Abstract
The occurrence of highly conserved amyloid-forming sequences in Candida albicans Als proteins (H. N. Otoo et al., Eukaryot. Cell 7:776–782, 2008) led us to search for similar sequences in other adhesins from C. albicans and Saccharomyces cerevisiae. The β-aggregation predictor TANGO found highly β-aggregation-prone sequences in almost all yeast adhesins. These sequences had an unusual amino acid composition: 77% of their residues were β-branched aliphatic amino acids Ile, Thr, and Val, which is more than 4-fold greater than their prevalence in the S. cerevisiae proteome. High β-aggregation potential peptides from S. cerevisiae Flo1p and C. albicans Eap1p rapidly formed insoluble amyloids, as determined by Congo red absorbance, thioflavin T fluorescence, and fiber morphology. As examples of the amyloid-forming ability of the native proteins, soluble glycosylphosphatidylinositol (GPI)-less fragments of C. albicans Als5p and S. cerevisiae Muc1p also formed amyloids within a few days under native conditions at nM concentrations. There was also evidence of amyloid formation in vivo: the surfaces of cells expressing wall-bound Als1p, Als5p, Muc1p, or Flo1p were birefringent and bound the fluorescent amyloid-reporting dye thioflavin T. Both of these properties increased upon aggregation of the cells. In addition, amyloid binding dyes strongly inhibited aggregation and flocculation. The results imply that amyloid formation is an intrinsic property of yeast cell adhesion proteins from many gene families and that amyloid formation is an important component of cellular aggregation mediated by these proteins.
Protein amyloids are characteristic of pathological conditions, including neurodegenerative diseases (4, 11, 17, 38). These protein aggregates can also occur naturally in adhesive bacterial curli (3), melanosomes (14), condensed peptide hormone arrays (24), as regulatory prions in yeast (2, 5), and fungal hydrophobins, which are nonantigenic coats to some fungi (1, 33, 39). Nevertheless, such natural occurrences are relatively few, considering the negative free energy for amyloid formation (28).
We have recently discovered that there are amyloid-forming sequences in the cell surface Als adhesins of Candida albicans. Cells that express these adhesins aggregate readily, and the aggregation has amyloid-like properties, including protein conformational shifting, surface birefringence, and ability to bind the amyloid-active dyes Congo red and amino-naphthalene sulfonic acid (ANS) (29). A five- to seven-residue sequence in Als1p, Als3p, and Als5p has extremely high potential for formation of β-aggregates, according to the protein state prediction program TANGO (13, 27, 31). Such β-aggregates include amyloids, which are ordered structures with paracrystalline regions of stacked parallel β-strands that are perpendicular to the long axis of micrometer-long fibrils. The strands are stabilized by interaction of identical sequences from many protein molecules (31, 32). Where TANGO analyses have shown that specific sequences have β-aggregate potentials greater than 20%, an insoluble β-aggregate state is likely to form. These β-aggregates nucleate formation of amyloids if the proteins can associate to form fibers (13, 27, 31). Sequences in the conserved 127-residue T region of Als1p, Als3p, and Als5p have β-aggregation potentials of >90% (27). An oligopeptide with this sequence, as well as 412- and 645-residue fragments of Als5p formed authentic amyloids, as determined by characteristic dye binding and fiber morphology. The amyloid-forming sequences were rich in the β-branched amino acids Thr, Val, and Ile. This amino acid composition is unusual among proteins in general, but is common in the Thr-rich mid-piece domains of yeast adhesins.
Yeasts display many cell-wall-bound adhesins that mediate colonial and biofilm interactions as well as host-pathogen binding (9, 21, 41). Such adhesins have a common mosaic structure. In general, the adhesins have N-terminal globular binding domains (often immunoglobulin-like or lectin-like), Thr-rich mid-piece sequences including tandem repeats, and 300- to 800-residue heavily glycosylated Ser and Thr-rich “stalk” domains near the C-terminal domain that extend the active regions from the surface of the wall. The adhesins are covalently cross-linked to wall polysaccharides through modified glycosylphosphatidylinositol (GPI) anchors and/or glycosyl esters of glutamic acid (9, 18).
Because the yeast adhesins share this common modular domain structure, we searched among known and putative yeast adhesins for sequences with high β-aggregation potential. We have found that many of these proteins share amyloid-forming sequences and amyloid-like behavior on activation.
MATERIALS AND METHODS
Throughout, protein and gene names are preceded by their species abbreviation: “Ca” for C. albicans and “Sc” for Saccharomyces cerevisiae.
Approximately 110 sequences of fungal and bacterial adhesins, other yeast cell wall proteins, and intracellular controls were screened in TANGO (http://tango.crg.es/) (13) with default settings for pH, ionic strength, and temperature. Test screenings at pH 5 and/or low ionic strength did not significantly alter the results. Because TANGO can only accommodate sequences of 500 residues or less, longer sequences were screened in segments of 500 residues with 50-residue overlaps. Control sequences included non-adhesin cell wall proteins, representative intracellular enzymes, and randomized sequences with the same amino acid composition as the test sequences. Regions with predicted β-aggregation occupancies of ≥30% were listed and analyzed.
Expression of CaAls5p1–1351.
We have not previously expressed a soluble version of CaAls5p that included the Ser/Thr-rich C-terminal stalk region. A version lacking the 68 C-terminal residues (with the GPI addition signal deleted) was produced by PCR using a forward primer 5′ACAACTACCAACTGCTAACACCAGATG3′ (the start codon is underlined) and reverse primer 5′TCGACCTTCAATAGCACTGTCTCCATTCA3′. The product was ligated into pYES2.1 TOPO-TA (Invitrogen), adding C-terminal V5 epitope and His6 tags. The insert was fully sequenced and found to have the predicted sequence. S. cerevisiae transformants were grown with galactose as the carbon source, and the secreted protein was purified by concentration and His-Trap chromatography in a procedure similar to that used for shorter versions (27). SDS gel electrophoresis of the purified protein showed that the V5 epitope was not reactive. Coomassie blue staining showed a positive band with an apparent size of >150 kDa, as expected for this highly glycosylated protein (data not shown). Precipitates spontaneously formed when the purified protein was stored at 4°C. These precipitates were collected and sonicated before being assayed for amyloid formation.
