore color strain CEA125 (w1 cnx1 pyrG1) with brown spore color strain CEA129 (r7 niaD2 pyrG1) to give green spore-forming diploid CEA131. Selection on chlorate-containing medium allowed the isolation of a strain unable to use nitrate as the sole source of nitrogen due to defects in both niaD alleles [CEA153 (w1/+ +/r7 cnx1/+ niaD4/niaD2 pyrG1/pyrG1)]. CEA153 was transformed with plasmid pNIpyr, a construct comprising a nonfunctional Aspergillus nidulans niaD gene disrupted by the imp160::pyrG transposon (which incorporates the A. nidulans pyrG gene within its terminal inverted repeats) to give strains CEA225 and CEA226. Both strains harbor single copies of the transposon cassette. Mobilization of impala160 in CEA225 and CEA226 can be detected by monitoring reconstitution of the niaD gene, as evidenced by the strain's ability to utilize nitrate as the sole nitrogen source.
Isolation of A. nidulans pyrG niaD− isolate PCAN002 was performed in two stages. A stable pyrG− derivative of A. nidulans A4 strain PCAN001 was selected from cultures exposed to 5-fluoroorotic acid and was confirmed by transforming with a plasmid cassette carrying the A. fumigatus pyrG gene. This strain was then plated on complete medium containing 0.6 M potassium chlorate to identify nitrate assimilation mutants. These were then screened for the presence of a mutation in the niaD gene as described earlier (3).
Transformation.
NdeI-digested pNIpyr was introduced into haploid strain PCAN002 using previously described methodologies. Briefly, protoplasts were generated by incubating mycelia from PCAN002 for 2 h at 30°C with 5% (wt/vol) Glucanex (Novozymes) in STC (sorbitol at 1.2 M, Tris at 200 mM, calcium chloride at 50 mM). Transformation was achieved by mixing 100 μl (ca. 1 × 107) of protoplasts with 1 to 5 μg of NdeI-digested pNIpyr and 1 ml of 60% polyethylene glycol 6000. The transformation mixture was combined with molten 1.2 M sorbitol in Vogel's minimal agar cooled to 50°C. Transformants were streaked to purity before storage at −80°C. PCAN005 was identified as a stable transformant carrying multiple copies of the niaD::imp160::pyrG cassette.
Assessment of transposition rates.
Mobilization of impala160 in CEA225, CEA226, and PCAN005 is evident in the restoration of function of the niaD gene and the subsequent ability of strains to grow on minimal-nitrate medium, MMNO3 (3). Preliminary studies with strains CEA225 and CEA226 indicated that transposon mobilization occurred while spore stocks were stored, so all stocks were used immediately after preparation and, with the exception of the −80°C stocks, never stored. To determine the transposition rates of freshly prepared spore stocks, diploid isolates CEA225 and CEA226 (which harbor single copies of the niaD::imp160::pyrG cassette) were streaked to purity and spores from 30 colonies of each strain were inoculated into separate tissue culture flasks containing SAB agar and allowed to grow for 3 days at 37°C. Spores were then either plated on MMNO3 agar or embedded in molten cooled 50°C MMNO3 agar. The mean frequency of transposition for each strain was determined by scoring colonies able to use nitrate as the sole nitrogen source, indicating excision of the imp160::pyrG transposon from the niaD marker gene. A total of 1 × 109 spores per isolate were plated at 5 × 107 per plate on minimal-nitrate medium.
The basal transposition rate was determined by incubating plates at 37°C for up to 10 days. Several variables were assessed to induce transposition. For assessment of the effect of low temperature on transposition, 1 × 109 spores were used to inoculate 400 ml of molten MMNO3 agar cooled to 50°C. Exactly 20 ml of molten agar was inoculated into a 9-cm petri dish and wrapped with Parafilm to prevent drying. Twenty plates were assessed for each time and temperature point. Plates were incubated at 4 to 37°C for up to 96 h before transfer to 37°C and incubation for a further 72 h. Emerging colonies were counted.
Expression analysis.
Approximately 5 × 107 conidia were inoculated into 50 ml of Vogel's minimal medium. Cultures were incubated for 14 h at 37°C with shaking at 220 rpm. Biomass was collected, and 0.3 g (wet weight) was used to inoculate the experimental 50-ml Vogel's minimal medium cultures. Cultures were incubated for 1 h, 2 h, 4 h, and 8 h at 10°C with shaking at 220 rpm, and as a control, a culture was incubated for 4 h at 37°C with shaking at 220 rpm. Total RNA was extracted using the FastRNA kit (QBIOgene) by following the manufacturer's instructions. Absence of contaminating DNA was confirmed by performing PCRs with control primers ACT470F and ACT960R on the extracted RNA. cDNA was synthesized from 1 μg RNA using an avian myeloblastosis virus reverse transcriptase kit (Promega) with random hexamers.
Expression levels of the impala transposase, the transposase of the native A. fumigatus transposon Aft1, and tubB encoding β-tubulin were determined by real-time PCR using primer sets shown in Table 1. Reactions were carried out in an iCycler thermal cycler fitted with an iQ real-time PCR detection system (Bio-Rad) running iCycler iQ Optical System software (version 3a; Bio-Rad). PCRs were performed in triplicate in 96-well plates with each 25-μl reaction mixture containing 12.5 μl of iQ SYBR green Super-mix (Bio-Rad), 5 pmol of each primer (Table 1), and 50 ng of template cDNA. PCR conditions were 3 min at 95°C and then 60 cycles of 30 s at 95°C, 30 s at 55°C, and 15 s at 72°C. Fluorescence was measured at the end of the annealing step. Average cycle thresholds were calculated, and the Pfaffl method (42) was used to calculate relative expression with respect to that of β-tubulin. Statistical significance was calculated using the Student t test (unpaired, unequal variance), and results with P values of <0.05 were deemed significantly different from the control.
Table 1.
PCR primers used in this study
| Primer | Sequence | Target |
|---|---|---|
| ACT470F | TGGTGTCACTCACGTTGTCC | Actin |
| ACT960R | TCATAGACGAGGGAGCAAGG | |
| RtbtubF | GCTCACTCTTTCCGTGCTGTCT | β-Tubulin |
| RtbtubR | AGCAGGTGAGGTAACGTCCATT | |
| RTimpF | GAGAATCTGTGGGCGTTGAT | impala |
| RTimpR | GCCTCCATTGCAGCTAAGAC | |
| RTaftF | GCCTGCACATTCATCACATC | Aft1 |
| RTaftR | AAGCCGCATCTTATCCTCAA | |
| UesAR1 | CGTCCTTTGGTGGAGGACCCG | UesA |
| UesAF1 | TAACTCAAGGATGAGAGGAT | |
| UesAF2 | GGACGATGACGATGAAGACC | |
| UesAF3 | AAGATGGGCCAGTAGGAGGT |
For analysis of the expression of UesA, cDNA was prepared as described above from AF293 cultures grown at 37°C for 20 h in MMNO3 liquid medium. PCR conditions were 3 min at 95°C and then 40 cycles of 30 s at 95°C, 60 s at 55°C, and 60 s at 72°C. Positive control reactions, to confirm that successful amplification could be achieved using the primer sets, were carried out with genomic DNA isolated from strain AF293.
