Abstract
Budding yeast (Saccharomyces cerevisiae) responds to iron deprivation both by Aft1-Aft2-dependent transcriptional activation of genes involved in cellular iron uptake and by Cth1-Cth2-specific degradation of certain mRNAs coding for iron-dependent biosynthetic components. Here, we provide evidence for a novel principle of iron-responsive gene expression. This regulatory mechanism is based on the modulation of transcription through the iron-dependent variation of levels of regulatory metabolites. As an example, the LEU1 gene of branched-chain amino acid biosynthesis is downregulated under iron-limiting conditions through depletion of the metabolic intermediate α-isopropylmalate, which functions as a key transcriptional coactivator of the Leu3 transcription factor. Synthesis of α-isopropylmalate involves the iron-sulfur protein Ilv3, which is inactivated under iron deficiency. As another example, decreased mRNA levels of the cytochrome c-encoding CYC1 gene under iron-limiting conditions involve heme-dependent transcriptional regulation via the Hap1 transcription factor. Synthesis of the iron-containing heme is directly correlated with iron availability. Thus, the iron-responsive expression of genes that are downregulated under iron-limiting conditions is conferred by two independent regulatory mechanisms: transcriptional regulation through iron-responsive metabolites and posttranscriptional mRNA degradation. Only the combination of the two processes provides a quantitative description of the response to iron deprivation in yeast.
Iron is a key trace element for virtually all organisms. It functions as an essential cofactor in central cellular processes such as respiration, DNA synthesis and repair, ribosome biogenesis, metabolite biosynthesis, and oxygen transport in vertebrates. Although iron is highly abundant, its bioavailability is low due to its poor solubility under ambient conditions. On the other hand, iron is a major source of reactive oxygen species and thus toxic at higher concentrations. Therefore, cells have developed sophisticated systems for ensuring a tightly regulated cellular iron homeostasis (23, 24, 35, 51, 53). In the budding yeast Saccharomyces cerevisiae, adaptation to iron-deprived conditions elicits a strong response at the level of mRNA. This response includes the downregulation of genes for components of mitochondrial respiration and the citric acid cycle; the remodeling of genes of biosynthetic pathways that include iron-dependent enzymes such as the synthesis of ergosterol, unsaturated fatty acids, biotin, purines, and several amino acids; and of genes encoding heme proteins or iron-sulfur (Fe/S) proteins (23, 35, 37, 38, 47). At the same time, genes involved in hexose acquisition, fermentation, and cellular iron acquisition are highly induced. Disruption of the mitochondrial iron-sulfur cluster (ISC) assembly and export systems elicits a similar response, indicating that both mitochondrial Fe/S protein synthesis activity and extracellular iron levels are key regulatory factors for the maintenance of cellular iron homeostasis in S. cerevisiae (2, 20).
Many of the iron-responsive genes that are induced upon iron deprivation are under the control of the iron-responsive transcription factors Aft1 and, to a minor extent, Aft2 (23, 35, 41, 42). The Aft1-Aft2-dependent yeast iron regulon comprises a set of ∼40 genes encoding products that function mostly in iron uptake at the cell surface and in intracellular iron transport (35, 41, 42, 47). Aft1 shuttles between the cytosol and nucleus in an iron-responsive manner and acts as a transcriptional activator (52, 54). The monothiol glutaredoxins Grx3 and Grx4, the regulatory proteins Fra1 and Fra2, and a signaling molecule produced and exported by the mitochondrial ISC systems are required for proper sensing of iron by Aft1 (4, 20, 26, 33, 43).
The iron regulon includes two members of a conserved family of tandem zinc finger-containing mRNA-binding proteins, Cth1 and Cth2, which promote mRNA decay (37, 38, 53). Under iron-limiting conditions, Cth1 and Cth2 bind to specific AU-rich elements (AREs) within the 3′ untranslated region (UTR) of many mRNAs encoding proteins involved in iron-dependent pathways. The Cth proteins recruit the Dhh1 RNA helicase, which interacts with multiple members of the general machinery of mRNA decay, thus promoting the 5′-to-3′ degradation of mRNAs at cytoplasmic P bodies (34). Virtually all genes that are repressed upon iron deprivation display an aberrant expression in cells lacking CTH1 and CTH2, demonstrating the global contribution of this posttranscriptional process to the regulation of iron homeostasis in S. cerevisiae (53). However, the extent of downregulation of most genes upon iron deprivation is not quantitatively explained by the roles of the Cth proteins. Hence, there seems to be another important level of regulation that so far cannot be explained by Aft1-Aft2-dependent transcriptional activation and/or Cth protein-dependent mRNA degradation.
In this work we have identified the molecular basis of the Cth-independent downregulation of iron-responsive genes during the adaptation of yeast to iron deprivation. By focusing our analyses on the LEU1 gene of the branched-chain amino acid biosynthesis pathway and on CYC1, encoding cytochrome c, we show that the downregulation of iron-dependent metabolic pathways is mediated by decreased levels of key regulatory metabolites, in these cases, α-isopropylmalate (α-IPM) and heme, respectively, under iron-limiting conditions. Decreased synthesis of α-isopropylmalate, a transcriptional coactivator of the transcription factor Leu3 regulating LEU1 expression (13, 25, 49), is caused by the loss of function of the Fe/S protein Ilv3 under iron limitation. Likewise, the levels of the iron-containing heme are adjusted in an iron-dependent fashion. Thus, downregulation of iron-responsive genes in response to iron deprivation involves two levels of regulation in S. cerevisiae: transcriptional adaptation through metabolites that are synthesized in an iron-dependent fashion and posttranscriptional mRNA degradation.
MATERIALS AND METHODS
Yeast strains, cell growth, and plasmids.
Yeast strains used in this study are listed in Table S1 in the supplemental material. Cells were cultivated in synthetic complete minimal medium containing all recommended supplements (SC) and 2% (wt/vol) glucose (SD) or 2% (wt/vol) raffinose (48). Repression of conditional Gal strains was performed as described in the corresponding literature (see Table S1). Plasmid-based reporter constructs for monitoring LEU1 expression were constructed by replacing the MET25 promoter and the CYC1 terminator of vector p416-MET25-luc2 harboring the firefly luc2 gene (32). Those for monitoring the expression of CYC1 were based on vector p416-MET25-hRluc harboring Renilla luciferase, hRluc, or p414-MET25-luc2 (32). For further details on plasmids used in this study, see Table S2 in the supplemental material. Constructs were verified by DNA sequencing and/or functional complementation of corresponding yeast mutants.
Reporter assays.
For analysis of iron-responsive gene expression using luciferase-based reporter constructs, cells were cultivated at 30°C overnight in minimal medium containing either 50 μM bathophenanthroline or 50 μM FeCl3. Cells were diluted in 5 ml of the same medium to an optical density at 600 nm (OD600) of 0.2, incubated until the optical density reached 0.5 to 1 (∼6 h), washed once in water, and frozen. Luciferase activities were determined using firefly and Renilla luciferase assay systems (Promega). Cells were suspended in 250 μl CCRL buffer (Renilla) or buffer A (firefly) (25 mM Tris, pH 7.8, 2 mM EDTA, 2 mM dithiothreitol, 10% glycerol, 0.1% Triton X-100) supplemented with phenylmethylsulfonyl fluoride (PMSF) and lysed by being vortexed with glass beads three times for 1 min each with intermediate cooling. Cell debris was removed by centrifugation, 5 μl yeast extract was mixed with 20 μl (25 μl for Renilla) substrate working solutions, and bioluminescence was recorded in an InfinTe M200 microplate reader (Tecan) with an integration time of 1 s. Protein concentrations were determined in microplates by the Bradford method (Bio-Rad). Standard deviations were calculated from duplicate measurements of at least 4 independent cells. Reporter assays using green fluorescent protein (GFP) were carried out as described previously (20, 32).