Expression of other proteins.
Soluble ScMuc1p1–1331 was purified from supernatants of cells expressing from plasmid pHis-PGK1-MUC1 in S. cerevisiae var. diastaticus strain YIY 345 (8). The secreted protein was dialyzed into phosphate-saline buffer, pH 7.4, and stored at 4°C. The Als1-expressing plasmid pADH-ALS1 was a gift from F. Yu, UCLA, and was expressed in W303-1B (36). ScFlo1p was expressed on the surface of strain BX24-2B, purchased from ATCC (Manassas, VA).
Peptides.
SNGIVIVATTRTV (CaAls1p positions 322 to 334 [CaAls1p322–334]; GenBank accession XM_712917.1; CaAls3p322–334, accession no. AAO72958.1; and CaAls5p322–334, accession no. O13368), HTAVTTGVTIITVTND (CaEap1p117–133, accession no. XP_71466.1), and TDETVIVIRTP (ScFlo1305–315 and other repeats, accession no. NP_009424), EVTTGVVVVTSEE (CaHwp1p380–392, accession no. EU477610.1; and CaRbt1p432–443, accession no. AF254142.1), and VTTVVSTTVVTT (ScMuc1p/Flo11p1031–1042, GenBank accession no. ABS87372.1) were synthesized by the Rockefeller University Proteomics Facility. The CaHwp1p and ScMuc1p peptides were insoluble in all tested solvents and were not purified or further studied. The purified CaAls, CaEap1p, and ScFlo1p peptides were suspended in hexafluoro-isopropanol, dried to a film, and then resuspended at 1.0 or 0.5 mg/ml in 10 mM Tris-EDTA buffer, pH 7.0, or phosphate-saline and stirred for periods up to several weeks at 4°C before being assayed for presence of amyloid (26).
Amyloid assays.
Congo red and thioflavin T binding assays for in vitro amyloids, as well as transmission electron microscopy (TEM) of negative-stained fibers were carried out as previously described (10, 27). Far-UV circular dichroism spectroscopy was carried out on a Chirascan spectrometer scanning from 180 to 260 nm. The amyloid-forming peptides and proteins were negative stained with uranyl acetate and examined under transmission electron microscopy (27). Birefringence and fluorescence microscopy of cellular aggregates were performed as previously described (29). Cells and cellular aggregates were treated with thioflavin T at 30 μM in Tris-EDTA buffer (10 mM each, pH 7.0), washed twice in the same buffer, and observed at 480 to 540 nm, with excitation at 425 to 440 nm.
Aggregation assays.
Aggregation assays for Als adhesins were carried out as previously described (15). Briefly, 108 S. cerevisiae cells expressing CaAls1p or CaAls5p were mixed with 106 magnetic beads covalently derivatized with heat-denatured bovine serum albumin (BSA) in 0.1 M sodium acetate buffer, pH 5.5 (15). The suspension was agitated gently for 45 min before microscopic observation.
Assays for flocculation mediated by ScFlo1p or ScMuc1p/Flo11p were carried out as described by Lo and Dranginis, using 3 × 107 cells/ml prewashed with EDTA to inhibit flocculation before assay. Flocculation was initiated by addition of 0.67 mM CaCl2; unless otherwise stated, the suspension was vortexed for 5 s, and the optical density at 600 nm (OD600) was monitored at 5-s intervals in a Spectronic 20 D+ spectophotometer.
RESULTS
Adhesin sequences.
We screened 70 extracellular proteins from fungi and bacteria for sequences with high β-aggregation potential in TANGO (13). Adhesins from seven tested C. albicans and S. cerevisiae adhesin gene families contained one, two, or three internal sequences that TANGO predicted to have very high frequency of β-aggregate states (Table 1). The frequency of adhesins with predicted amyloid-forming sequences increased to 19 of 20 when all paralogous loci were included, although not all paralogs are listed in Table 1 (27). Thus, the results in Table 1 represent several unrelated adhesin gene families: CaHWP/RBT, CaEAP1, and CaEPE1, as well as ScFLO1, ScMUC1 (alternately designated ScFLO11), and ScAGA1/FIG2, as well as CaALS. The exceptional adhesin was ScSag1p. In several cases, the sequence and position of the β-aggregation-prone sequences were conserved among paralogs (CaALS, CaHWP/RBT, and ScFLO1 gene families) (Table 1) (27).
Table 1.
β-Aggregation-prone sequences in yeast adhesins
| Proteina | β-Aggregation sequenceb | % β-Aggregation | Ile, Val, and Thr content (%) |
|---|---|---|---|
| C. albicans | |||
| Als1c | IVIVA | 90 | 80 |
| Als5c | IVIVA | 93 | 80 |
| Eap1 | AYTTTVITV | 70 | 78 |
| VTTGVTIITVT | 90 | 91 | |
| TVITV | 36 | 100 | |
| Ece1 | IIGIIMGIL | 65 | 56 |
| VIQIIMSIV | 66 | 60 | |
| Hwp1 | VTTGVIVIT | 82 | 89 |
| TGVVVVT | 98 | 86 | |
| Hwp2 | AIVVT | 42 | 80 |
| Rbt1 | GVVVV | 58 | 80 |
| VTTGVVVVT | 75 | 89 | |
| Sap3 | LTVVI | 50 | 80 |
| S. cerevisiae | |||
| Aga1 | TILVTIT | 86 | 88 |
| ILLF | 39 | 25 | |
| Fig2 | TWVVI | 68 | 80 |
| LVLSTVT | 38 | 57 | |
| Flo1d | TVIVI | 42 | 100 |
| TVIVI | 43 | 100 | |
| TLVTVT | 34 | 100 | |
| Muc1 | VVSTTV | 75 | 83 |
| VTTAVTTTVV | 52 | 90 | |
| Sag1 | None |
Accession numbers: CaAls1, XM_712917.1; CaAls5p, O13368; CaEap1, XP_71466.1; CaEce1p, DQ465883.1; CaHwp1, EU477610.1; CaHwp2, XP_711600; CaRbt1, AF254142.1; CaSap3, L22358.1; ScAga1, P32323; ScFig2, P25653; ScFlo1, NP_009424; ScMuc1/Flo11, ABS87372; ScSag1p, NP_012537.
