Abstract
Aspergillus fumigatus has three zinc transporter-encoding genes whose expression is regulated by both pH and the environmental concentration of zinc. We have previously reported that the zrfA and zrfB genes of A. fumigatus are transcribed at higher levels and are required for fungal growth under acidic zinc-limiting conditions whereas they are dispensable for growth in neutral or alkaline zinc-limiting media. Here we report that the transporter of the zinc uptake system that functions in A. fumigatus growing in neutral or alkaline environments is encoded by zrfC. The transcription of zrfC occurs divergently with respect to the adjacent aspf2 gene, which encodes an immunodominant antigen secreted by A. fumigatus. The two genes—zrfC and aspf2—are required to different extents for fungal growth in alkaline and extreme zinc-limiting media. Indeed, these environmental conditions induce the simultaneous transcription of both genes mediated by the transcriptional regulators ZafA and PacC. ZafA upregulates the expression of zrfC and aspf2 under zinc-limiting conditions regardless of the ambient pH, whereas PacC represses the expression of these genes under acidic growth conditions. Interestingly, the mode of action of PacC for zrfC-aspf2 transcription contrasts with the more widely accepted model for PacC function, according to which under alkaline growth conditions PacC would activate the transcription of alkaline-expressed genes but would repress the transcription of acid-expressed genes. In sum, this report provides a good framework for investigating several important aspects of the biology of species of Aspergillus, including the repression of alkaline genes by PacC at acidic pH and the interrelationship that must exist between tissue pH, metal availability in the host tissue, and fungal virulence.
In all organisms, zinc is required by a huge number of proteins, for which it plays a catalytic, cocatalytic, and/or structural role (2). Indeed, organisms are equipped with a plethora of zinc homeostasis-maintaining proteins, including membrane-embedded zinc transporters, zinc-chelating proteins, and at least one zinc-responsive transcriptional activator.
Among fungi, zinc homeostasis has mainly been characterized at both the genetic and biochemical levels in Saccharomyces cerevisiae (12). This yeast is able to grow in a broad pH range (between 2.4 and 8.6), although its growth rate is strongly influenced by the environmental pH. The kinetics of S. cerevisiae growth is not affected between pH 3.5 and 6.0 because it prefers acidic media (25), and all investigations of yeast zinc-homeostasis have been performed using acidic media such as those described by Zhao and Eide (56, 57). However, exposure of yeast to alkaline pH represents a stressing situation that limits its nutrient uptake ability when coupled to a proton gradient (44), thereby significantly lowering its growth rate in alkaline media. In addition, an alkaline pH favors the formation of insoluble metallic complexes, which lowers the availability of most metal ions, including iron, copper, and zinc (24). To improve its growth in alkaline media, S. cerevisiae has thus evolved a regulatory mechanism that increases the expression of several genes encoding proteins that are involved in both iron and copper homeostasis (20, 21, 36). Similarly, other genes that are to some extent related to zinc homeostasis, such as FET4 and PHO84, which respectively encode the low-affinity Fe2+/Cu2+/Zn2+ transporters (48) and high-affinity phosphate transporters (18), also increase their expression under alkaline conditions (21, 36), even though they are only relevant for zinc uptake under anaerobic and low-phosphate growth conditions, respectively (18, 48). Nevertheless, the expression of genes directly related to zinc homeostasis in yeast growing in acidic media, including that of ZRT1 and ZRT2 (56, 57), does not change upon exposure to alkaline pH, as may be deduced from large-scale expression analyses (20, 21, 36). Accordingly, yeast growth in an aerobic and phosphate-replete, alkaline medium may be improved by supplementing the medium with iron and copper but not with zinc (35). Therefore, although this has not been formally demonstrated, most zinc required by yeast to grow in alkaline media is expected to be taken up by Zrt1p and/or Zrt2p, as happens in yeast growing in acidic media (56, 57).