Miscellaneous methods.
The following published methods were used: manipulation of DNA and PCR (44), transformation of yeast cells (16), isolation of yeast mitochondria and postmitochondrial supernatant (7), immunostaining (19), Northern blotting (20), and determination of enzyme activities of isopropylmalate isomerase, Leu1, and aconitase (32); dihydroxy-acid dehydratase, Ilv3 (9); and ferrochelatase (3). Error bars represent the standard errors of the means (SEM) (n ≥ 4).
RESULTS
No significant role of Aft1 in downregulation of iron-responsive genes.
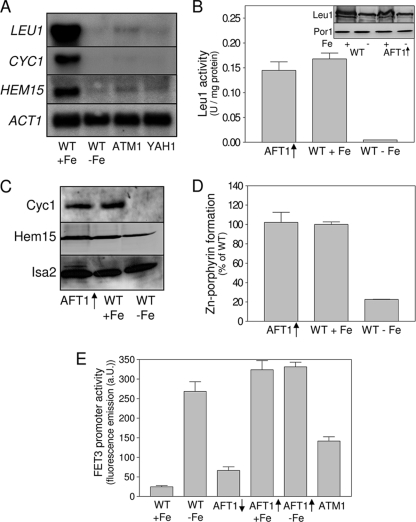
In order to analyze the mechanisms underlying the downregulation of iron-responsive genes upon iron deprivation, we analyzed the transcription of two representative genes involved in amino acid biosynthesis and respiration. LEU1, encoding the Fe/S protein isopropylmalate isomerase of the leucine biosynthesis pathway, was chosen because the gene displays the strongest downregulation upon iron starvation in several strain backgrounds (20, 37, 47). CYC1, encoding cytochrome c, was selected because it displays the strongest iron responsiveness of all genes involved in respiration (20, 37, 47). Northern blot analysis confirmed the downregulation of these genes after 16 h of cultivation under iron-limiting conditions, while HMX1, a member of the yeast iron regulon, was strongly induced (Fig. 1A and data not shown [20]). Upon depletion of two components of the mitochondrial ISC assembly and export systems, Yah1 and Atm1, transcription of both LEU1 and CYC1 was modified by a margin similar to that in iron-deprived wild-type cells, confirming the central role of the mitochondrial ISC systems in the regulation of cellular iron homeostasis (20, 43). Enzyme activities of Leu1 were more than 20-fold reduced upon iron deprivation, as expected from the loss of its Fe/S cluster, and yet protein levels of Leu1 were reduced only ∼2-fold upon iron deprivation (Fig. 1B). Cytochrome c levels of mitochondria from iron-starved cells dropped below the level of detection (Fig. 1C).
Fig. 1.
Aft1 alone plays no significant role in the downregulation of LEU1, CYC1, and HEM15. (A) Northern blot analysis of LEU1, CYC1, and HEM15 transcription. Total RNA was isolated from exponentially growing wild-type cells (WT) cultivated under iron-replete conditions in the presence of 50 μM FeCl3 (+Fe) or under iron-deprived conditions in the presence of 50 μM bathophenanthroline for 16 h (−Fe) and from Atm1- or Yah1-depleted Gal-ATM1 (ATM1) or Gal-YAH1 (YAH1) cells grown under iron-replete conditions in synthetic complete minimal (SD) medium. RNA was separated on agarose gels, blotted onto nylon membranes, and hybridized with 32P-labeled probes. ACT1 served as a loading control (20). (B) Cell extracts of wild-type cells cultivated for 16 h in the presence of 50 μM FeCl3 (+Fe) or 50 μM bathophenanthroline (−Fe) and wild-type cells overproducing Aft1p from vector p424MET3-AFT1 (AFT1↑) were analyzed for Leu1 activity. The inset shows an immunostaining for Leu1 and Por1 in wild-type and Aft1-overproducing cells cultivated under iron-replete and iron-deprived conditions. (C) Mitochondria were isolated from iron-replete, iron-starved, or Aft1-overproducing wild-type cells (AFT1↑). Protein levels of cytochrome c (Cyc1) and ferrochelatase (Hem15) were assessed by immunostaining. An immunostaining for Isa2 served as a loading control. (D) Ferrochelatase activities of mitochondria were determined by monitoring the insertion of Zn2+ into protoporphyrin IX. (E) The wild-type strain (W303A), the Atm1-depleted Gal-ATM1 strain (ATM1), and the W303 wild-type strain overproducing Aft1 from the methionine-regulated vector p424MET3-AFT1 harboring reporter plasmid pFET3-GFP were cultivated under iron-replete and iron-depleted conditions. Aft1-overproducing cells were cultivated under repressive conditions in the presence of methionine (AFT1↓) or under inducing conditions in the absence of methionine (AFT1↑). FET3 promoter activities were determined by measuring the GFP-specific fluorescence emission of logarithmically grown cells. Error bars indicate the standard errors of the means (n ≥ 4).
Since under iron starvation conditions the iron-responsive transcription factor Aft1 is activated and the iron regulon, including the CTH1 and CTH2 genes, is induced, we tested whether Aft1 induction might be responsible for the observed decrease in LEU1 and CYC1 expression. In cells overproducing Aft1 (i.e., under constitutive induction of the iron regulon), Leu1 displayed wild-type activities and cytochrome c was present at wild-type levels under iron-replete conditions (Fig. 1B and C). A similar picture was observed for HEM15 encoding ferrochelatase. While HEM15 mRNA levels (20), protein levels, and ferrochelatase activities were reduced in iron-deprived yeast cells, they remained virtually unchanged in cells overproducing Aft1 (Fig. 1D). In contrast, mRNA levels of FET3, a member of the iron regulon, were strongly induced upon overproduction of Aft1 (Fig. 1E). These data suggest that the transcription factor Aft1 alone plays no significant role in the iron-responsive expression of LEU1, CYC1, or HEM15, despite an induction of the mRNA-degrading proteins Cth1 and Cth2. The fact that the overproduction of Aft1 is not associated with any obvious mitochondrial defects or with the downregulation of a significant number of genes in DNA microarray analyses confirms these observations (20, 42).
Dual regulation of the iron-dependent expression of LEU1.