Sequence of amino acids with β-aggregation potential of >30%, as predicted by TANGO.
Sequences are also present in other CaAls proteins (27).
Similar sequences are also present in ScLG-Flo, ScFlo5p, and ScFlo9p (9).
Conserved, highly β-aggregation-prone sequences were also present in Als homologs from Candida dubliniensis and C. tropicalis, as well as in some orthologs from Debaryomyces hanseni. There was also a similar β-aggregation-prone sequence in a predicted GPI-anchored protein from Aspergillus niger (data not shown). Several bacterial adhesins also had β-aggregation-prone sequences similarly rich in these aliphatic β-branched residues: Borrelia burgdorferi OspC, Streptococcus gordonii CshA, and Streptococcus mutans AtlA (data not shown). CaSap3, a GPI-anchored protease with a similar sequence, is also included in Table 1.
These adhesin β-aggregation-prone sequences had an unusual composition: they were highly enriched for the β-branched aliphatic amino acids Ile, Thr, and Val (Table 1). The widespread occurrence of these potentially amyloid-forming sequences, their unusual composition, and their conservation across paralogs led us to test whether they could in fact form amyloids.
Soluble Als5p forms amyloids.
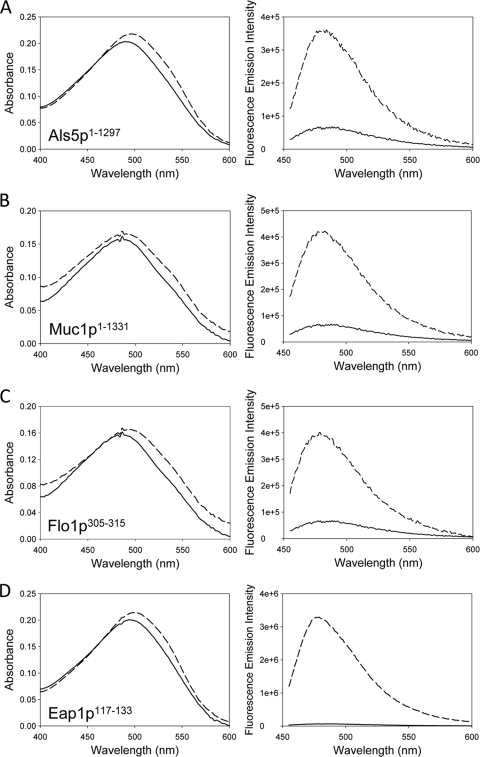
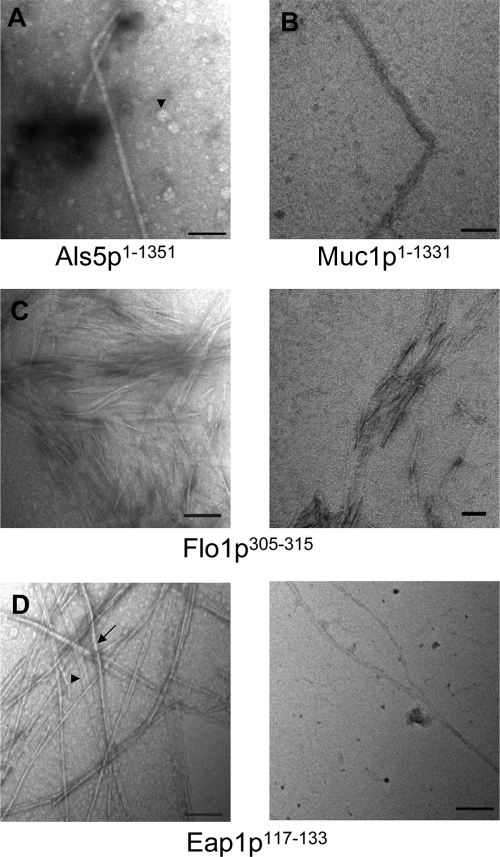
We have demonstrated that 13-, 414-, and 647-residue fragments of CaAls5p adhesins form amyloid fibers under physiological conditions (27). We repeated these experiments with a soluble version derived from construct expressing CaAls5p1–1351, which lacks only the nucleotides encoding the C-terminal 68 amino acid residues. Like the smaller fragments, the purified protein rapidly precipitated from neutral buffer at submicromolar concentrations. Like other amyloids, the precipitate enhanced and red-shifted the absorbance spectrum of Congo red and enhanced the fluorescence of thioflavin (Fig. 1A) (10, 26, 27). Negative-stain transmission electron microscopy showed uniform fibers 14 nm in diameter (Fig. 2A). The fibers were smooth, with uniform diameter, and appeared to be composed of smaller fibrils of a few nm in diameter. There were also amorphous and discoidal aggregates throughout the field (arrowhead). These are similar to structures seen with shorter fragments of CaAls5p (27).
Fig. 1.