Screening for essential genes.
Spores of diploid strains CEA225 and CEA226 were harvested from 3-day-old SAB agar flasks inoculated with a small number (ca. 1 × 103) of spores from a −80°C stock. Spores were used immediately to prevent transposon mobilization and propagation of such mutants during storage. Approximately 1 × 109 spores were used to inoculate 400 ml of molten MMNO3 agar cooled to 50°C. Molten agar was inoculated into a 9-cm petri dish and wrapped with Parafilm to prevent drying. To initiate transposition, plates were incubated at 10°C for 72 h before transfer to 37°C and incubation for a further 72 h. Emerging colonies were picked to a single well of a deep 96-well plate containing MMNO3 agar, allowed to sporulate by incubation at 37°C for 3 days, and then harvested. To avoid the possibility that parental-type haploids would outgrow slow-growing mutant-type haploids and hence lead to the identification of false positives, rather than use point inoculation (as previously described [14]), we spread diploid spores at low density into SAB agar culture flasks supplemented with 5 mM uridine and uracil and 1.2 μg/ml of the microtubule inhibitor benomyl and incubated them for 5 days at 37°C. To further ensure that sufficient haploid derivatives (evident because of their spore color) were screened, spores were only harvested from flasks showing more than 100 independent haploid colonies (Fig. 1) and replica plated onto Vogel's medium with and without supplemental uridine and uracil, each at 5 mM. Inability of haploid strains to propagate on medium without supplementation was deemed to be due to the insertion of imp160::pyrG at a locus critical for growth or formation of spores. Spores from strains with this phenotype were replated on SAB agar supplemented with 5 mM uridine and uracil and 1.2 μg/ml benomyl. Strains showing haploid sectors which were unable to form spores were eliminated from the data set.
Sequence determination.
Flanking sequence tags (FSTs) corresponding to genomic sequences bordering the 5′ end of imp160::pyrG were determined by adaptation of a two-step PCR strategy developed by Chun and coworkers (2) as described elsewhere (14).
The location of the transposon insertion was defined by comparing the sequence tags to the publicly available A. fumigatus (A1163) genome database using the BLAST facility at the Central Aspergillus Data REpository (CADRE; www.cadre-genomes.org.uk) (32). The insertion sites in all of the 96 diploid isolates were confirmed by performing a standard PCR on amplicons across the transposon boundary. The regions identified by the sequence tags were subjected to similarity searches using various BLAST algorithms (National Center for Biotechnology Information nonredundant protein and expressed genomic tag sequence databases, available at www.ncbi.nlm.nih.gov/BLAST) in order to identify putative coding sequences.
RESULTS
High rates of transposition redundancy impede the identification of essential genes.
In a previous study (14), we employed the imp160::pyrG transposon to generate a library of insertional mutants and screened 2,386 heterozygous diploid strains, which resulted in the identification of 20 essential genes. To extend our coverage of the essential gene set in A. fumigatus, we screened a further 12,769 heterozygous diploid strains. A total of 120 isolates (0.9%) from this collection exhibited a haploid lethal phenotype indicative of the insertion of impala at an essential locus. Characterization of the flanking sequence was possible for all of the 120 isolates; however, these were found to correspond to only 13 independent loci (Table 2).
Table 2.
Characterization of the genes at the impala160::pyrG insertion site for all of the 96 strains identified in this studya
| Functional group and strain | Nearest annotated gene to insertion site | Distance (bp) upsteam of nearest geneb | S. cerevisiae namec | Spectrumd | Selectivitye | Essential in S. cerevisiae/N. crassaf |
|---|---|---|---|---|---|---|
| Amino acid biosynthesis | ||||||
| M7-D04 | Afua_1g13740 | 0 | Aro1 | +++ | ++++ | Yes/unknown |
| M35-G03 | Afua_2g07570 | 0 | Pro1 | +++ | +++h | Yes/unknown |
| M43-H04 | Afua_3g11710 | 0 | Lys1 | +++ | ++++ | Yes/unknown |
| M18-D11 | Afua_4g11240 | 85 | Lys2 | +++ | ++++ | Yes/unknown |
| M14-F03 | Afua_6g09070 | 0 | Met22 | ++ | ++++ | Yes/yes |
| Cellular homeostasis M53-G04 | Afua_5g02370 | 150 | Tfp1 | ++++ | − | No/yes |
| Cellular organization | ||||||
| M34-B08 | Afua_1g13100 | 0 | Nup188 | − | ++++ | No/yes |
| M26-A02 | Afua_2g03310 | 0 | Nsp1 | − | +++ | Yes/yes |
| M22-E01 | Afua_4g09740 | 0 | Cct8 | +++ | ++ | Yes/unknown |
| M3-A02 | Afua_6g04740 | 163 | Act1 | ++++ | − | Yes/unknown |
| F8-E06 | Afua_1g10740 | 0 | Gim5 | ++ | ++ | No (benomyl sensitive)/no |
| M34-A03 | Afua_6g04740 | 0 | Act1 | ++++ | − | Yes/unknown |
| Cofactor biosynthesis | ||||||
| M50-H02 | Afua_2g05820 | 308 | Fmn1 | + | +++ | Yes/unknown |
| M46-D10 | Afua_3g08470 | 49 | Zwf1 | +++ | + | Yes/unknown |
| Cytokinesis | ||||||
| M6-A08 | Afua_2g03640 | 0 | Iqg1 | − | +++ | Yes/yes |
| Chromosomal organization F5-F09 | Afua_1g13790 | 145 | Hht1 | ++++ | − | No/unknown |
| DNA replication | ||||||
| M5-F04 | Afua_2g09060 | 0 | Cdc54 | +++ | ++ | Yes/unknown |
| M24-G09 | Afua_2g12250 | 0 | Rfc5 | +++ | + | Yes/yes |
| M37-B09 | Afua_2g16600 | 180 | Pol3 | ++++ | + | Yes/yes |
| M25-C12 | Afua_6g05040 | 0 | Rfc4 | ++++ | − | Yes/yes |
| M34-F11 | Afua_2g16710 | 0 | No orthologue (S. pombe CDC27) | − | +++ | NAg/yes |