For a detailed analysis of LEU1 gene expression, we used luciferase-based reporter assays. The 900-bp segment immediately upstream of the start codon of LEU1 was inserted in front of the firefly luciferase gene of the yeast vector p416MET25-luc2 (32). Cell extracts of W303A wild-type cells harboring the resulting vector pLEU1-Luc2 displayed significant amounts of luciferase-based luminescence under iron-replete conditions, and these were reduced 10-fold upon cultivation under iron-limiting conditions in the presence of the iron chelator bathophenanthroline for 24 h (Fig. 2A). In cells depleted of components of the mitochondrial ISC assembly and export systems, Ssq1, Yah1, and Atm1, a ∼4-fold reduction of luciferase activity was observed. These data are similar to those of the Northern blot analysis (Fig. 1A). Recent work has shown that terminators play an important role in iron-responsive gene expression in that they contain AU-rich elements (AREs) as binding sites for the RNA-binding proteins Cth1 and Cth2, which induce mRNA degradation under iron-limiting conditions (34, 37, 38). Corresponding potential AREs for Cth1 and Cth2 are present in the 3′ UTR of LEU1 90 and 110 bp downstream of the stop codon (not shown). In addition, the CYC1 terminator present in vector pLEU1-Luc2 is involved in Cth1/2-mediated mRNA decay (38). Replacement of the CYC1 terminator in vector pLEU1-Luc2 by a 470-bp region immediately downstream of the LEU1 stop codon (resulting in vector pLEU1/LEU1-term) gave a twofold increase in expression (compare Fig. 2A and B), and a strong, 25-fold downregulation was observed upon iron deprivation. No significant change in LEU1 expression was observed upon overproduction of Aft1, confirming that this transcription factor plays no significant role in LEU1 expression. The LEU1 terminator conferred iron responsiveness on the MET25 promoter, which shows no iron regulation per se, that was twofold stronger than that obtained with the terminator of CYC1 (Fig. 2C). This demonstrates that the LEU1 terminator is involved in iron-responsive gene expression. Finally, luciferase expression under the control of the LEU1 promoter and terminator was studied in cells lacking the RNA-binding proteins Cth1 and Cth2 (Fig. 2D). Under iron-replete conditions, deletion of CTH1 and CTH2 had little effect. Under iron-limiting conditions, deletion of CTH1 had little effect while that of CTH2 increased LEU1 expression by a factor of 2. In the cth1Δ/cth2Δ double deletion mutant, expression under iron-limiting conditions increased to almost 50% of the value under iron-replete conditions. On the one hand these data indicate that Cth1 and Cth2 cooperate in inducing the degradation of the LEU1 mRNA upon iron deprivation. On the other hand, they show that Cth1 and Cth2 do not fully account for the decreased expression under iron-depleted conditions and hence point toward an important role of the LEU1 promoter in this process. We therefore analyzed the reason for the iron dependence of the LEU1 promoter in more detail.
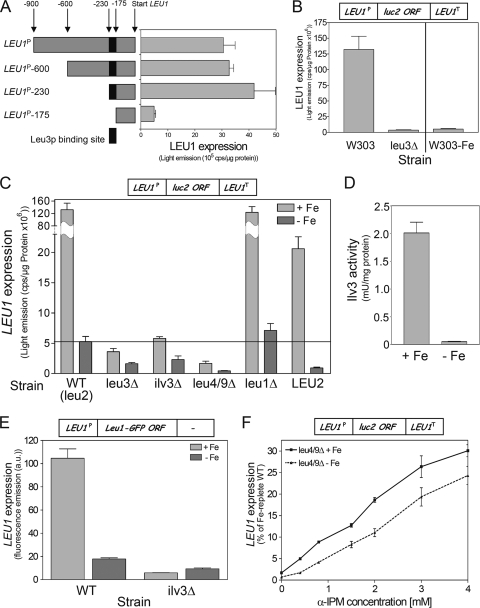
Fig. 2.
Both promoter and terminator mediate the iron-responsive expression of LEU1. (A) W303A wild-type cells cultivated for 24 h in the presence of 50 μM FeCl3 (+Fe) or 50 μM bathophenanthroline (−Fe) and Atm1-, Ssq1-, or Yah1-depleted Gal-ATM1, Gal-SSQ1, and Gal-YAH1 cells harboring reporter plasmid pLEU1-Luc2 (containing the CYC1 terminator; see box on top) were grown to mid-log phase. The luciferase-derived luminescence of clarified cell extracts was quantified. (B) Wild-type (W303A) and Aft1-overproducing cells (AFT1↑) harboring reporter plasmid pLEU1/LEU1-term were cultivated for 24 h under iron-replete and iron-depleted conditions, and the luciferase activity of cell extracts was quantified. (C) luc2 transcript levels from the MET25 promoter under iron-replete and iron-limiting conditions were determined in wild-type (W303) cells. The reporter plasmids harboring the luciferase gene contained either no terminator (pMET25-Luc2, none), or the 3′ UTRs (term) of LEU1 (pMET25/LEU1-term) or CYC1 (p416MET25-luc2), respectively. (D) Wild-type (BY4742) and the respective cth1Δ, cth2Δ, and cth1Δ/cth2Δ (cth1/2Δ) deletion strains were transformed with the reporter plasmid pLEU1/LEU1-term. Cells were cultivated under iron-replete and iron-depleted conditions for 24 h, and luc2 transcript levels were determined. Error bars indicate the SEM (n ≥ 4). ORF, open reading frame.
Several genes of the biosynthesis pathway for the branched-chain amino acids leucine, isoleucine, and valine are under the control of the transcriptional activator Leu3 (25). A single Leu3 binding site is predicted between positions −175 and −230 upstream of the start codon of the LEU1 open reading frame (Fig. 3A) (12, 13, 29, 30). Promoter deletions from the 5′ side up to −230 bp did not significantly affect LEU1 expression, while deletions that removed the putative Leu3 binding site resulted in a collapse of expression to basal transcription levels (Fig. 3A). Deletion of LEU3 results in a strong decline of LEU1 expression, confirming that LEU1 is under the dominant control of a single transcription factor, similarly to other genes of the branched-chain amino acid biosynthetic pathway (Fig. 3B) (13, 25). Moreover, remnant LEU1 expression levels in leu3Δ cells were in the same range as those found in iron-deprived wild-type cells (Fig. 3B). These results indicate that the lack of another metabolite attenuates the ability of Leu3 to activate LEU1 upon iron deficiency.
Fig. 3.
Iron-responsive transcription of LEU1 is mediated by cellular levels of α-isopropylmalate. (A) Wild-type cells (BY4742) were transformed with luciferase-based reporter plasmids harboring truncated versions of the LEU1 promoter as indicated (left), and luciferase activities of cell extracts were quantified (right). (B) LEU1 expression was determined in wild-type cells (W303) cultivated under iron-replete and iron-depleted (−Fe) conditions for 24 h and in the leu3Δ deletion strain (BY4742) harboring reporter plasmid pLEU1/LEU1-term (see box on top). (C) LEU1 expression was determined in strains (W303) carrying deletions of the indicated genes of the biosynthetic pathway for leucine, isoleucine, and valine; a W303 strain harboring a wild-type LEU2 allele (LEU2); and the leu3Δ strain (BY4742) (Fig. 4C). Cells were transformed with plasmid pLEU1/LEU1-term (box on top) and cultivated under iron-replete and iron-depleted conditions for 24 h. (D) Enzyme activities of the yeast dihydroxy-acid dehydratase Ilv3 were determined in mitochondria isolated under anaerobic conditions from wild-type cells cultivated under iron-replete and iron-depleted conditions. (F) Wild-type (BY4741) and ilv3Δ cells harboring a genomic copy of a terminatorless LEU1 gene with a C-terminally fused GFP were cultivated under iron-replete and iron-depleted conditions for 24 h. Promoter activities were determined by measuring the GFP-specific fluorescence emission of logarithmically grown cells. (F) LEU1 expression was determined in leu4/9Δ cells cultivated in SD medium supplemented with increasing amounts of α-IPM under iron-replete or iron-depleted conditions. Cells contained reporter plasmid pLEU1/LEU1-term. Error bars indicate the SEM (n ≥ 4). ORF, open reading frame; WT, wild type.