Congo red absorbance (left column) and thioflavin T fluorescence (right column) of suspended adhesion proteins and peptides. Control spectra are solid lines, and spectra taken in the presence of aggregates are dashed. Congo red spectra show aggregate-dependent enhancement and red-shifting. Thioflavin T fluorescence in increased 2- to 30-fold in the presence of the aggregates: CaAls5p1–1351 (A), ScMuc1p1–1331 (B), ScFlo1p305–315 (C), and Eap1p117–133 (D).
Fig. 2.
Negative-stain transmission electron microscopy of fibers. Bars are 100 nm in length. (A) CaAls5p1–1351. The arrowhead shows a less-structured aggregate, and apparent protofibrils are seen in the lower region of the long fiber. (B) ScMuc1p1–1331. Individual fibrils are visible in the upper part of the fiber. (C) ScFlo1p305–315. Note many smaller wavy fibrils in the background. (D) Eap1p117–133. There are both fibers (arrow) and ribbons (arrowhead) visible.
ScMuc1p1–1331 forms amyloids.
Soluble ScMuc1p was collected from supernatants of cells expressing the protein from plasmid pHis-PGK1-MUC1 (8). The secreted protein was dialyzed into phosphate-saline buffer, pH 7.4, and stored at 4°C. Within a few days, precipitates formed as previously observed (8). These protein suspensions increased fluorescence of thioflavin T and increased absorbance and red-shifted Congo red solutions (Fig. 1B). Electron microscopy showed short fibers of 5 nm in diameter and a 15-nm diameter fiber with the appearance of a braided rope (Fig. 2B).
Synthetic peptides of the high β-aggregate potential sequence form amyloids.
Peptides were synthesized corresponding to the high β-aggregate potential sequences of C. albicans Eap1p and S. cerevisiae ScFlo1p, each sequence flanked with non-amyloid-forming natural sequence residues at each end (see Materials and Methods). The peptides were suspended in neutral buffer and assayed for amyloid formation.
The peptide from ScFlo1p also formed amyloids. Stirred suspensions showed circular dichroism (CD) spectra characteristic of β-aggregation. Congo red absorbance was slightly increased and red-shifted (Fig. 1C). In all trials, the absorbance increased and shifted with stirring and incubation at 4°C. Thioflavin T fluorescence increased over 48 h of stirring and showed 2-fold enhancement after several months. The fiber morphology was ribbon-like, with typical fibers of 2.7 nm in diameter clearly braided into larger ropes and aggregates (Fig. 2C). A dense mat of these small “proto-fibrils” is clearly visible in the right-hand micrograph.
CD spectra of the CaEap1p peptide showed no secondary structure in fresh suspensions, but developed minima characteristic of β-sheets (215 nm) and β-aggregation (235 nm) within 48 h and persisted over 2 months of incubation (data not shown). Within 48 h of suspending the peptide, Congo red absorbance spectra showed increased absorbance and red-shifting characteristic of amyloid formation (Fig. 1D). Similarly, thioflavin T fluorescence emission increased three- to 4-fold, also characteristic of amyloids (Fig. 1). Electron microscopy showed 3.5- to 7-nm-diameter fibers in braided structures characteristic of amyloids (Fig. 2D) (42).
Cell adhesion amyloids in vivo.
To obtain evidence as to whether or not amyloid formation is present in vivo, we looked for amyloids in intact cells. S. cerevisiae cells expressing CaAls5p are markedly birefringent in polarization microscopy during aggregation (i.e., they show light and dark regions when examined between crossed Polaroid filters), whereas nonexpressing cells or nonaggregated cells are not as birefringent (29). Similarly, aggregated C. albicans cells are birefringent under conditions that maximize expression of CaAls1p, but show less birefringence when unaggregated (29) or when CaAls protein expression is minimal (unpublished data).
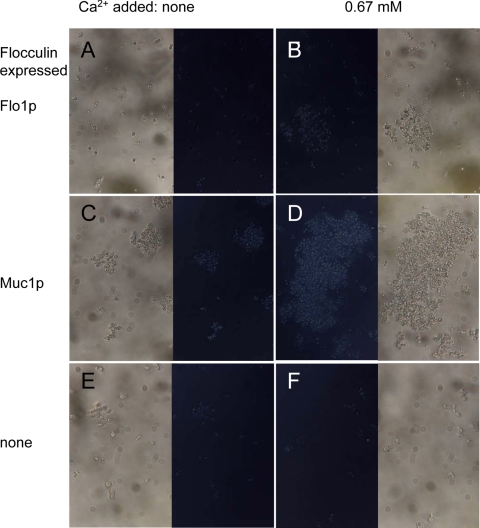
When we examined cells expressing ScMuc1p or ScFlo1p flocculins, we also saw birefringence, a characteristic of ordered structures like amyloids (Fig. 3A and C). Cells expressing ScFlo1p were slightly more birefringent than W303-1B (Fig. 3A and E) and became more birefringent upon initiation of flocculation by addition of Ca2+ (Fig. 3B and F). For cells expressing ScMuc1p, the birefringence was minimal in the absence of Ca2+ and increased when flocculation was induced with Ca2+ (Fig. 3C and D). Thus, like the CaAls adhesins, the ScFlo1p and ScMuc1p flocculins showed increased birefringence upon cell aggregation.
Fig. 3.
Birefringence of cells expressing flocculins. S. cerevisiae cells were analyzed between polarizing filters with a 20× objective under bright-field conditions. The paired micrographs show identical fields between parallel (outer images) and crossed polarizing filters (central images) in the absence (left) or presence (right) of 0.67 mM Ca2+. The strains were BX24-2B (A and B), YIY345/pHis-PGK1-MUC1 (C and D), and W303-1B (E and F).