On the basis of previous yeast investigations, we began to study zinc homeostasis in the filamentous fungus Aspergillus fumigatus. We first showed that the ZrfA/ZrfB zinc uptake system of A. fumigatus, which somewhat resembles the Zrt1/Zrt2 zinc uptake system of S. cerevisiae, functions to obtain zinc mainly from acid, zinc-limiting environments (45). However, in contrast to S. cerevisiae, Aspergillus species exhibit strong metabolic versatility and show similar growth rates over a pH range between 3.0 and 10.5 (51). In addition, A. fumigatus is a pathogen able to grow extensively and invade lung tissue, resulting in invasive aspergillosis, which is a life-threatening infectious disease and a major cause of mortality among immunocompromised patients (34). Like many environments in which A. fumigatus may grow, lung tissue is a slightly alkaline zinc-limiting medium in which Zn2+ ions are tightly bound to high-affinity zinc-binding proteins (38). However, the zinc transporters ZrfA and ZrfB of A. fumigatus are not required for fungal growth under neutral or alkaline zinc-limiting conditions (45). Accordingly, a system other than that encoded by the zrfA and zrfB genes must exist in A. fumigatus to enable this organism to obtain zinc during growth in neutral or alkaline environments. Indeed, scrutiny of the A. fumigatus genome revealed that it actually has eight genes encoding proteins (ZrfA to ZrfH) of the ZIP (Zrt-like, Irt-like Protein) family of zinc transporters usually involved in zinc uptake from the environment (8). However, apart from the zrfA and zrfB genes, which encode the acid ZrfA-ZrfB zinc uptake system (45), only the promoter region of zrfC has cis-acting sequences for both the PacC and ZafA transcriptional regulators that respectively regulate pH and zinc homeostasis in Aspergillus species (28, 31). In this work we demonstrate that zrfC does indeed encode a transporter devoted to obtaining zinc from alkaline zinc-limiting media.
MATERIALS AND METHODS
Strains, media, and culture conditions.
All fungal strains used in this work are listed in Table 1. The A. fumigatus strains were routinely grown on either potato dextrose agar (PDA) supplemented with 100 μM Zn2+ or complete AMMH agar medium, which is the base Aspergillus minimal medium supplemented with 1.0 ml/liter of complete Hunter's trace-element solution and 15 g/liter agar (1). Conidia from A. fumigatus strains grown in either medium were used to inoculate the synthetic dextrose ammonium EDTA (SDAE) (pH 4.4) or synthetic dextrose nitrate EDTA (SDNE) (pH 7.5) zinc-limiting agar media (1, 27). These media were converted into zinc-replete agar media upon supplementation with Zn2+ at the concentrations specified for each experiment using a stock sterile solution of 100 mM ZnSO4·7H2O in ultrapure water. The liquid SDA and SDN zinc-limiting media were prepared in the same way as the SDAE and SDNE zinc-limiting agar media but with the omission of both EDTA and agar. Prewarmed SDA and SDN liquid media were inoculated to a density of 5 × 105 spores/ml and incubated at 37°C with shaking at 200 rpm. Both SDAE and SDNE agar media were spotted with 103 conidia per strain. Fungal growth reached a maximum after 5 and 3 days of incubation at 37°C in SDAE and SDNE, respectively, regardless of the zinc supplement added to the medium. Thus, SDAE and SDNE agar plates inoculated with conidia from A. fumigatus were incubated at 37°C for 5 and 3 days, respectively, before pictures were taken.
Table 1.