The Leu3 transcription factor requires α-isopropylmalate (α-IPM), a metabolic intermediate of the branched-chain amino acid biosynthesis and the substrate of Leu1, as a transcriptional coactivator (25, 49). In order to study the effect of α-IPM on LEU1 expression, we took advantage of strains carrying deletions in individual genes of the branched-chain amino acid biosynthesis pathway. Both Ilv3 and Leu4/9 enzymes are required for the production of α-IPM in this pathway (see Fig. 4C). In ilv3Δ and leu4/9Δ cells LEU1 expression was reduced to basal levels similar to those seen in leu3Δ and iron-deprived wild-type cells (Fig. 3C). In addition, the iron responsiveness of LEU1 was strongly reduced in these cells. Deletion of LEU1, which lies downstream of α-IPM synthesis in the pathway for leucine, showed wild-type expression from the pLEU1/LEU1-term plasmid. LEU2 is inactivated in virtually all common yeast laboratory strains. Complementation of the leu2 mutation in W303 wild-type cells with a wild-type genomic LEU2 copy reduced LEU1 expression under iron-replete conditions ∼9-fold, presumably by restoring the metabolic flux through the pathway and thus decreasing the levels of α-IPM (see Fig. 4C). Iron deprivation of LEU2 cells reduced LEU1 mRNA levels ∼22-fold, which is very close to the levels observed in the leu2 background of our W303 wild-type cells. Apparently, the leu2 background of common yeast laboratory strains amplifies the expression of LEU1 without significantly changing its iron-responsive regulation. Taken together, these observations demonstrate a central activating role of α-IPM for LEU1 expression and are consistent with earlier data for other genes of branched-chain amino acid biosynthesis (25).
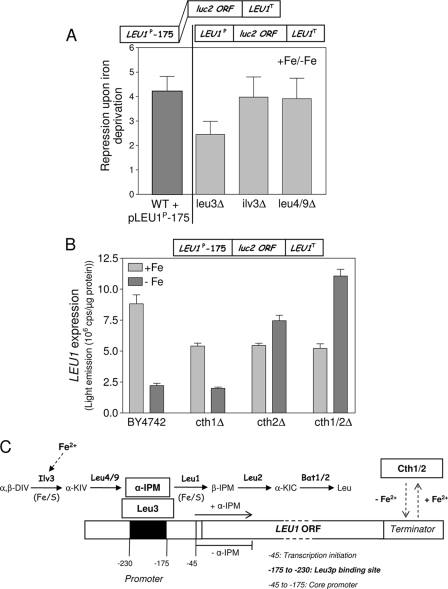
Fig. 4.
Dual regulation of the iron responsiveness of LEU1. (A) The ratio of LEU1 expression under iron-replete conditions to that under iron-depleted conditions in the indicated strains as derived from Fig. 3C (right panel) was compared with that of a LEU1 core promoter construct (pLEU1P-175/LEU1-term) in W303 wild-type cells. (B) Expression levels of the LEU1 core promoter were determined under iron-replete (+Fe) and iron-depleted (−Fe) conditions in wild-type cells (BY4742) and the cth1Δ, cth2Δ, and cth1Δ/cth2Δ (cth1/2Δ) deletion strains harboring reporter plasmid pLEU1P-175/LEU1-term (box on top). (C) Model for the iron-responsive expression of LEU1 in S. cerevisiae. Abbreviations: α,β-DIV, α,β-dihydroxyisovalerate; α-KIV, α-ketoisovalerate; β-IPM, β-isopropylmalate; α-KIC, α-ketoisocaproate. WT, wild type; ORF, open reading frame.
Branched-chain amino acid biosynthesis contains two iron-dependent enzymes, the Fe/S proteins Ilv3 (mitochondrial dihydroxy-acid dehydratase) and Leu1 (9, 25) (Fig. 4C). Similarly to Leu1 (Fig. 1B), the activity of Ilv3 was virtually undetectable in wild-type cells upon iron depletion (Fig. 3D). Thus, an iron-starved wild-type cell likely contains low levels of α-IPM and in this respect behaves similarly to an ilv3Δ strain. This conclusion easily explains why LEU1 expression levels in iron-deprived cells are similar to those of mutants that are impaired in the production of α-IPM or that lack the transcription activator Leu3 (Fig. 3C). In order to verify the iron-responsive transcription of LEU1 independently of the LEU1 terminator, we took advantage of a yeast strain that contains a chromosomal LEU1-GFP gene fusion replacing the LEU1 terminator region with an in-frame GFP/HIS3 cassette. This strain was previously used in a systematic protein localization study with S. cerevisiae (22). The GFP-specific fluorescence of this strain was ∼6-fold reduced upon cultivation under iron-deprived conditions below that in iron-replete conditions (Fig. 3E). Upon deletion of ILV3 in this strain, the fluorescence emission strongly declined to basal levels similar to those under iron deprivation and showed virtually no iron response. Finally, in order to directly show the regulatory role of α-IPM in LEU1 expression under iron-limiting conditions, α-IPM was added to leu4/9Δ cells that are unable to synthesize α-IPM (Fig. 4C). Under both iron-replete and iron-depleted conditions, added α-IPM increased LEU1 expression in virtually identical concentration-dependent fashions that were almost linear up to the maximal tested amount of the inducer (Fig. 3F). We therefore conclude that the iron-dependent synthesis of α-IPM fully explains the iron-responsive transcription of LEU1.
Transcription, however, was not alone responsible for the iron-dependent modulation of LEU1, since a small (∼4-fold) iron response was retained in cells that are unable to activate the LEU1 promoter via α-IPM and Leu3 (i.e., in ilv3Δ, leu4/9Δ, and leu3Δ cells) (Fig. 3C and 4A). Likely, this residual iron response was conferred by the LEU1 terminator alone. If so, this response should be similar to that of a truncated LEU1 gene that cannot be activated by Leu3. In order to test this conclusion, we took advantage of a LEU1 core promoter construct, pLEU1P-175/LEU1term, which lacks the Leu3 activator domain (Fig. 3A). Indeed, iron depletion reduced the expression of the LEU1 core promoter construct in wild-type cells by virtually the same margin (∼4-fold) as that for the full-length LEU1 promoter in ilv3Δ and leu4/9Δ cells (Fig. 4A). This reduction was quantitatively similar to that observed for the iron-indifferent MET25 promoter upon insertion of the LEU1 terminator (5.8-fold) (Fig. 2C). In addition, deletion of CTH2 completely abolished the iron responsiveness of the LEU1 core promoter construct (Fig. 4B). In the cth1Δ/cth2Δ double mutant, expression was even higher upon iron deprivation than under iron-replete conditions. Most likely, the residual iron response of LEU1 in cells that are unable to activate LEU1 transcription is quantitatively conferred by Cth1-Cth2 function in LEU1 mRNA degradation. Clearly, the iron-responsive expression pattern of the LEU1 core promoter in cells lacking Cth1 and/or Cth2 differed from that of the full promoter construct (Fig. 2D).
Taken together, these data show that the iron-responsive expression of LEU1 can be quantitatively explained by a combined regulation through both the promoter and the terminator (Fig. 4C). LEU1 transcription is controlled by the activity of Leu3, which is modulated by cellular levels of α-IPM. These are low under iron-limiting conditions due to the inactivation of the Fe/S protein Ilv3, which is essential for α-IPM production. The LEU1 terminator contains information for posttranscriptional mRNA degradation facilitated by the two mRNA-binding proteins Cth1 and Cth2 (53). Quantitatively, transcriptional regulation contributes about sixfold (Fig. 3E) and posttranscriptional mRNA decay contributes fourfold to the iron responsiveness of LEU1, respectively (Fig. 4A). Assuming that both mechanisms are cumulative, a 24-fold downregulation of LEU1 upon iron deprivation is expected, which is close to the experimental data (25-fold effect in Fig. 2B). Thus, the combination of the two regulatory mechanisms of metabolite-linked iron-responsive transcription and posttranscriptional mRNA degradation provides a quantitative description of the iron-responsive expression of LEU1.