| DNA transcription | ||||||
| M28-E02 | Afua_1g05830 | 0 | Mot1 | ++ | +++ | Yes/yes |
| M39-D11 | Afua_1g05830 | 176 | Mot1 | ++ | +++ | Yes/yes |
| M12-H01 | Afua_1g13910 | 79 | Rad3 | ++++ | + | Yes/yes |
| M26-A03 | Afua_1g14740 | 0 | Toa1 | + | ++++ | Yes/yes |
| M34-E12 | Afua_2g06060 | 0 | Hfi1 | − | ++++ | No/yes |
| M25-D11 | Afua_2g10870 | 330 | Rrn11 | − | ++++ | Yes/unknown |
| F3-A04 | Afua_6g08330 | 163 | No orthologue (S. pombe SSR4) | − | ++++ | NA/yes |
| M35-F01 | Afua_6g14190 | 0 | No orthologue (S. pombe SFC9) | − | ++++ | NA/no |
| Lipid biosynthesis | ||||||
| M32-D10 | Afua_5g02450 | 71 | Erg20 | ++++ | ++ | Yes/yes |
| M5-D04 | Afua_6g12390 | 0 | Lcb1 | ++ | +++ | Yes/unknown |
| Mitochondrial function and maintenance | ||||||
| M45-C06 | Afua_2g05510 | 118 | Atp17 | − | ++++ | No/no |
| M33-D12 | Afua_2g12370 | 0 | Ynl213c | − | ++++ | No/no |
| M45-F09 | Afua_3g08080 | 0 | Mrpl49 | − | ++++ | No/yes |
| F4-B03 | Afua_5g12820 | 81 | Imp1 | + | +++ | No/unknown |
| M26-A01 | Afua_6g04420 | 0 | Mmm1 | + | ++++ | No/no |
| M43-H12 | Afua_6g04910 | 0 | Nam2 | ++ | +++ | No/unknown |
| M41-A08 | Afua_6g06890 | 56 | Mrpl40 | − | ++++ | No/yes |
| M35-B05 | Afua_7g04670 | 65 | Atp11 | + | ++++ | No/no |
| Protein modification | ||||||
| M23-F05 | Afua_2g06150 | 0 | Pdi1 | ++ | +++ | Yes/unknown |
| M1-B12 | Afua_2g11240 | 0 | Alg7 | ++ | ++ | Yes/yes |
| M28-H05 | Afua_3g07170 | 0 | Gpi2 | + | +++ | Yes/yes |
| M8-A04 | Afua_4g07800 | 186 | Ram2 | + | +++ | Yes/unknown |
| F3-G06 | Afua_4g11280 | 0 | Gpi18 | − | ++++ | Yes/yes |
| M53-G05 | Afua_5g08370 | 180 | Doa1 | + | +++ | No/yes |
| M40-C12 | Afua_6g02460 | 0 | Gna1 | ++ | +++ | Yes/no |
| M55-E02 | Afua_5g12680 | 0 | No orthologue (S. pombe Cul4B) | − | +++ | NA/no |
| Protein trafficking | ||||||
| M30-C10 | Afua_1g03940 | 77 | Srp68 | − | ++++ | Yes/unknown |
| M25-F02 | Afua_2g01570 | 0 | Aps1 | ++++ | − | No/unknown |
| M43-H11 | Afua_2g13450 | 0 | Ydl058w/Uso1 | − | +++ | Yes/no |
| M41-A03 | Afua_3g08840 | 0 | Cop1 | +++ | + | Yes/yes |
| M42-D04 | Afua_4g04630 | 0 | Yip1 | ++ | +++ | Yes/unknown |
| M20-G06 | Afua_4g07110 | 37 | Pep12 | − | ++++ | No/yes |
| M1-E02 | Afua_5g13120 | 272 | Bcp1 | + | ++++ | Yes/unknown |
| M19-G08 | Afua_6g04060 | 0 | Cog1 | − | ++++ | No/unknown |
| M21-A02 | Afua_6g12830 | 331 | Sec24 | +++ | +++ | Yes/no |
| M17-G07 | See text | NA | SRP RNA | N/A | NA | Yes/unknown |
| Ribosome biogenesis | ||||||
| M57-D01 | Afua_1g04840 | 0 | Mrd1 | + | ++ | Yes/yes |
| F10-B01 | Afua_1g04840 | 0 | Mrd1 | + | ++ | Yes/yes |
| M28-D01 | Afua_1g13200 | 133 | Urb2 | − | ++++ | Yes/unknown |
| M24-E04 | Afua_2g01330 | 0 | Rrp46 | + | ++++ | Yes/yes |
| M35-D08 | Afua_2g01330 | 124 | Rrp46 | + | ++++ | Yes/yes |
| M3-B06 | Afua_2g08860 | 0 | Rrp45 | + | +++ | Yes/yes |
| M18-H10 | Afua_2g10300 | 69 | Rps17a | − | −h | Yes (duplicated)/unknown |
| M2-C10 | Afua_2g11810 | 0 | Rrp12 | + | ++++ | Yes/unknown |
| F1-G02 | Afua_2g16040 | 0 | Rrp5 | − | ++++ | Yes/unknown |
| M19-G01 | Afua_3g12300 | 136 | Rpl22a | + | ++ | Yes (duplicated)/unknown |
| F8-D10 | Afua_4g07570 | 68 | Mak11 | − | ++++ | Yes/yes |
| F11-E02 | Afua_4g07730 | 0 | Rpl11a | − | ++h | Yes (duplicated)/yes |
| M44-A04 | Afua_5g07390 | 0 | Sqt1 | − | ++++ | Yes/unknown |
| RNA modification | ||||||
| M24-F01 | Afua_2g08550 | 0 | Ess1 | ++ | + | Yes/unknown |
| M22-A08 | Afua_3g12290 | 0 | Dib1 | ++++ | − | Yes/unknown |
| M37-B01 | Afua_6g13410 | 0 | Rse1 | + | + | Yes/yes |
| M46-A11 | Afua_8g04420 | 0 | Pta1 | − | ++++ | Yes/unknown |
| F11-A07 | Afua_3g10450 | 0 | No orthologue (U2 associated SR140-human) | − | ++++ | NA/unknown |
| Signaling | ||||||
| M44-E07 | Afua_1g13420 | 0 | Rrd2 | ++ | +++ | Yes (duplicated)/no |
| M35-G09 | Afua_2g02510 | 0 | Kog1 | ++ | ++ | Yes/yes |
| M7-F08 | Afua_6g13300 | 145 | Gsp1 | ++++ | − | No/yes |
| t-RNA synthesis | ||||||
| M19-H07 | Afua_1g09690 | 0 | Trl1 | + | ++++ | Yes/unknown |
| M43-B06 | Afua_5g03560 | 0 | Gus1 | +++ | ++ | Yes/unknown |
| F11-C03 | Afua_5g05920 | 0 | Grs1 | ++++ | ++ | Yes/yes |
| M32-F03 | Afua_6g12630 | 0 | Cdc60 | +++ | ++ | Yes/unknown |
| Unknown function | ||||||
| M31-E09 | Afua_3g10070 | 103 | No orthologue (S. pombe PCH1) | − | +++h | NA/unknown |
| M22-E03 | Afua_7g00510 | 280 | No orthologue | − | ++++ | NA/unknown |
| M23-B04 | Afua_5g07770 | 0 | No orthologue | − | ++++ | NA/unknown |
| M24-H06 | Afua_3g06490 | 0 | No orthologue | − | ++++ | NA/no |
| M35-G12 | Afua_7g04390 | 63 | No orthologue | − | ++++ | NA/unknown |
| M36-C01 | Afua_2g11320 | 0 | No orthologue | − | ++++ | NA/no |
| M53-D02 | Afua_6g06810 | 0 | No orthologue | − | ++++ | NA/no |
| No gene assigned— distance from gene | ||||||
| M28-B09 | Afua_7g05340 | 850 | ||||
| F7-E07 | Afua_6g04610 | 412 | ||||
| M55-G01 | Afua_1g14100 | 588 | ||||
| F7-G05 | Afua_1g04190 | 485 | ||||
| M6-F08 | Afua_1g05240 | 1,456 | ||||
| No gene assigned— insertion lies between genes | ||||||
| M42-H05 | Afua_5g10740 | 179, US of Afu5g10740; 199, US of Afu5g10750 | ||||
| M22-C06 | Afua_2g14030 | 220, US of Afu2g14030; 295, US of Afu2g14040 |
Strains with names starting with the letter F were isolated from screen 1. Each gene has, where possible, been allocated to a primary functional group based on information from the nearest BLAST hit identified or a known function defined in A. fumigatus. The location of the insertion site is given with respect to the predicted ATG start codon of the nearest gene as defined by annotations at CADRE.