Amyloids bind thioflavin T and greatly enhance its fluorescence, but the dye does not inhibit amyloid formation at low concentrations (Fig. 1) (10). Therefore, we stained intact yeast cells with thioflavin T and inspected them by fluorescence microscopy. Figure 4 shows that few cells of S. cerevisiae strain W303-1B bound to the beads (panel A). When CaAls5p or CaAls1p was expressed in these cells, they aggregated well in the presence of beads, but with no increase in background fluorescence (Fig. 4B and C). When the aggregated cells were stained with thioflavin T, the aggregated cells fluoresced brightly (Fig. 4D to F). Cells concentrated by centrifugation were less bright than aggregated cells (Fig. 4G to I). Under these conditions, thioflavin T did not inhibit CaAls-mediated binding to BSA-coated beads or cell-to-cell aggregation (Fig. 4, compare panels B versus E and C versus F). Thus, thioflavin T specifically stained cells expressing CaAls proteins, and fluorescence was brighter in aggregated cells.
Fig. 4.
Thioflavin T (ThT) staining of cells expressing CaAls proteins. Shown are paired bright-field and fluorescence micrographs of S. cerevisiae W303-1B transformed with the empty vector (EV; no insert) or vectors encoding CaAls5p or CaAls1p. (A to C) Designated cells were aggregated with BSA-coated beads. Bright-field micrographs in the top row show dark spherical 2.8-μm beads interspersed with gray-colored yeast cells, which are spheroidal and larger. The bottom row shows fluorescence of the same field. (D to F) The indicated cells were aggregated with beads and then stained with thioflavin T. Bright-field micrographs are in the top row, and fluorescence of the corresponding field is shown below. (G to I) The indicated cells were concentrated by centrifugation and stained with thioflavin T. The fluorescence micrographs are on top, with the corresponding bright-field images shown below.
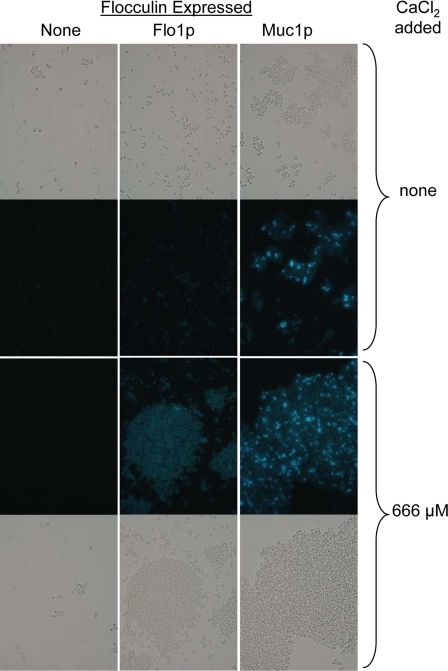
There were similar results for cells expressing the flocculins ScFlo1p or ScMuc1p (Fig. 5). Thioflavin T stained the cells, and the staining was brighter after Ca+2-induced flocculation than in nonflocculating samples. Therefore, thioflavin T fluorescence was seen in S. cerevisiae cells expressing any of the tested amyloid-forming proteins, and the fluorescence was greater in the aggregates than in nonaggregated cells.
Fig. 5.
Thioflavin T staining of S. cerevisiae cells expressing ScFlo1p or ScMuc1p. Indicated strains were stained and visualized under bright-field (top and bottom rows), with matched fields for thioflavin T fluorescence in the middle two rows. Flocculation was induced with added Ca2+ for the images in the bottom two rows: left column, W303-1B; middle column, strain BX24-2B; right column, strain YIY345.
Effects of amyloid-binding dyes on cellular aggregation.
The ability of yeast adhesins to form amyloids raised the question of whether amyloid formation has functional consequences. In the case of CaAls5p, the amyloid-binding dyes Congo red or ANS decreased yeast cell aggregation (29). Therefore, we determined whether such dyes would have similar effects on aggregation mediated by the highly expressed adhesin CaAls1p. In aggregation assays with magnetic beads coated with heat-denatured BSA, the dyes Congo red (1 mM), thioflavin S (1.5 mM), and thioflavin T (1.5 mM) attenuated cell-to-cell aggregation of cells expressing CaAls5p or CaAls1p (Fig. 6) (29). At these dye concentrations, cells expressing CaAls1p retained their ability to bind to the beads (Fig. 6F, I, and L). At higher concentrations, all binding was abolished (not shown). For the cells expressing CaAls5p, Congo red and thioflavin S abolished all binding, but cell-to-bead binding persisted in 1.5 mM thioflavin T (Fig. 6I). Therefore, amyloid-binding dyes attenuated aggregation caused by CaAls1p as well as CaAls5p, and the effective concentrations were in the low-mM range.
Fig. 6.
Aggregation assays with S. cerevisiae cells expressing CaAls proteins. Strain W303-1B cells carrying an empty vector or expressing the designated protein were aggregated with heat-denatured BSA-coated magnetic beads, and the beads and adherent cells were separated and examined by light microscopy (×40 magnification). Dark spherical 2.8-μm beads are interspersed with the gray-colored cells, which are spheroidal and slightly larger. Assays were carried out in the presence of amyloid-binding dyes as indicated: CR, Congo red; ThT, thioflavin T; and ThS, thioflavin S.
We also tested the effect of amyloid-binding dyes on flocculation of S. cerevisiae cells expressing the flocculins ScFlo1p or ScMuc1p. Such cells flocculate, or form large aggregates, in the presence of Ca2+ ions (9, 23). Congo red, which binds to and disrupts amyloids (12), inhibited flocculation caused by either flocculin at concentrations as low as 30 μM, with half-maximal inhibition at 0.5 mM (Table 2). Thioflavin S was similarly potent, and completely inhibited the flocculation reaction for both proteins (Table 2 and Fig. 7A to D). The dyes reduced both the rate at which the cells flocculated (the initial slope) and the amount of flocculation (final decrease in OD) (Table 2 and Fig. 7A). The half-maximal inhibitory concentrations of thioflavin S were 45 μM for ScMuc1p-mediated flocculation and 100–200 μM for the ScFlo1-mediated reaction (Fig. 7B and Table 2). Congo red showed half-maximal inhibition at about 500 μM. ANS had little effect on flocculation, and high concentrations of thioflavin T mediated a more rapid and extensive aggregation (Table 2). These dyes do not inhibit amyloid formation in many cases (12).