Hap-mediated iron-dependent expression of CYC1.
The above data show that the iron-dependent remodeling of levels of a key regulatory molecule of a biochemical pathway confers an iron-responsive gene expression pattern. In S. cerevisiae, the iron-containing cofactor heme functions as a key regulatory metabolite during the adaptation to hypoxic conditions (6, 55, 56). Since heme levels are low in iron-deprived cells, it was tempting to speculate whether heme also functions as a regulatory molecule in iron-responsive expression of genes involved in respiration. In order to explore this possibility, we analyzed the CYC1 gene encoding the heme protein cytochrome c, which is transcribed in an iron-responsive manner (36). Apart from the roles of Cth1 and Cth2 in CYC1 mRNA degradation, little is known concerning the mechanism of the iron responsiveness of CYC1 (38). The CYC1 promoter harbors two upstream activating sequences, UAS1 and UAS2, which are controlled by the transcriptional activator Hap1 and the Hap2-5 complex (17) (Fig. 5A). This tandem structure is shared by other genes involved in respiration (40, 45). While the activating molecule of the Hap2-5 complex is unknown, Hap1 requires heme for transcriptional activity (6, 55, 56).
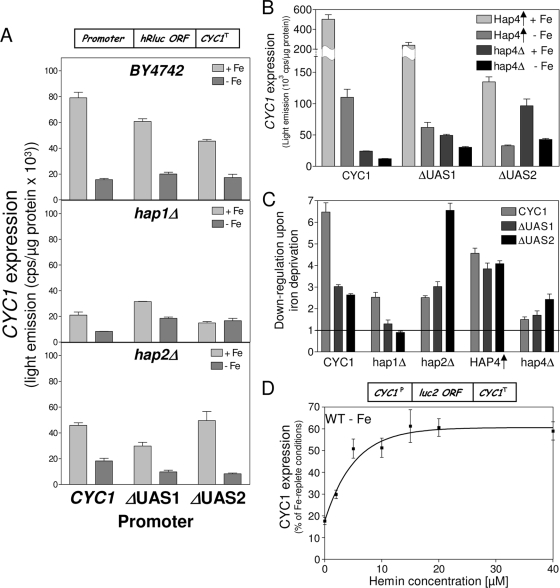
Fig. 5.
The iron responsiveness of CYC1 is mediated by transcription and Cth1/Cth2-dependent mRNA degradation. (A) Model for the CYC1 promoter (17). (B and C) The wild-type strain (BY4742) and the cth1Δ, cth2Δ, and cth1Δ/cth2Δ (cth1/2Δ) isogenic deletion strains were transformed with the reporter plasmid pΔUAS1/2-hRluc (B) or pCYC1-hRluc (C) (boxes on top). Cells were cultivated under iron-replete (+Fe) and iron-depleted (−Fe) conditions in the presence of raffinose for 24 h, and the Renilla luciferase level was determined in cell extracts. Error bars indicate the SEM (n ≥ 4). ORF, open reading frame.
In order to quantify the contribution of the CYC1 terminator to the iron-responsive expression of CYC1, we employed a luciferase reporter construct, pΔUAS1/2-hRluc, which lacks the two upstream activating domains (Fig. 5A). The analysis was carried out in raffinose-containing medium in order to avoid glucose repression. In BY4742 wild-type and cth1Δ cells, only a 1.3-fold-reduced expression of pΔUAS1/2-hRluc was observed upon iron deprivation (Fig. 5B). This effect was surprisingly small, as the CYC1 terminator is a known target of both Cth proteins (38). In cth2Δ cells, the expression of pΔUAS1/2-hRluc was twofold higher under iron-limiting conditions than under iron-replete conditions and increased to >3-fold in cth1Δ/cth2Δ cells. Higher expression under iron-limiting than under iron-replete conditions in cth1Δ/cth2Δ cells seems to be a common theme for core promoters of ARE-containing genes, as this effect was also observed for LEU1 (Fig. 4B). Most likely, this induced expression reflects an increased mRNA stability that allows an increased number of translations in cth1Δ/cth2Δ cells. A promoter construct, pCYC1-hRluc, harboring the complete CYC1 core promoter, including the two upstream activation sequence (UAS) regions (Fig. 5A), showed a fourfold-higher expression level in BY4742 wild-type cells than did the activatorless core promoter construct pΔUAS1/2-hRluc (Fig. 5C). In addition, a ∼6-fold downregulation was observed for the full CYC1 promoter construct upon iron deprivation of wild-type cells, in contrast to the weak effect seen with pΔUAS1/2-hRluc. Thus, the promoter appears to be the dominant element for the iron-responsive expression of CYC1. In cth1Δ cells CYC1 was expressed similarly to wild-type cells, while the deletion of CTH2 alone or in combination with CTH1 increased the expression under iron-replete conditions and yet attenuated the downregulation of CYC1 under iron-limiting conditions. The fact that the deletion of CTH2 induced a stronger deregulation under iron deprivation of all constructs tested than did that of CTH1 confirms the dominant role of Cth2 in posttranscriptional mRNA decay upon long-term iron deprivation (38). Taken together, these data show that the iron responsiveness of CYC1 is explained by a combination of transcriptional effects involving promoter elements and by mRNA degradation via the Cth proteins. The former mechanism is the dominant effect.
We attempted to identify the reason for the iron responsiveness of CYC1 transcription by analyzing the roles of the two activators Hap1 and Hap2-5. The iron responsiveness of the CYC1 promoter was confined to the two upstream activating domains UAS1 and UAS2, as a construct harboring the 1-kb 5′ upstream region of the CYC1 open reading frame displayed an expression pattern in wild-type cells almost identical to that of the reporter construct pCYC1-hRluc above, which included only UAS1 and UAS2 (not shown). We therefore concentrated our analysis on these two promoter regions and, in addition to wild-type cells, employed cells in which HAP1, HAP2, or HAP4 was deleted or HAP4 was overexpressed (Fig. 6A and B). The extent of remodeling between iron-depleted and iron-replete conditions is presented in Fig. 6C. Removal of UAS1 and that of UAS2 reduced CYC1 expression in BY4742 wild-type cells under iron-replete conditions by similar margins but did not significantly affect the expression level upon iron deprivation (Fig. 6A, top panel). Thus, similarly to LEU1, a reduced transcriptional activation of CYC1 goes along with a diminished iron responsiveness. We then studied the role of Hap1 in iron-responsive transcription of CYC1 which binds to the UAS1 region (6, 18, 55, 56). Most laboratory yeast strains harbor a transposon insertion at the HAP1 locus that replaces the C-terminal 12 amino acids with a 30-amino-acid random sequence (14). In BY4742, CYC1 expression dropped fourfold upon complete deletion of the HAP1 open reading frame (Fig. 6A, middle panel). A similar effect was seen in the W303 background (not shown), confirming previous findings that indicate that the natural hap1 mutant of S288c-derived strains retains at least limited function (14). Iron deprivation of BY4742 hap1Δ cells reduced CYC1 expression by a factor of 2.5, which is >2-fold less than that in the wild type (Fig. 6A, middle panel). Iron responsiveness of CYC1 was reduced further in hap1Δ cells (to 1.7-fold) upon removal of the first activating domain, UAS1 (Fig. 6A, middle panel). Strikingly, a reporter construct, pΔUAS2-hRluc, which lacks the second activation domain of CYC1, which is not targeted by Hap1, displayed virtually no iron-dependent expression in hap1Δ cells (Fig. 6A, middle panel, and 6C). This observation clearly demonstrates that the Hap1 transcription factor contributes significantly to the iron-responsive expression of CYC1. Likely, the impact of Hap1 in this process is much stronger in strains that harbor a wild-type HAP1 allele.