Fungal strains used in this study
| Microorganism | Strain | Genotype | Reference or source |
|---|---|---|---|
| A. fumigatus | CEA17 | pyrG1 (auxotrophic pyrG−) | 11 |
| AF10 | pyrG1 ΔzrfA::neo ΔzrfB::hisG-pyrG-hisG | 45 | |
| AF14 | Prototrophic wild-type (isogenic to CEA17) | 45 | |
| AF15 | pyrG1 ΔzrfA::neo ΔzrfB::hisG (auxotrophic pyrG−) | 45 | |
| AF17 | pyrG1 ΔzafA::hisG-pyrG-hisG | 28 | |
| AF52 | pyrG1 ΔzrfC::lacI (auxotrophic pyrG−) | This study | |
| AF56R | pyrG1 zafA::pyrG-hisG | 28 | |
| AF58 | pyrG1 pacCΔ1598→2215::pyrG (phenotype pacCC) | 1 | |
| AF60 | pyrG1 pacCΔ1598→1855::pyrG (phenotype pacC+/−) | 1 | |
| AF251 | pyrG1 ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI-pyrG-lacI | This study | |
| AF252 | pyrG1 ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI-pyrG-lacI | This study | |
| AF2511 | pyrG1 ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI (auxotrophic pyrG−) | This study | |
| AF431 | pyrG1 ΔzrfC::lacI-pyrG-lacI | This study | |
| AF432 | pyrG1 ΔzrfC::lacI-pyrG-lacI | This study | |
| AF731 | ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI [zrfCP → zrfC] | This study | |
| AF751 | ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI [zrfAP → zrfA] | This study | |
| AF761 | ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI [zrfBP → zrfB] | This study | |
| AF791 | ΔzrfA::neo ΔzrfB::hisG ΔzrfC::lacI [zrfCP → zrfCΔ13–622] | This study | |
| AF801 | ΔzrfC::lacI [zrfCPR → zrfC] | This study | |
| AF811 | pyrG1 Δaspf2::lacI-pyrG-lacI | This study | |
| AF812 | pyrG1 Δaspf2::lacI-pyrG-lacI | This study | |
| AF861 | pyrG1 Δaspf2::lacI (auxotrophic pyrG−) | This study | |
| AF881 | Δaspf2::lacI [aspf2P → aspf2] | This study | |
| AF891 | Δaspf2::lacI [aspf2PR → aspf2] | This study | |
| S. cerevisiae | DY1457 | MATα ade6 can1 his3 leu2 trp1 ura3 | 57 |
| ZHY3 | MATα ade6 can1 his3 leu2 trp1 ura3 zrt1::LEU2 zrt2::HIS3 | 57 |
Yeast strains were routinely grown at 28°C in YEPD agar medium (10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose, 20 g/liter agar) supplemented with 1 mM Zn2+. Yeast transformant strains derived from the ZHY3 strain were selected on SDA agar supplemented with 1 mM Zn2+ and a suitable complement supplement dropout mixture (CSM from Q-BIOgene). The SDA-buffered (SDAB) zinc-limiting agar medium used for yeast growth was the synthetic yeast nitrogen base with ammonium sulfate (5 g/liter) and without amino acids, dextrose, phosphate, Cu, Zn, or Fe (catalog no. 4028-812; Q-BIOgene) and supplemented with the appropriate complement supplement dropout mixture, 20 g/liter dextrose, 10 μM FeCl3·6H2O, 2 μM CuSO4·5H2O, and a 100 mM concentration of either KH2PO4 (for pH 4.4) or the KH2PO4-K2HPO4 buffer (for pH 7.5) and 20 g/liter agar.
In all cases, to prevent contamination of the media by traces of Zn2+ contained in salts, ultrapure compounds of the highest quality (obtained from Merck) were used. All solid media were prepared by adding agar to the salt solution before sterilization by autoclaving, whereas the glucose solution used to make up the media was autoclaved separately from the other components.
Standard molecular biology procedures.
DNA manipulations were performed following standard molecular biology protocols (32). DNA and RNA from A. fumigatus were purified and analyzed by Southern and Northern blotting, respectively, as described previously (1). The plasmids and oligonucleotides used and constructed during the course of this study are listed and briefly described in Tables S1 and S2, respectively, in the supplemental material. The probes used for Northern analyses were as follows: an EcoRI-KpnI fragment of 697 bp obtained from plasmid pZRF1g for zrfA; a StuI-SacI fragment of 929 bp from pZRF24g for zrfB; a BglII-SmaI fragment of 1,583 bp from pZRF30 for zrfC; an XbaI-BamHI fragment of 954 bp from pASPF23 for aspf2; and a HpaI-SmaI fragment of 1,605 bp from pPAC2 for pacC. We used an EcoRI-EcoRI fragment of 926 bp that carries part of the γ-actin gene (actG) coding sequence (obtained from the pACTF1 plasmid) as a loading and quality control in most of the Northern assays.
Cloning of the zrfC and aspf2 cDNAs of A. fumigatus.
The cDNA of zrfC was obtained by reverse transcription (RT)-PCR using oligonucleotides JA299 (for retrotranscription) and JA60 and JA54 (both for PCR), and a 1.6-kb cDNA fragment was subcloned into pGEM-T Easy (Promega) to generate plasmid pZRF30 and sequenced.