Table 2.
Effect of amyloid-binding dyes on flocculation
| Dye and flocculin expressed | Rate of flocculation (% of control) | Extent of flocculation (% of control) |
|---|---|---|
| ANS (1.0 mM) | ||
| Flo1p | 109 | 107 |
| Muc1p | 205 | 122 |
| Congo red (0.50 mM) | ||
| Flo1p | 49 | 59 |
| Muc1p | 60 | 60 |
| Thioflavin S (0.19 mM) | ||
| Flo1p | 49 | 45 |
| Muc1p | 0 | 6 |
| Thioflavin T (5 mM) | ||
| Flo1p | 384 | 239 |
| Muc1p | 227 | 191 |
Fig. 7.
Effects of thioflavin S on flocculent strains of S. cerevisiae. (A) Flocculation assays in the presence of increasing concentrations of thioflavin S. (A) Strain BX24-2B expressing ScFlo1p flocculating in the presence of CaCl2 (667 μM); (B) strain YIY345 expressing ScMuc1p flocculating in the presence of CaCl2 (667 μM); (C) dose-response analysis of effect of thioflavin S on rates of ScMuc1p-mediated flocculation; (D) dose-response analysis of effect of thioflavin S on rates of ScFlo1p-mediated flocculation; (E) growth inhibition assay. Serial dilutions of the indicated strains were grown on the indicated media.
Growth of yeast in the presence of Congo red results in inhibition of wall assembly, because the dye interferes with formation of polysaccharide fibrils (20, 30). It was unlikely that this effect was inhibiting flocculation, because the effective inhibitory concentrations were low and the dyes were present only during the flocculation assay itself, and not during wall biogenesis. Nevertheless, we tested whether the inhibitory dyes inhibited growth of S. cerevisiae. Congo red (30 μM) and thioflavin T (5 mM) inhibited growth of the flocculating strains in cell dilution growth assays (data not shown). In contrast, 190 μM thioflavin S was not growth inhibitory (Fig. 7E). Therefore, thioflavin S did not affect growth, but had potent antiaggregation effects for interactions mediated by CaAls1p, CaAls5p, ScFlo1p, and ScMuc1p. In general, there was no correlation between their growth inhibition and their effects in aggregation assays: some dyes were cytotoxic but did not inhibit aggregation, and others inhibited aggregation but were not cytotoxic.
DISCUSSION
Amyloid sequences in yeast adhesins.
We conclude that many families of adhesins of ascomycetous yeasts have sequences that can and do form amyloids under physiological conditions involving concentration and pH (13, 27). Supporting evidence showed that several of these adhesins formed amyloids in vivo and that amyloid formation was an integral part of cellular aggregation reactions.
Peptide or protein sequences from four different adhesin families formed insoluble amyloids at low concentrations and neutral pH (Fig. 1 and 2). These results validated the TANGO predictions for these sequences. For each, amyloid formation was rapid and voluminous: the large proteins precipitated rapidly, making spectroscopy of purified proteins difficult within a few days of isolation (data not shown). These adhesin sequences thus appeared to have a uniform ability to rapidly form amyloid when soluble, at neutral pH or native acidic pH, and at low (nM to μM) concentrations. These concentrations are lower than those typically found on cell surfaces (9, 35).
The exceptional adhesin without a high β-aggregation potential sequence was S. cerevisiae mating adhesin α-agglutinin, Sag1p. Nevertheless, mating requires the Sag1 ligand a-agglutinin, including the anchorage subunit ScAga1p, which has two strong β-aggregation potential sequences and spontaneously aggregates when purified (35). Therefore, even though ScSag1p does not have a strong β-aggregation sequence, mating requires a protein that has these sequences (9, 16).
Amyloid sequences in nonadhesin cell wall proteins.
TANGO also identified high β-aggregation potential sequences in nonadhesin surface proteins. These included some proteases: Yapsins from S. cerevisiae and Saps from C. albicans (Table 1) (data not shown). Other yeast cell wall proteins also had potential amyloid sequences, including the alkaline phosphatases ScPho8p, ScPho10p, and ScPho13p, the invertase ScSuc2p, and the transglycosylase ScGas1p, although as a class, the amino acid compositions were not enriched for the β-branched aliphatic amino acids. Remarkably, there were no predicted amyloid-forming sequences in structural cell wall proteins, including ScCwp1p, ScCwp2p, and ScPir1p, ScTir1p, ScDan1p, or ScDan4p. Thus, the potential amyloid-forming sequences were found primarily in proteins with adhesin or enzyme activity, and the unusual composition was present mostly in the adhesins.
Amino acid composition.
The adhesins have a high frequency of the β-aggregation-prone sequences, and the amino acid composition of these sequences was highly biased in a way uncommon for amyloid-forming sequences in general (13, 22, 32). The amino acids Ile, Thr, and Val constituted 77% of the adhesin TANGO high β-aggregation regions (Table 1). In contrast, these three residues constitute 18% of the S. cerevisiae proteome and are enriched to 31% in wall proteins (7). These β-branched aliphatic amino acids were much less frequent in high β-aggregation-potential sequences from intracellular proteins and random sequences. High TANGO β-aggregation potential sequences from yeast cell surface proteins that are not adhesins had 36% Ile, Thr, and Val residues, and in 62 sequences with high β-aggregation potential from intracellular proteins, these residues were only 26%. Thus, the enrichment of Ile, Thr, and Val was a unique property of the yeast adhesin sequences and may contribute to their unusual ability to rapidly form amyloids under physiological conditions.