Fig. 6.
Hap1 and Hap4 are involved in the iron-responsive expression of CYC1. (A) Wild-type (BY4742) cells and the hap1Δ and hap2Δ isogenic deletion strains were transformed with reporter plasmid pCYC1-hRluc, pΔUAS1-hRluc, or pΔUAS2-hRluc carrying the indicated CYC1 promoters (Fig. 5A). Cells were cultivated under iron-replete (+Fe) and iron-depleted (−Fe) conditions for 24 h, and the Renilla luciferase-derived luminescence was determined. (B) Expression of the different CYC1 promoter constructs was determined in hap4Δ and wild-type cells overproducing Hap4 (Hap4↑) as described for panel A. (C) The ratio of the iron-responsive expression was calculated from the quotas of the expression of the different CYC1 promoter constructs under iron-replete and iron-deprived conditions for the indicated strains in panels A and B. (D) Wild-type cells expressing the C. elegans heme importer CeHRG4, which harbored the reporter plasmid pCYC1-Luc2, were cultivated under iron-depleting conditions and supplemented with increasing amounts of hemin for 16 h, and the luciferase-derived luminescence was determined. Error bars indicate the SEM (n ≥ 4). ORF, open reading frame.
Finally, we analyzed whether the inactivation of the Hap2-5 complex, the second activator that binds predominantly to the UAS2 activator element, influences the iron-responsive expression of CYC1 (11, 31). In hap2Δ cells that lack the core component of the Hap2-5 complex, CYC1 expression was twofold lower than that in wild-type cells and also displayed a 2.5-fold-lower iron response (Fig. 6A, bottom panel). In addition, the iron response of the pΔUAS1-hRluc construct was also ∼2-fold lower in hap2Δ cells than in wild-type cells, demonstrating that the Hap2-5 complex is contributing to the iron response of CYC1. However, the fact that the pΔUAS1-hRluc construct that is not regulated by Hap1 retained an iron response in hap2Δ cells at all indicates that further unknown transcription factors contribute to the iron responsiveness of CYC1 that is conferred via UAS2. Remarkably, for the pΔUAS2-hRluc construct, an almost-wild-type (sixfold) downregulation was observed in hap2Δ cells upon iron limitation (Fig. 6A, bottom panel, and 6C). This observation that a truncated CYC1 promoter construct that is activated only by Hap1 via UAS1 displayed an iron response similar to that of the full CYC1 promoter confirms the prominent role of Hap1 in this process. The wild-type iron responsiveness of the pΔUAS2-hRluc construct was not observed in hap4Δ cells that lack the principal activator component of the Hap2-5 complex, suggesting that an inactivated Hap2-5 complex occupies the promoter and by this interferes with the activation via Hap1. Nevertheless, both expression and iron responsiveness of the full CYC1 promoter were reduced by similar margins in hap4Δ and hap1Δ cells (Fig. 6B and C). This suggests that the two independent Hap activator systems contribute equally to global expression and iron responsiveness of CYC1. Overproduction of Hap4 increased CYC1 expression 20-fold over that in hap4Δ cells, as expected. Expression of the pΔUAS1-hRluc construct was increased 4.8-fold and that of pΔUAS2-hRluc was increased 1.4-fold over that in hap4Δ cells. At the same time, the iron response of the full CYC1 promoter also increased from 2-fold in hap4Δ cells to 4.5-fold in Hap4-overproducing cells, confirming the involvement of the Hap2-5 complex in the iron regulation of CYC1. Hap4 overproduction also increased the iron responsiveness of the pΔUAS1-hRluc construct 2.2-fold over that in hap4Δ cells and that of pΔUAS2-hRluc 1.6-fold (Fig. 6B and C). Thus, the effect of Hap4 on the iron responsiveness of CYC1 is transmitted via both activating domain UAS2 and, to a lesser extent, UAS1. The fact that this Hap4-dependent modulation was mediated via both upstream activating domains is in accordance with previous findings (28).
Taken together, these data demonstrate that the transcriptional activators Hap1 and Hap4 play decisive roles for the iron-responsive expression of CYC1. The heme-dependent activation of Hap1 likely explains how Hap1 contributes to the downregulation of CYC1 upon iron limitation. Heme, an iron-containing cofactor, is sparse under conditions of iron deprivation. At the same time, the activity of ferrochelatase, the enzyme that catalyzes the last step in heme synthesis, is significantly reduced under these conditions (Fig. 1D). Under our experimental conditions, heme levels were ∼20-fold reduced in wild-type cells cultivated in the presence of bathophenanthroline (not shown). In order to directly show the involvement of heme in transcriptional adaptation to iron-deprived conditions, hemin was added to iron-deprived wild-type cells that expressed the heme importer CeHRG4 from Caenorhabditis elegans (39). Hemin increased CYC1 expression in a concentration-dependent fashion up to a maximum of ∼60% of that observed under iron-replete conditions (Fig. 6D). No significant induction was seen when hemin was added to cells cultivated under iron-replete conditions (data not shown). In addition, the activity of aconitase, an Fe/S protein, increased only marginally in iron-deprived cells upon feeding with hemin, indicating that the iron that is imported into the cells by heme was not converted into a biologically available form in large amounts (data not shown). The observed induction of CYC1 was therefore mainly due to increasing levels of intracellular hemin. Thus, the lack of activation of Hap1 due to diminishing levels of intracellular heme plays a decisive role in the transcriptional adaptation of respiratory genes to iron-deprived conditions.
DISCUSSION
S. cerevisiae responds to iron deprivation with a massive adaptive remodeling of mRNA transcript levels that includes genes for components of mitochondrial respiration and the citric acid cycle and genes involved in the biosynthetic pathways for ergosterol and unsaturated fatty acids, biotin, purines, and several amino acids (20, 24, 35, 37, 38, 47). The majority of genes that are downregulated upon iron deprivation are members of metabolic processes that contain iron-dependent enzymes. It was thus suggested that yeast adapts to iron deprivation by minimizing dispensable iron-dependent processes in order to liberate and spare iron for more-essential tasks (24, 35, 53). The mechanisms underlying this iron sparing, however, are not fully understood. In this work, we have studied the contribution of transcription to the iron responsiveness of two representative genes, LEU1 and CYC1, which are strongly downregulated upon iron deprivation. Previous work focused on the central role of posttranscriptional mRNA degradation of iron-responsive genes upon iron deprivation involving the mRNA-binding proteins Cth1 and Cth2 (37, 38). Our current analysis identifies a novel mechanism of transcriptional regulation of iron-responsive genes that involves iron-dependent metabolites. This regulatory mechanism provides a second level of control in the adaptation of S. cerevisiae to iron-deprived conditions that operates in addition to the previously reported posttranscriptional mechanism. The sum of the two pathways allows the quantitative description of iron-responsive gene expression.