A value of 0 indicates that the insertion site is within the gene; US, upstream.
The standard gene name of the nearest BLAST hit from S. cerevisiae is given, or where no orthologue exists and where possible, an appropriate alternative is given.
Spectrum (defined by the percent identity of the nearest BLAST hit in C. albicans) scores: ++++, >60%; +++, 50 to 60%; ++, 40 to 50%; +, 30 to 40%; −, <30% or absent.
Selectivity (defined by the percent identity of the nearest BLAST hit in human) scores: ++++, <30% or absent; +++, 30 to 40%; ++, 40 to 50%; +, 50 to 60%; −, >60%.
Where possible, the essentiality of the nearest appropriate BLAST hits from S. cerevisiae and N. crassa is given. This information is taken from the data sets at the Saccharomyces Genome Database and the list of knockouts given by the Neurospora genome project at http://www.dartmouth.edu/∼neurosporagenome/index.html. Neurospora knockout strains which are only available as heterokaryons are deemed to be essential.
NA, not applicable.
Partial alignment used.
Previous studies have shown that the imp160::pyrG transposon always integrates within a TA dinucleotide (14). Given that it displays no other sequence-specific requirement for integration, it was thought that the mutation redundancy must therefore be caused by a series of single transposition events occurring within spore stocks, being amplified during storage and therefore appearing as apparent multiple single events in our insertional library. Attempts were therefore made to reduce or eliminate this source of mutation redundancy.
Transposition of imp160 is low temperature dependent.
Our preliminary analysis revealed that the transposition rates of the imp160::pyrG transposon in isolates CEA225 and CEA226 were 1.8 and 2.0/109 spores after 3 days of growth at 37°C, respectively. These rates are 4 orders of magnitude lower than the previously published niaD reversion rate for our strains (14). To investigate the possibility that impala transposition is somehow induced, strains CEA225 and CEA226 were exposed to several stresses known to induce transposition in other organisms. These included high levels of CuSO4 (21), carbon and nitrogen starvation (35, 36), heat shock at a range of temperatures between 45 and 70°C (16), cold shock between 20 and 30°C (26, 44), and exposure to sub-MIC levels of antimicrobial agents (in our case, amphotericin and itraconazole) (47). Only incubation at 20°C for 2 days resulted in a significant increase in the number of transposition events from 3 ± 0.37/109 to 8 ± 0.83/109 spores (P < 0.05), suggesting that transposition rates could be induced by incubation at low temperatures.
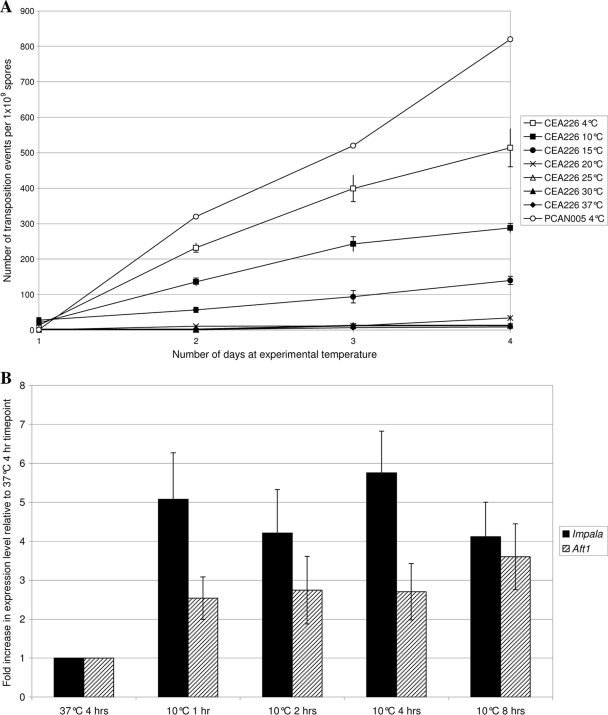
To further understand the effect of low temperature on transposition, the transposition rate was assessed at 4, 10, 15, 20, 30, and 37°C. The rate of transposition of impala was seen to be approximately linear over time at all temperatures (Fig. 2A). Although the transposition frequency was stable between 37 and 25°C, the rate increased dramatically at temperatures below 20°C and was at its highest at 4°C, where it reached 128 revertants/109 spores/day, 200 times more than that seen at 37°C.
Fig. 2.
Response of impala transposition rates to low temperatures. (A) Number of transposition events obtained per 1 × 109 spores upon exposure to low temperatures for increasing lengths of time. Each data point is the mean of three independent experiments, each collating the data from the plating of 1 × 109 spores, with the exception of PCAN005, where data from only one experiment are shown. Error bars indicate standard errors. (B) Expression levels of the impala (filled) and Aft1 (hatched) transposase genes in response to cold shock at 10°C relative to the levels seen at 37°C. The results presented show mean values from four separate experiments and have been normalized to β-tubulin gene expression. Error bars indicate standard errors.
To confirm that the revertants observed were generated as a result of independent excision-reintegration events, FSTs of 16 diploid isolates obtained from the 4°C study were amplified using a two-stage PCR approach developed by Chun et al. (2). All 16 isolates showed different amplification patterns, indicating that the insertion sites differed (data not shown). Sequencing of the PCR products confirmed this result, as all 16 isolates harbored different FSTs. Although current predictions indicate that coding sequence represents 50% of the A. fumigatus genome, only three (19%) of the isolates had insertions at coding loci. Also, a disproportionate number of insertions (n = 8 [50%]) were found in regions immediately upstream (within 350 bp) of predicted ATG sites, suggesting that impala has a preference for insertion into promoters rather than either coding or noncoding regions. These data suggest that impala insertion is not completely random.