Ile, Thr, and Val residues have aliphatic β-branched side chains that greatly restrict backbone conformation and have high β-strand potential (6). These residues are very hydrophobic, bulky, and have side-chain interactions that stabilize the β-sheets in amyloids. These properties are what we might expect in sequences whose primary purpose is to form amyloids. In contrast, the adhesin sequences had very few aromatic residues, which are the major category of β-aggregation- and amyloid-prone sequences in other proteins. Thus, the β-aggregation-prone sequences in the adhesins are also biased against aromatic residues. We suggest that the unusual composition of the adhesin amyloid sequences leads to the unusually facile amyloid formation that these peptides and proteins display.
These sequences are strongly conserved in the CaAls, ScFlo, and CaHwp/Rbt gene families (Table 1) (27). Such sequence conservation among paralogs is unusual in evolution, because paralogs generally diversify in function and therefore diverge faster than orthologs (25). Therefore, the result supports our previous observation of positive selection for amyloid sequences in the CaALS gene family (27).
A role for amyloid formation in cell adhesion.
Our results strongly support a functional role for amyloid formation in yeast cell adhesion. We have demonstrated that diverse yeast adhesins can form amyloids under native conditions of pH and at concentrations that are lower than those found in vivo. Yeast cells themselves showed surface birefringence and binding of thioflavin T (39), both characteristics of amyloids, and in at least the cases of CaAls- and ScMuc1-mediated aggregation, these characteristics increased in aggregates relative to nonaggregated cells.
The aggregation reactions were inhibited or potentiated by dyes that bind to amyloids. Notably, thioflavin S inhibited each aggregation reaction at μM concentrations and was not toxic or growth inhibitory to the cells. In the flocculation reactions, all the inhibitory compounds were active at concentrations that were well below those reported for most haptenic oligosaccharides, which bind to the lectin-like domains in ScFlo1p and ScMuc1p (9, 19, 34). Moreover, thioflavin T, which is used to monitor amyloid formation because it often does not inhibit amyloid formation (10, 12), actually potentiated flocculation. Therefore, for each type of adhesion assayed here, amyloid-binding compounds affected the aggregation reactions at low concentrations.
Some sequelae of amyloid formation are predictable from thermodynamic considerations. Formation of a multimeric aggregate of adhesins at the cell surface will increase the avidity of the adhesins by increasing local adhesin concentration (Fig. 8). Such “bundling” increases the probability that a ligand that dissociates from one adhesin molecule will rapidly bind to another adhesion molecule in the same cluster. The measured result is a marked decrease in the macroscopic dissociation rate, koff, and a correspondingly smaller dissociation constant, KD (9, 35). A commonly cited example of this phenomenon is the distinction between antibody affinity (the measured dissociation constant KD for a monomeric Fab), and its avidity (the measured KD for the intact multimeric IgG or IgM molecule). Therefore, amyloid formation like that illustrated in Fig. 8 can greatly increase the intercellular binding strength by increasing avidity. The apparent increase in the amyloid state on aggregation (Fig. 3 to 5) and the inhibition of aggregation by amyloid inhibitory dyes (Fig. 6 and 7) imply that amyloids form between adhesion molecules on contacting cells (Fig. 3 to 7) (29). Such intercellular amyloids would be covalently anchored to the walls of apposed cells, and so would strengthen intercellular adhesive bonds.
Fig. 8.
Model for forming multimers of flocculins through amyloid formation. Each monomeric flocculin is covalently anchored into the cell wall. The monomers are clustered on the cell surface through the interactions of amyloid-forming sequences.
Conclusions.
We have demonstrated that many adhesins from budding yeasts contain amyloid-forming sequences that have unusual composition and are conserved in paralogous gene families. The sequences form amyloids under native conditions at low concentrations. In the case of several adhesins, these amyloids are functional: amyloid inhibitors attenuate CaAls-, ScFlo1p-, and ScMuc1p-mediated cellular aggregation (29). (Note that the ScFlo1 amyloid sequence appears five times in the referenced sequence [NP_009424], including once in each 90-residue repeat. Adhesive activity increases with the number of these repeats, so there is a correlation between the number of amyloid-forming sequences and the adhesive strength of the intercellular bonds [37, 40].) Thus, amyloid formation may be more widespread than previously thought as a mechanism for cell-to-cell interactions, as well as their well-characterized role in Gram-negative bacteria (3).
ACKNOWLEDGMENTS
We thank Kyeng “Joe” Lee (Department of Biology and Medical Laboratory Technology, Bronx Community College) and Roland Hosein (Department of Biology, Brooklyn College) for valuable assistance with the preparation and visualization of negatively stained electron microscopy samples. We also thank Lesley Davenport (Department of Chemistry, Brooklyn College) for use of the Spex Fluorolog fluorimeter and diode array spectrometer; Francise Lamothe, Kiara Gomez, and Jordan Klein for assistance with flocculation experiments; and Atina Silkovic for help with aggregation experiments.