The biosynthetic pathway for branched-chain amino acids serves as an excellent example of a pathway that is subjected to iron sparing, as it contains two Fe/S proteins, isopropylmalate-isomerase (Leu1) and dihydroxy-acid dehydratase (Ilv3), that have no other known function outside the biosynthesis of isoleucine, leucine, and valine. (25). Leu1 is the most abundant Fe/S protein in S. cerevisiae (15), and consequently, LEU1 displays the strongest downregulation upon iron starvation of all genes in several strain backgrounds (20, 37). Other iron-responsive genes of this pathway include LEU2, BAT1, BAT2, ILV3, ILV5, and BAP2 (20, 37, 38, 47). The transcriptional activator Leu3 plays a central role in gene expression of the branched-chain amino acid biosynthesis, and several genes that are regulated by Leu3 are also iron responsive, including GDH1, encoding glutamate dehydrogenase involved in nitrogen assimilation (21, 25, 47). Leu3 is activated by the metabolite α-isopropylmalate, an intermediate of this pathway and the substrate of Leu1 (25, 49). Our data show that LEU1 transcription is dominantly regulated by Leu3 and that LEU1 expression in iron-starved wild-type cells is reduced to basal levels that are similar to those of leu3Δ cells or cells that are unable to synthesize α-IPM. In addition, LEU1 displays only limited iron responsiveness in cells lacking Leu3. These observations identify Leu3 as an indirect iron-responsive transcription factor. The mechanism of the iron regulation via Leu3 is intimately linked to its activation by α-IPM. Under iron-deprived conditions, the Fe/S protein Ilv3, which is crucial for α-IPM production (25), is inactive due to the lack of its cofactor and the ILV3 mRNA is the target of the Cth proteins (38). Thus, the downregulation of Leu3-dependent genes under iron deprivation is achieved through the back door via the iron-dependent modulation of cellular levels of a key regulatory metabolic intermediate, α-IPM, thus linking iron availability to metabolism (25).
In a similar fashion, the expression of lysine biosynthesis genes in S. cerevisiae depends on the inducer α-aminoadipate semialdehyde, a metabolic intermediate that activates a single transcription factor, Lys14 (8). Its synthesis essentially requires the Fe/S enzymes aconitase and homoaconitase, which operate early in the pathway. Further examples of biosynthetic pathways that include both iron-dependent enzymes and iron-responsive genes are those for methionine (Met3), glutamate (Aco1 and Glt1), ergosterol (Erg3, Erg35, and Ncp1), and ubiquinone (Coq7). More indirect examples include pathways with enzymes containing lipoate or biotin, cofactors whose synthesis depends on the Fe/S proteins Lip5 and Bio2, respectively (10). For example, the genes of the glycine cleavage complex, a lipoate-containing enzyme, are downregulated upon iron deprivation (47). The common theme of all these examples is the inactivation of iron-containing enzymes. This global inactivation is seen upon iron deprivation and upon defects in mitochondrial Fe/S protein maturation. Thus, transcriptional effects caused by the inactivation of iron-containing enzymes likely contribute to the significant overlap in the transcriptional response of yeast cells to these two situations (2, 20).
Most genes that are downregulated upon iron deprivation are targets of the mRNA-binding proteins Cth1 and Cth2, which promote mRNA degradation upon iron limitation (34, 37, 38, 53). Nevertheless, the effects of Cth1 and Cth2 are too small to quantitatively explain the adaptation to iron deprivation. In addition, Cth1- and Cth2-dependent mRNAs are not significantly downregulated upon activation of Aft1, despite the fact that CTH1 and CTH2 are strongly induced in Aft1-up cells or upon Aft1 overproduction (42). Consequently, cells overproducing Aft1 show little biochemical evidence for the massive metabolic remodeling that is taking place upon iron starvation, except for the induction of the genes of the yeast iron regulon (20, 42, 47). As shown here, LEU1 contains all the hallmarks of a gene targeted by the Cth proteins. Its 3′ downstream region contains potential binding sites for Cth1 and Cth2 and confers iron responsiveness on an iron-indifferent promoter. Consequently, LEU1 mRNA accumulates in cells lacking CTH1 and CTH2 in a cooperative manner (38). The role of the terminator in the iron responsiveness of LEU1 becomes evident in cells that are unable to activate Leu3 or with LEU1 core promoter constructs that lack the Leu3 activating domain. These effects are relatively small (∼4-fold), consistent with the systematic studies of Cth1 and Cth2 (37, 38). Yeast mutants lacking the LEU1 terminator retain a strong iron-responsive LEU1 expression that is completely lost by inactivating Leu3. Thus, in the absence of a terminator, the iron responsiveness of LEU1 expression is conferred exclusively by Leu3-dependent transcription. Vice versa, the Cth proteins dominate the iron-regulated expression of LEU1 when Leu3 is inactive. We conclude that α-IPM–Leu3-dependent transcription of LEU1 adds a second important level of iron-responsive regulation, and only the combination of the effects conferred by both transcriptional and posttranscriptional regulation provides a quantitative portrayal of the iron responsiveness of LEU1.
CYC1, encoding cytochrome c, is a further example of an iron-responsive gene that is subjected to dual iron regulation by transcriptional and posttranscriptional mechanisms. Several metabolic processes that involve heme-containing enzymes are repressed upon iron deprivation, including respiration (20, 37, 38, 47). In S. cerevisiae, heme is used as a sensor for oxygen and functions both as a coactivator and, occasionally, as a transcriptional repressor (5, 27, 55, 56). Our observation that CYC1 expression can be induced under iron-limiting conditions by feeding hemin demonstrates the crucial role of this cofactor for the iron responsiveness of heme-containing pathways, as suggested previously (24, 35). In S. cerevisiae, CYC1 expression is dominated by the transcriptional activator Hap1 and the Hap2-5 complex (11, 17, 46, 55). This dual regulation is shared by several other genes involved in respiration (40, 45, 55). We show that both activator systems contribute significantly to the iron responsiveness of CYC1. The heme-activated transcription factor Hap1 plays an important role in the adaptation of S. cerevisiae to hypoxia. It is therefore no surprise that most genes of the Hap1 regulon are repressed under both hypoxic and iron-deprived conditions, two conditions that result in low levels of heme (1, 37, 50, 55, 56). In this context, in cells depleted of components of the mitochondrial ISC assembly systems, many genes of the Hap1 regulon are repressed, and yet surprisingly HAP1 itself is induced (20). The fact that these cells are heme deficient underscores the finding that low heme levels are the primary physiological signal that provides the connection between the downregulation of genes of the Hap1 regulon upon hypoxia or iron deprivation and that in cells with defective mitochondrial ISC systems.
Taken together, in S. cerevisiae the regulation of those iron-responsive genes that are downregulated upon iron deprivation is conferred by two independent and likely additive mechanisms: transcriptional regulation and Cth1-Cth2-dependent posttranscriptional mRNA degradation. On the transcriptional level, the iron-responsive expression is achieved through the back door via key regulatory metabolites that are involved in activation of central transcription factors rather than directly by iron (25). Upon iron deprivation, the levels of these metabolites are reduced due to the inactivation of iron-dependent enzymes required for their biosynthesis or, in the case of heme, due to the lack of iron. The low levels of iron-dependent metabolites are then directly interpreted through diminished transcriptional activation. This indirect iron-responsive regulation likely operates in most pathways involving iron-dependent enzymes and thus easily explains the strong overlap of the transcriptomes of iron-deprived yeast cells and yeast with defects in the maturation of cellular Fe/S proteins (20). In addition, it leaves little room for additional transcription factors that sense iron by a direct mechanism similar to the Aft1 transcription factor in S. cerevisiae.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Mahnke and U. Koert for providing 2-3-dihydroxy-3-methylbutyrate.