To see whether temperature induction of transposition was unique to A. fumigatus, the same plasmid used to generate strains CEA225 and CEA226 (pNIrpyr) was used to transform an A. nidulans pyrG− niaD− (PCAN002) strain, resulting in a pyrG+ niaD− isolate harboring the niaD::imp160::pyrG cassette (PCAN005). No revertants were obtained from 109 spores after 7 days of incubation at 37°C on nitrate medium, whereas an average of 820 revertants/109 spores were obtained after 4 days of incubation at 4°C, followed by transfer to 37°C for 3 days (Fig. 2A). Again, an approximately linear relationship between the time of incubation at 4°C and the transposition rate was observed, indicating that low-temperature induction is not unique to A. fumigatus (Fig. 2A).
In an attempt to explain the increase in the transposition rate of impala at low temperatures, real-time PCR was used to quantify the levels of transposase mRNA in A. fumigatus strain CEA226. Mycelia from 37°C starter cultures were pooled and transferred to 10°C, and samples were taken at 1, 2, 4, and 8 h posttransfer. A control culture was incubated at 37°C for 4 h. Expression of the transposase gene could be detected in the 37°C cultures at levels equivalent to 0.3 times the β-tubulin control. In comparison, expression levels in cultures incubated at 10°C were approximately fivefold higher (Fig. 2B). To assess if this phenomenon was unique to the impala transposase, the expression of the native multicopy Aft1 transposase was assessed in the same way. Expression of the Aft1 transposase gene at 37°C was relatively high (ca. 1.4 times the level of the β-tubulin gene). Cultures incubated at 10°C showed approximately threefold higher levels of expression than the 37°C control. The increased transposition rate seen here is therefore likely, at least in part, to be due to increased levels of transposase or perhaps the presence of an alternative transcript.
Screening for essential genes by haploidization of somatic diploids.
Using a modified method based on the use of low-temperature-induced transposon mutagenesis, we generated a collection of 4,369 heterologous A. fumigatus pyrG+ niaD+ revertants from CEA226 by selection on nitrate-minimal medium lacking uridine and uracil. Haploidization of these revertants, followed by screening on selective (minimal medium lacking uridine and uracil) and nonselective (minimal medium supplemented with uridine and uracil) media, revealed 88 strains whose haploid derivatives were unable to grow on selective medium, indicating that an insertion into a gene essential either for growth or for the development of asexual spores had occurred. Microscopic analysis of haploid colonies from all 88 strains growing on the haploidization medium revealed 5 that produced a large number of haploid sectors that were unable to form asexual spores. These were eliminated from further analysis.
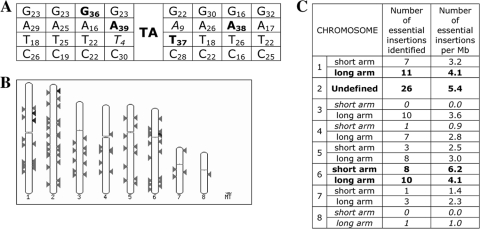
FSTs were obtained for all of the 96 diploid strains (13 from the first screen plus 83 from the second screen) whose haploid derivatives were unable to grow in the absence of uridine and uracil, and the location of the transposon insertion was identified. Determination of the exact imp160::pyrG insertion site was possible for all of the 96 essential loci. Alignment of the FSTs confirmed the absolute requirement for the TA dinucleotide at the point of insertion, but in addition, our results suggested that the presence of a thymine immediately 5′ or an adenine immediately 3′ of the target dinucleotide is rare and the presence of a 5′ adenine and/or a 3′ thymine is preferred (Fig. 3A). By mapping the BLAST hits from the FSTs to the karyotype map of AF293, we were able to show how the essential loci are dispersed throughout the genome. Essential loci have been identified from all eight chromosomes, although some clustering is evident, particularly toward the telomere on the long arm of chromosome 6 and the centromere on the short arm of chromosome 6 and within a central region of the long arm of chromosome 1. A larger-than-expected number of essential loci were also identified on chromosome 2 (Fig. 3B and C). This may indicate that several clusters of essential genes exist within the A. fumigatus genome, as is known to be the case for other organisms (20, 30); however, we cannot rule out the possibility that nonrandom distribution of the transposon is responsible for the clustering observed.
Fig. 3.
Transposition of the impala mobile element in A. fumigatus. (A) Summary of the sequences flanking 96 impala insertion sites. Each insertion occurs at a TA dinucleotide; the frequency of each flanking nucleotide is shown as a subscript. Nucleotides representing ≥35% of the total for that locus are in bold; those representing ≤15% are in italics. (B) Schematic representation of the locations of the impala insertion sites for the 96 essential hits on a karyotype map based on the AF293 genome given by the output of the CADRE BLAST search. Black arrowheads mark regions (ca. 25 kb) where a single insertion has occurred, and white arrowheads show regions where two insertion events have occurred. (C) Tabular representation of the nuclear karyotype map showing the frequency of insertion with respect to each chromosome arm. Those locations overrepresented are in bold; those underrepresented are in italics.
The regions identified by the sequence tags were subjected to similarity searches using various BLAST algorithms in order to identify putative coding sequences. Fifty-five of the isolates identified had insertions within open reading frames, and 34 had insertions in promoter regions of predicted genes (within 350 bp of the predicted start codon), while the remaining 7 had insertions >400 bp upstream of a predicted gene. The genes identified by this study are listed in Table 2. For two of the strains in which insertions had occurred in promoters, it was not possible to tell which gene was affected since the promoter regions were shared by two genes.
Isolates with insertions >400 bp from a predicted gene were subjected to further analysis. The mutation in isolate 17G07 is located 580 bp upstream from its nearest gene (Afua_1g10420), and a 280-bp feature showing high levels of conservation with other aspergilli was identified at the insertion site. Separately, a group identified this feature as an ncRNA (22), and we have subsequently confirmed, by comparison with sequences from the Signal Recognition Particle Database (http://rnp.uthct.edu/rnp/SRPDB/SRPDB.html), that it is the homologue of the signal recognition particle RNA.
The mutation in strain M6-F08 was located 1,456 bp upstream of the nearest annotated gene. We have identified a small but highly conserved DNA sequence at this site (ca. 115 bp; see Fig. S1 in the supplemental material) which appears to be restricted to genomes of the aspergilli. No obvious open reading frame can be discerned, but analysis of this region using reverse transcription-PCR suggests that it is expressed (see Fig. S2 in the supplemental material). The conserved sequence is flanked at its 5′ end by a GA-rich region and at its 3′ end by a CT-rich region; however, the functional significance of this is unknown. It is possible that this is an ncRNA; we have named it uesA (unknown essential sequence A).
The integration sites in strains F7-E07 (412 bases upstream of Afua_6g04610), M55-G01 (588 bases upstream of Afua_1g14100), and F7-G05 (485 bases upstream of Afua_1g04190) are also conserved across several species of aspergilli (see Fig. S1 in the supplemental material) and are similar in architecture to that of M6-F08 in that they are flanked by poly(T) chains at the 3′ end and GA-rich regions at the 5′ end. In all of these mutants, however, the insertion sites lie upstream of known S. cerevisiae essential genes (rpc19, tam41, and pab1). We have named these regions uesB, uesC, and uesD.