This work was supported by the NIGMS-SCORE Program under grants S06 GM 70758 and SC1 GM 83756 to Brooklyn College.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S. R., Clavaud C., Paris S., Brakhage A. A., Kaveri S. V., Romani L., Latge J. P. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 2.Alberti S., Halfmann R., King O., Kapila A., Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart M. M., Chapman M. R. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barten D. M., Albright C. F. 2008. Therapeutic strategies for Alzheimer's disease. Mol. Neurobiol. 37:171–186 [DOI] [PubMed] [Google Scholar]
- 5.Baxa U., Cassese T., Kajava A. V., Steven A. C. 2006. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv. Protein Chem. 73:125–180 [DOI] [PubMed] [Google Scholar]
- 6.Chou P. Y., Fasman G. D. 1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 47:45–148 [DOI] [PubMed] [Google Scholar]
- 7.Coronado J. E., Attie O., Epstein S. L., Qiu W. G., Lipke P. N. 2006. Composition-modified matrices improve identification of homologs of Saccharomyces cerevisiae low-complexity glycoproteins. Eukaryot. Cell 5:628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas L. M., Li L., Yang Y., Dranginis A. M. 2007. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 6:2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dranginis A. M., Rauceo J. R., Coronado J. E., Lipke P. N. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71:282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisert R., Felau L., Brown L. R. 2006. Methods for enhancing the accuracy and reproducibility of Congo red and thioflavin T assays. Anal. Biochem. 353:144–146 [DOI] [PubMed] [Google Scholar]
- 11.Elgersma R. C., Mulder G. E., Kruijtzer J. A., Posthuma G., Rijkers D. T., Liskamp R. M. 2007. Transformation of the amyloidogenic peptide amylin(20-29) into its corresponding peptoid and retropeptoid: access to both an amyloid inhibitor and template for self-assembled supramolecular tapes. Bioorg. Med. Chem. Lett. 17:1837–1842 [DOI] [PubMed] [Google Scholar]
- 12.Feng B. Y., Toyama B. H., Wille H., Colby D. W., Collins S. R., May B. C., Prusiner S. B., Weissman J., Shoichet B. K. 2008. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 4:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22:1302–1306 [DOI] [PubMed] [Google Scholar]
- 14.Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. 2006. Functional amyloid formation within mammalian tissue. PLoS Biol. 4:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaur N. K., Klotz S. A. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65:5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo B., Styles C. A., Feng Q., Fink G. R. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C. J., Lin C. Y., Haataja L., Gurlo T., Butler A. E., Rizza R. A., Butler P. C. 2007. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress-mediated beta cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes 58:2016–2027 [DOI] [PubMed] [Google Scholar]
- 18.Klis F. M., Boorsma A., De Groot P. W. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202 [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi O., Hayashi N., Kuroki R., Sone H. 1998. Region of Flo1 proteins responsible for sugar recognition. J. Bacteriol. 1998:6503–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecka M., Gabriel M. 1992. The influence of congo red on the cell wall and (1-3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 158:115–126 [DOI] [PubMed] [Google Scholar]
- 21.Linder T., Gustafsson C. M. 2008. Molecular phylogenetics of ascomycotal adhesins—a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 45:485–497 [DOI] [PubMed] [Google Scholar]
- 22.Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. 2004. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 342:345–353 [DOI] [PubMed] [Google Scholar]
- 23.Lo W. S., Dranginis A. M. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178:7144–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. 2009. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nei M., Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 26.Nilsson M. R. 2004. Techniques to study amyloid fibril formation in vitro. Methods 34:151–160 [DOI] [PubMed] [Google Scholar]
- 27.Otoo H. N., Lee K. G., Qiu W., Lipke P. N. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Kahng B., Hwang W. 2009. Thermodynamic selection of steric zipper patterns in the amyloid cross-beta spine. PLoS Comput. Biol. 5:e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauceo J. M., Gaur N. K., Lee K. G., Edwards J. E., Klotz S. A., Lipke P. N. 2004. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect. Immun. 72:4948–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roncero C., Duran A. 1985. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rousseau F., Schymkowitz J., Serrano L. 2006. Protein aggregation and amyloidosis: confusion of the kinds? Curr. Opin. Struct. Biol. 16:118–126 [DOI] [PubMed] [Google Scholar]
- 32.Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., Madsen A. O., Riekel C., Eisenberg D. 2007. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447:453–457 [DOI] [PubMed] [Google Scholar]
- 33.Scholtmeijer K., de Vocht M. L., Rink R., Robillard G. T., Wosten H. A. 2009. Assembly of the fungal SC3 hydrophobin into functional amyloid fibrils depends on its concentration and is promoted by cell wall polysaccharides. J. Biol. Chem. 284:26309–26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shankar C. S., Umesh-Kumar S. 1994. A surface lectin associated with flocculation in brewing strains of Saccharomyces cerevisiae. Microbiology 140:1097–1101 [DOI] [PubMed] [Google Scholar]
- 35.Shen Z. M., Wang L., Pike J., Jue C. K., Zhao H., de Nobel H., Kurjan J., Lipke P. N. 2001. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J. Biol. Chem. 276:15768–15775 [DOI] [PubMed] [Google Scholar]
- 36.Sheppard D. C., Yeaman M. R., Welch W. H., Phan Q. T., Fu Y., Ibrahim A. S., Filler S. G., Zhang M., Waring A. J., Edwards E. J., Jr 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480–30489 [DOI] [PubMed] [Google Scholar]
- 37.Smukalla S., Caldara M., Pochet N., Beauvais A., Guadagnini S., Yan C., Vinces M. D., Jansen A., Prevost M. C., Latge J. P., Fink G. R., Foster K. R., Verstrepen K. J. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorrentino G., Bonavita V. 2007. Neurodegeneration and Alzheimer's disease: the lesson from tauopathies. Neurol. Sci. 28:63–71 [DOI] [PubMed] [Google Scholar]
- 39.Teertstra W. R., van der Velden G. J., de Jong J. F., Kruijtzer J. A., Liskamp R. M., Kroon-Batenburg L. M., Muller W. H., Gebbink M. F., Wosten H. A. 2009. The filament-specific Rep1-1 repellent of the phytopathogen Ustilago maydis forms functional surface-active amyloid-like fibrils. J. Biol. Chem. 284:9153–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verstrepen K. J., Jansen A., Lewitter F., Fink G. R. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verstrepen K. J., Klis F. M. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5–15 [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Smith D. R., Jones J. W., Chapman M. R. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 282:3713–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]