We dedicate this paper to G. Kohlhaw.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 593, Gottfried-Wilhelm Leibniz program, and GRK 1216), von Behring-Röntgen Stiftung, Max-Planck Gesellschaft, and Fonds der chemischen Industrie.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/cgi/content/full/9/3/460/DC1.
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Becerra M., Lombardia-Ferreira L. J., Hauser N. C., Hoheisel J. D., Tizon B., Cerdan M. E. 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43:545–555 [DOI] [PubMed] [Google Scholar]
- 2.Belli G., Molina M. M., Garcia-Martinez J., Perez-Ortin J. E., Herrero E. 2004. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279:12386–12395 [DOI] [PubMed] [Google Scholar]
- 3.Camadro J. M., Labbe P. 1988. Purification and properties of ferrochelatase from the yeast Saccharomyces cerevisiae. Evidence for a precursor form of the protein. J. Biol. Chem. 263:11675–11682 [PubMed] [Google Scholar]
- 4.Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., Kaplan J. 2004. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279:29513–29518 [DOI] [PubMed] [Google Scholar]
- 5.Crisp R. J., Adkins E. M., Kimmel E., Kaplan J. 2006. Recruitment of Tup1p and Cti6p regulates heme-deficient expression of Aft1p target genes. EMBO J. 25:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Defranoux N., Gaisne M., Verdiere J. 1994. Functional analysis of the zinc cluster domain of the CYP1 (HAP1) complex regulator in heme-sufficient and heme-deficient yeast cells. Mol. Gen. Genet. 242:699–707 [DOI] [PubMed] [Google Scholar]
- 7.Diekert K., de Kroon A. I., Kispal G., Lill R. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37–51 [DOI] [PubMed] [Google Scholar]
- 8.Feller A., Dubois E., Ramos F., Pierard A. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14:6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint D. H., Emptage M. H., Finnegan M. G., Fu W., Johnson M. K. 1993. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J. Biol. Chem. 268:14732–14742 [PubMed] [Google Scholar]
- 10.Fontecave M., Ollagnier-de-Choudens S., Mulliez E. 2003. Biological radical sulfur insertion reactions. Chem. Rev. 103:2149–2166 [DOI] [PubMed] [Google Scholar]
- 11.Forsburg S. L., Guarente L. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3:1166–1178 [DOI] [PubMed] [Google Scholar]
- 12.Friden P., Schimmel P. 1988. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol. Cell. Biol. 8:2690–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friden P., Schimmel P. 1987. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol. Cell. Biol. 7:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaisne M., Becam A. M., Verdiere J., Herbert C. J. 1999. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36:195–200 [DOI] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 16.Gietz R. D., Woods R. A. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96 [DOI] [PubMed] [Google Scholar]
- 17.Guarente L., Lalonde B., Gifford P., Alani E. 1984. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 36:503–511 [DOI] [PubMed] [Google Scholar]
- 18.Guarente L., Mason T. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32:1279–1286 [DOI] [PubMed] [Google Scholar]
- 19.Harlow E., Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Hausmann A., Samans B., Lill R., Muhlenhoff U. 2008. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 283:8318–8330 [DOI] [PubMed] [Google Scholar]
- 21.Hu Y., Cooper T. G., Kohlhaw G. B. 1995. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol. Cell. Biol. 15:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 23.Kaplan C. D., Kaplan J. 2009. Iron acquisition and transcriptional regulation. Chem. Rev. 109:4536–4552 [DOI] [PubMed] [Google Scholar]
- 24.Kaplan J., Ward D. M., Crisp R. J., Philpott C. C. 2006. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim. Biophys. Acta 1763:646–651 [DOI] [PubMed] [Google Scholar]
- 25.Kohlhaw G. B. 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumanovics A., Chen O. S., Li L., Bagley D., Adkins E. M., Lin H., Dingra N. N., Outten C. E., Keller G., Winge D., Ward D. M., Kaplan J. 2008. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 283:10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwast K. E., Burke P. V., Staahl B. T., Poyton R. O. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. U. S. A. 96:5446–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalonde B., Arcangioli B., Guarente L. 1986. A single Saccharomyces cerevisiae upstream activation site (UAS1) has two distinct regions essential for its activity. Mol. Cell. Biol. 6:4690–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Clarke N. D. 2002. Rationalization of gene regulation by a eukaryotic transcription factor: calculation of regulatory region occupancy from predicted binding affinities. J. Mol. Biol. 323:1–8 [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Lee C. K., Granek J. A., Clarke N. D., Lieb J. D. 2006. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 16:1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNabb D. S., Pinto I. 2005. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 4:1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molik S., Lill R., Muhlenhoff U. 2007. Methods for studying iron metabolism in yeast mitochondria. Methods Cell Biol. 80:261–280 [DOI] [PubMed] [Google Scholar]
- 33.Ojeda L., Keller G., Muhlenhoff U., Rutherford J. C., Lill R., Winge D. R. 2006. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281:17661–17669 [DOI] [PubMed] [Google Scholar]
- 34.Pedro-Segura E., Vergara S. V., Rodriguez-Navarro S., Parker R., Thiele D. J., Puig S. 2008. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 283:28527–28535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philpott C. C., Protchenko O. 2008. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Protchenko O., Philpott C. C. 2003. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278:36582–36587 [DOI] [PubMed] [Google Scholar]
- 37.Puig S., Askeland E., Thiele D. J. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110 [DOI] [PubMed] [Google Scholar]
- 38.Puig S., Vergara S. V., Thiele D. J. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. 2008. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453:1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramil E., Agrimonti C., Shechter E., Gervais M., Guiard B. 2000. Regulation of the CYB2 gene expression: transcriptional co-ordination by the Hap1p, Hap2/3/4/5p and Adr1p transcription factors. Mol. Microbiol. 37:1116–1132 [DOI] [PubMed] [Google Scholar]
- 41.Rutherford J. C., Bird A. J. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 3:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutherford J. C., Jaron S., Winge D. R. 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636–27643 [DOI] [PubMed] [Google Scholar]
- 43.Rutherford J. C., Ojeda L., Balk J., Muhlenhoff U., Lill R., Winge D. R. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135–10140 [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45.Schneider J. C., Guarente L. 1991. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 11:4934–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuller H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139–160 [DOI] [PubMed] [Google Scholar]
- 47.Shakoury-Elizeh M., Tiedeman J., Rashford J., Ferea T., Demeter J., Garcia E., Rolfes R., Brown P. O., Botstein D., Philpott C. C. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41 [DOI] [PubMed] [Google Scholar]
- 49.Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258:1143–1145 [DOI] [PubMed] [Google Scholar]
- 50.Ter Linde J. J., Steensma H. Y. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19:825–840 [DOI] [PubMed] [Google Scholar]
- 51.Theil E. C., Goss D. J. 2009. Living with iron (and oxygen): questions and answers about iron homeostasis. Chem. Rev. 109:4568–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueta R., Fujiwara N., Iwai K., Yamaguchi-Iwai Y. 2007. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vergara S. V., Thiele D. J. 2008. Post-transcriptional regulation of gene expression in response to iron deficiency: co-ordinated metabolic reprogramming by yeast mRNA-binding proteins. Biochem. Soc. Trans. 36:1088–1090 [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi-Iwai Y., Ueta R., Fukunaka A., Sasaki R. 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277:18914–18918 [DOI] [PubMed] [Google Scholar]
- 55.Zhang L., Hach A. 1999. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zitomer R. S., Lowry C. V. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.