Finally, the insertion identified from mutant clone M28-B09 is also in a region devoid of an annotated coding sequence, 850 bp from the nearest annotated gene. The region of this insertion site showed exceptionally high levels of sequence conservation with A. flavus but not with any other fungal genome. This may indicate the presence of an essential gene limited to A. fumigatus and A. flavus. We have named this region uesE.
DISCUSSION
Novel properties of the imp160::pyrG transposon.
The mobilization of the imp160::pyrG transposon in a range of filamentous fungi has been previously described (10, 14, 19, 40); however, the mechanism by which this is regulated was not known until now. At standard laboratory culture temperatures (30 to 37°C), the imp160::pyrG construct is almost inactive in A. fumigatus and apparently completely inactive in A. nidulans. We have shown that transposition can be activated by exposing cultures to low temperatures and deactivated again by raising the temperature. Modulation of transposition activity by environmental factors was described recently for the transposable elements Aft1 of A. fumigatus, crawler of A. oryzae, and OPHIO1 of Ophiostoma ulmi (1, 16, 39). crawler, which shows high levels of sequence identity to impala, is activated in the presence of low millimolar levels of copper sulfate, while both crawler and Aft1 have increased levels of transposition in response to heat shock. When activated, crawler, like impala, shows elevated levels of expression (ca. 2.5-fold); however, significant amounts of transcript are still present even in the absence of induction. The increase in transposition cannot be solely attributed to changes in transcription, since mRNA can be detected at relatively high levels even when the transposon activity is low. Therefore, it is likely that transposition is additionally affected by an as-yet-undefined posttranscriptional control mechanism.
A major issue surrounding the use of fungal transposons has been the perception that they integrate into “hot spots” and rarely into coding regions. Despite sequencing the insertion sites of over 200 niaD revertants (many of them detailed in this study), we did not identify a single insertion site hot spot. This is in contrast to a similar study using the mimp1 transposon (a small element activated by the impala transposase) where analysis of 91 transposition events resulted in insertion into 66 independent loci, with 1 locus being identified 19 times (11). Our analysis of 16 impala insertions does, however, suggest that integration within promoter regions is preferred. Our result is mirrored by the study of Dufresne et al. (11), where 29% of the insertions occurred within a region 500 bp upstream of predicted genes. The preference for promoter regions may be due to these stretches of DNA being generally free of nucleosomes (28), and the absence of nucleosomes may provide the transposon greater physical access to the DNA. The demonstration of a mechanism for transposon control and clarification of the hot spot issue make imp160::pyrG a suitable agent for genome-wide transposon mutagenesis.
Essential gene identification in A. fumigatus.
Several methodologies have been employed in the past to identify essential genes in fungi. The most comprehensive of these was the whole-genome knockout study of S. cerevisiae which, from the most recent data, has enabled the identification of 1,179 essential genes (Saccharomyces Genome Database [http://www.yeastgenome.org]) (15). A number of groups have made efforts to identify essential genes in pathogenic fungi as a means of identifying drug targets, the most notable of which has been in C. albicans, where a system termed GRACE (gene replacement and conditional expression) was used to assess the homologues of S. cerevisiae essential genes, allowing the identification of 567 essential Candida genes (45). Recent attempts to transfer this technology to A. fumigatus proved to be more difficult, due primarily to difficulties, at the time, in generating directed mutants (18). In that study, only 35 essential genes were described. Both of these studies relied heavily on the essential gene set from S. cerevisiae, and hence, all of the genes identified had previously been described as essential for viability. In contrast, the transposon-based technology used in our study requires no foreknowledge of gene essentiality nor does it require predictions of the locations of a gene. This fact has allowed us to identify genes critical for viability which are either absent or not essential in the S. cerevisiae genome, a remarkable 43% of the genes identified.
The observation that only about half of the A. fumigatus genes identified here have essential homologues in S. cerevisiae echoes the results of a comparative study of S. cerevisiae and C. albicans in which only 61% of the essential genes were found to be shared (45). The differences between the S. cerevisiae and A. fumigatus essential gene sets can be attributed to those which are apparently absent from S. cerevisiae (n = 12), those that are duplicated in S. cerevisiae (n = 5), and those that have homologues but not essential partners (n = 19). The majority of the final group are genes required for mitochondrial maintenance and function (n = 8), highlighting the differences in the importance of respiration between the obligate aerobe A. fumigatus and S. cerevisiae, which can grow anaerobically.
In a number of cases, genes were identified which have been previously recognized as potential drug targets. For example, Afua_4g07800 is the homologue of S. cerevisiae RAM2, which encodes the common alpha subunit of both the farnesyltransferase and geranylgeranyltransferase, for which a number of inhibitors have been identified (27). Also, Afua_6g12390 encodes Lcb1, a component of serine palmitoyltransferase, and sphingofungins B and C (isolated from A. fumigatus) and lipoxamycin (isolated from Streptomyces) all specifically target serine palmitoyltransferase and are potent antifungal agents (33, 52). The mutant bank identified here also contains a number of potentially novel targets. For example, Afua_2g08550 is a homolog of the S. cerevisiae ESS1 gene that encodes a pin1-type peptidyl-prolyl isomerase (PPIase). PPIases function by switching peptidyl-prolyl bonds between their cis and trans conformations. The conformation of peptidyl-prolyl bonds is important, as they are differentially recognized by a series of other proteins, including proteases such as chymotrypsin A and phosphatases such as PP2A (23). One of the critical functions described for Ess1p is in the modulation of activity of the RNA polymerase II large subunit, switching its ability to bind basal promoters (25). Quite significant differences exist between the fungal and mammalian ESS1 enzymes, and PPIases are known to be responsive to small-molecule inhibitors (e.g., cantharidin targets the PPIase PTPA). To date, this target seems to have received very little attention in anti-infective drug discovery. Another target of potential interest is Afua_2g09060, which encodes Cdc54, a subunit of the MCM complex which is required for “melting” of DNA prior to replication (5, 29). Cdc54 shares less than 50% identity with its mammalian homologues, which may be sufficient to allow selective targeting of the fungal enzyme above its human counterpart and encodes an essential ATPase domain, a function which can be used as a basis for the development of a high-throughput screen.
By definition, the loss of function of an essential gene leads to complete abolition of growth. This strict definition of essentiality is, however, difficult to prove experimentally. The screening approach undertaken by Hu et al. (18), for example, defines essentiality as a severe impairment of growth when a gene is downregulated using the A. fumigatus niiA promoter. A study of gene fitness in Streptococcus pneumoniae defines “possible essential” genes as those with a relative fitness score of 0 (the wild-type fitness score is 1); however, their study was unable to distinguish slow-growing mutants with fitness scores of <0.53 from those with a score of 0 (49). The caveat to the definition of essentiality described here is that our ability to distinguish between genes which are essential and those which cause extreme growth retardation is limited by the length of time we allow haploids to appear on benomyl medium. In our study, mutations which result in the inability of a haploid to grow after 5 days are deemed to be essential.
Interestingly, our essential set included homologues of the glycosylphosphatidylinositol (GPI) biosynthesis pathway genes GPI2 and GPI18, which are responsible for the first step of GPI anchor biosynthesis (24). The presence of these genes in our data set was not expected, as although GPI anchors are required for hyphal and conidial wall stability, they are apparently dispensable for viability in A. fumigatus (31). It is clear from a study by Li et al. (31) that GPI mutants exhibit many growth defects. It could be that loss of GPI biosynthesis is synthetically lethal with the brown and white spore color mutations present in our diploid; however, a more likely scenario is that GPI biosynthesis mutants are hypersensitive to benomyl. Notwithstanding this, GPI biosynthesis is still an attractive target for the development of antifungal agents. The Eisai R&D Management Co. Ltd. has recently published evidence that small molecules can target this process in C. albicans, leading to a positive therapeutic outcome in a mammalian infection model (34). Interestingly and apparently in contradiction to the dispensability of GPI biosynthesis described by Li et al. (31), many of the compounds detailed in this patent application also possessed anti-Aspergillus activity.
Essential genes as drug targets.
The need for new antifungal agents with high levels of efficacy and low toxicity is pressing. The combination of the increase in resistance to the azoles (17, 46) and the emergence of resistance to the echinocandins (12) suggests that the focus of drug discovery should be the identification of compounds with entirely novel modes of action. Targets should have a suitable spectrum and selectivity profile; i.e., they should lack close mammalian homologues and be reasonably well conserved across the pathogenic fungi, and to enable the screening of large numbers of potential inhibitors, a route to the development of a high-throughput screen should be considered. Our study presents a large number of essential genes from A. fumigatus, including at least one ncRNA, which may be considered to be new starting points for target-based drug discovery efforts, as well as providing a robust method by which this data set can be increased.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/cgi/content/full/9/3/438/DC1.
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Bouvet G. F., Jacobi V., Plourde K. V., Bernier L. 2008. Stress-induced mobility of OPHIO1 and OPHIO2, DNA transposons of the Dutch elm disease fungi. Fungal Genet. Biol. 45:565–578 [DOI] [PubMed] [Google Scholar]
- 2.Chun K. T., Edenberg H. J., Kelley M. R., Goebl M. G. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233–240 [DOI] [PubMed] [Google Scholar]
- 3.Cove D. J. 1976. Chlorate toxicity in Aspergillus nidulans—selection and characterization of chlorate resistant mutants. Heredity 36:191–203 [DOI] [PubMed] [Google Scholar]
- 4.da Silva Ferreira M. E., Kress M. R. V. Z., Savoldi M., Goldman M. H. S., Hartl A., Heinekamp T., Brakhage A. A., Goldman G. H. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey M. J., Indiani C., O'Donnell M. 2003. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 278:4491–4499 [DOI] [PubMed] [Google Scholar]
- 6.Denning D. W., Brookman J. L., Rickers A., Birch M. 8January2003. Mutant bank. European patent application EP1272610. [Google Scholar]
- 7.Denning D. W. 2001. Chronic forms of pulmonary aspergillosis. Clin. Microbiol. Infect. 7Suppl. 2:25–31 [DOI] [PubMed] [Google Scholar]
- 8.Denning D. W., O'Driscoll B. R., Hogaboam C. M., Bowyer P., Niven R. M. 2006. The link between fungi and severe asthma: a summary of the evidence. Eur. Respir. J. 27:615–626 [DOI] [PubMed] [Google Scholar]
- 9.Denning D. W., O'Driscoll B. R., Powell G., Chew F., Atherton G. T., Vyas A., Miles J., Morris J., Niven R. M. 2009. Randomized controlled trial of oral antifungal treatment for severe asthma with fungal sensitization: the Fungal Asthma Sensitization Trial (FAST) study. Am. J. Respir. Crit. Care Med. 179:11–18 [DOI] [PubMed] [Google Scholar]
- 10.de Queiroz M. V., Daboussi M. J. 2003. Impala, a transposon from Fusarium oxysporum, is active in the genome of Penicillium griseoroseum. FEMS Microbiol. Lett. 218:317–321 [DOI] [PubMed] [Google Scholar]
- 11.Dufresne M., van der Lee T., Ben M'Barek S., Xu X. D., Zhang X., Liu T. G., Waalwijk C., Zhang W., Kema G. H. J., Daboussi M. J. 2008. Transposon-tagging identifies novel pathogenicity genes in Fusarium graminearum. Fungal Genet. Biol. 45:1552–1561 [DOI] [PubMed] [Google Scholar]
- 12.Erjavec Z., Kluin-Nelemans H., Verweij P. E. 2009. Trends in invasive fungal infections, with emphasis on invasive aspergillosis. Clin. Microbiol. Infect. 15:625–633 [DOI] [PubMed] [Google Scholar]
- 13.Firon A., Beauvais A., Latge J. P., Couve E., Grosjean-Cournoyer M. C., d'Enfert C. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 161:1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firon A., Villalba F., Beffa R., d'Enfert C. 2003. Identification of essential genes in the human fungal pathogen Aspergillus fumigatus by transposon mutagenesis. Eukaryot. Cell 2:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Arkin A. P., Astromoff A., El Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Guldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kotter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C. Y., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y. H., Yen G., Youngman E., Yu K. X., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 16.Hey P., Robson G., Birch M., Bromley M. 2008. Characterisation of Aft1 a Fot1/Pogo type transposon of Aspergillus fumigatus. Fungal Genet. Biol. 45:117–126 [DOI] [PubMed] [Google Scholar]
- 17.Howard S. J., Cerar D., Anderson M. J., Albarrag A., Fisher M. C., Pasqualotto A. C., Laverdiere M., Arendrup M. C., Perlin D. S., Denning D. W. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W. Q., Sillaots S., Lemieux S., Davison J., Kauffman S., Breton A., Linteau A., Xin C. L., Bowman J., Becker J., Jiang B., Roemer T. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua-Van A., Pamphile J. A., Langin T., Daboussi M. J. 2001. Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264:724–731 [DOI] [PubMed] [Google Scholar]
- 20.Hurst L. D., Pal C., Lercher M. J. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5:299–310 [DOI] [PubMed] [Google Scholar]
- 21.Ikeda K., Nakayashiki H., Takagi M., Tosa Y., Mayama S. 2001. Heat shock, copper sulfate and oxidative stress activate the retrotransposon MAGGY resident in the plant pathogenic fungus Magnaporthe grisea. Mol. Genet. Genomics 266:318–325 [DOI] [PubMed] [Google Scholar]
- 22.Jöchl C., Rederstorff M., Hertel J., Stadler P. F., Hofacker I. L., Schrettl M., Haas H., Huttenhofer A. 2008. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 36:2677–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordens J., Janssens V., Longin S., Stevens I., Martens E., Bultynck G., Engelborghs Y., Lescrinier E., Waelkens E., Goris J., Van Hoof C. 2006. The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 281:6349–6357 [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita T., Inoue N. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632–638 [DOI] [PubMed] [Google Scholar]
- 25.Kops O., Zhou X. Z., Lu K. P. 2002. Pin1 modulates the dephosphorylation of the RNA polymerase IIC-terminal domain by yeast Fcp1. FEBS Lett. 513:305–311 [DOI] [PubMed] [Google Scholar]
- 26.Kretschmer P. J., Cohen S. N. 1979. Effect of temperature on translocation frequency of the Tn3 element. J. Bacteriol. 139:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane K. T., Beese L. S. 2006. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J. Lipid Res. 47:681–699 [DOI] [PubMed] [Google Scholar]
- 28.Lee W., Tillo D., Bray N., Morse R. H., Davis R. W., Hughes T. R., Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235–1244 [DOI] [PubMed] [Google Scholar]
- 29.Lei M. 2005. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr. Cancer Drug Targets 5:365–380 [DOI] [PubMed] [Google Scholar]
- 30.Lercher M. J., Urrutia A. O., Hurst L. D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31:180–183 [DOI] [PubMed] [Google Scholar]
- 31.Li H., Zhou H., Luo Y. M., Ouyang H. M., Hu H. Y., Jin C. 2007. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 64:1014–1027 [DOI] [PubMed] [Google Scholar]
- 32.Mabey J. E., Anderson M. J., Giles P. F., Miller C. J., Attwood T. K., Paton N. W., Bornberg-Bauer E., Robson G. D., Oliver S. G., Denning D. W. 2004. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 32:D401–D405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandala S. M., Frommer B. R., Thornton R. A., Kurtz M. B., Young N. M., Cabello M. A., Genilloud O., Liesch J. M., Smith J. L., Horn W. S. 1994. Inhibition of serine palmitoyl-transferase activity by lipoxamycin. J. Antibiot. 47:376–379 [DOI] [PubMed] [Google Scholar]
- 34.Matsukura M., Inoue S., Tanaka K., Murai N., Shirotori S. 20September2007. Novel heteroaryl ring substituted pyridine derivative useful in pharmaceuticals as antifungal agent for preventing Candida infection and Aspergillus infection. European patent application EP07828273 [Google Scholar]
- 35.Morillon S., Springer M., Lesage P. 2000. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5766–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naas T., Blot M., Fitch W. M., Arber W. 1995. Dynamics of is related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198–207 [DOI] [PubMed] [Google Scholar]
- 37.Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., Berriman M., Abe K., Archer D. B., Bermejo C., Bennett J., Bowyer P., Chen D., Collins M., Coulsen R., Davies R., Dyer P. S., Farman M., Fedorova N., Fedorova N., Feldblyum T. V., Fischer R., Fosker N., Fraser A., Garcia J. L., Garcia M. J., Goble A., Goldman G. H., Gomi K., Griffith-Jones S., Gwilliam R., Haas B., Haas H., Harris D., Horiuchi H., Huang J., Humphray S., Jimenez J., Keller N., Khouri H., Kitamoto K., Kobayashi T., Konzack S., Kulkarni R., Kumagai T., Lafton A., Latge J. P., Li W. X., Lord A., Majoros W. H., May G. S., Miller B. L., Mohamoud Y., Molina M., Monod M., Mouyna I., Mulligan S., Murphy L., O'Neil S., Paulsen I., Penalva M. A., Pertea M., Price C., Pritchard B. L., Quail M. A., Rabbinowitsch E., Rawlins N., Rajandream M. A., Reichard U., Renauld H., Robson G. D., de Cordoba S. R., Rodriguez-Pena J. M., Ronning C. M., Rutter S., Salzberg S. L., Sanchez M., Sanchez-Ferrero J. C., Saunders D., Seeger K., Squares R., Squares S., Takeuchi M., Tekaia F., Turner G., de Aldana C. R. V., Weidman J., White O., Woodward J., Yu J. H., Fraser C., Galagan J. E., Asai K., Machida M., Hall N., Barrell B., Denning D. W. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 38.Ninomiya Y., Suzuki K., Ishii C., Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogasawara H., Obata H., Hata Y., Takahashi S., Gomi K. 2009. Crawler, a novel Tc1/mariner-type transposable element in Aspergillus oryzae transposes under stress conditions. Fungal Genet. Biol. 46:441–449 [DOI] [PubMed] [Google Scholar]
- 40.Olszewska A., Krol K., Weglenski P., Dzikowska A. 2007. Arginine catabolism in Aspergillus nidulans is regulated by the rrmA gene coding for the RNA-binding protein. Fungal Genet. Biol. 44:1285–1297 [DOI] [PubMed] [Google Scholar]
- 41.Patterson T. F., Kirkpatrick W. R., White M., Hiemenz J. W., Wingard J. R., Dupont B., Rinaldi M. G., Stevens D. A., Graybill J. R. 2000. Invasive aspergillosis—disease spectrum, treatment practices, and outcomes. Medicine 79:250–260 [DOI] [PubMed] [Google Scholar]
- 42.Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeifer F., Blaseio U. 1990. Transposition burst of the Ish27 insertion element family in Halobacterium halobium. Nucleic Acids Res. 18:6921–6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roemer T., Jiang B., Davison J., Ketela T., Veillette K., Breton A., Tandia F., Linteau A., Sillaots S., Marta C., Martel N., Veronneau S., Lemieux S., Kauffman S., Becker J., Storms R., Boone C., Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50:167–181 [DOI] [PubMed] [Google Scholar]
- 46.Snelders E., van der Lee H. A. L., Kuijpers J., Rijs A. J. M. M., Varga J., Samson R. A., Mellado E., Donders A. R. T., Melchers W. J. G., Verweij P. E. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomich P. K., An F. Y., Clewell D. B. 1980. Properties of erythromycin-inducible transposon-Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torrado J. J., Espada R., Ballesteros M. P., Torrado-Santiago S. 2008. Amphotericin B formulations and drug targeting. J. Pharm. Sci. 97:2405–2425 [DOI] [PubMed] [Google Scholar]
- 49.van Opijnen T., Bodi K. L., Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vermes A., Guchelaar H. J., Dankert J. 2000. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46:171–179 [DOI] [PubMed] [Google Scholar]
- 51.Wagner C., Graninger W., Presterl E., Joukhadar C. 2006. The echinocandins: comparison of their pharmacokinetics, pharmacodynamics and clinical applications. Pharmacology 78:161–177 [DOI] [PubMed] [Google Scholar]
- 52.Zweerink M. M., Edison A. M., Wells G. B., Pinto W., Lester R. L. 1992. Characterization of a novel, potent, and specific inhibitor of serine palmitoyltransferase. J. Biol. Chem. 267:25032–25038 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.