The cDNA of aspf2 was obtained by RT-PCR using oligonucleotides JA8 (for retrotranscription) and JA166 and JA167 (both for PCR), and a 0.96-kb cDNA fragment was subcloned into pGEM-T Easy to generate plasmid pASPF23 and sequenced.
Site-directed mutagenesis.
Mutagenesis of all pH response (PR) sites in the zrfC-aspf2 promoter region was performed sequentially by PCR using pairs of complementary oligonucleotides that contained the desired mutation as the primers and the plasmid that carried the DNA fragment to be mutated as the template. The pairs of oligonucleotides were used to mutate each PR site as follows: the pair JA222 and JA223 for PR1; JA216 and JA217 for PR2; JA224 and JA225 for PR3; JA226 and JA227 for PR4; JA220 and JA221 for PR5; and JA212 and JA213 for PR6. Pfu Turbo DNA polymerase (Roche) was used to replicate both plasmid strands with high fidelity. Following temperature cycling (15 cycles), the product was treated with DpnI to remove the parental DNA template and was used to transform Escherichia coli DH5α. The mutations were confirmed by sequencing.
Construction of the plasmids used for S. cerevisiae transformation.
The ZHY3 yeast strain was transformed with the centromeric plasmids pMC5-HSET (16), pZHA1, pZHA2, pZSF30, and pZSF310, which respectively carry the ZRT1, zrfA, zrfB, zrfC, and zrfCΔN genes under the control of the ZRT1 promoter of S. cerevisiae (55). To construct the plasmids pZSF30 and pZSF310, the ZRT1 promoter was obtained by PCR using genomic DNA from the S. cerevisiae DY1457 strain as the template and the oligonucleotides JA61 and JA62 as primers, digested with SphI and BglII, and ligated to pZRF30 digested with the same restriction enzymes to generate pZRFS3. The SacI-HpaI fragment of 2.18 kb of this plasmid was used to replace the SacI-HpaI fragment in pMC5-HSET to generate pZSF30. A PCR fragment of 1.09 kb obtained by PCR using pZRF30 as the template and the oligonucleotides JA165 and JA54 as primers was digested with BglII and HpaI and used to replace the BglII-HpaI fragment in pZSF30 to generate pZSF310. All DNA fragments obtained by PCR were sequenced. Plasmids pZSF30 and pZSF310 were used to transform the yeast ZHY3 strain.
Construction of plasmids used for A. fumigatus transformation.
To obtain a zrfCΔ null mutant and a triple zrfAΔzrfBΔzrfCΔ null mutant, the plasmid pZRF35 was constructed. This plasmid carries a transforming DNA designed to delete a 1.86-kb NheI-XmnI fragment containing the entire coding sequence of zrfC. A DNA fragment of 4.2 kb (upstream of zrfC) and one of 1.72 kb (downstream from zrfC) were obtained by high-fidelity PCR using genomic DNA from the A. fumigatus AF14 strain as the template and oligonucleotide pair JA59 and JA186 and oligonucleotide pair JA96 and JA97 as primers, respectively; the resulting DNA fragments were subcloned into pGEM-T Easy to generate the pASPF24 (4.2 kb) and pZRF33 (1.72 kb) plasmids, respectively. A 1.86-kb HpaI-NheI fragment obtained from plasmid pASPF24, containing the upstream region to zrfC, was ligated into the pPYRG4 plasmid digested with SmaI and NheI, containing the lacI-pyrG-lacI cassette, to generate plasmid pZRF34. Next, a 1.7-kb SpeI-XmnI fragment obtained from plasmid pZRF33 was ligated into pZRF34 digested with SpeI and StuI to generate plasmid pZRF35.
The plasmids pZRF19, pZRF27, pZRF313, and pZRF317 were constructed to reintroduce zrfA, zrfB, zrfC, and zrfCΔ13→622, respectively, at the pyrG locus of the A. fumigatus AF2511 strain by use of the plasmid pPYRGQ3 as a shuttle. The pPYRGQ3 plasmid was designed in our laboratory for specific reversion of the pyrG1 mutation (C756T) in A. fumigatus CEA17 (50) or in any pyrG− CEA17-derivative strain and to select pyrG+ prototrophic strains bearing one DNA fragment of interest at the pyrG locus. This plasmid carries the coding sequence of a wild-type pyrG gene that lacks 141 bp at its 5′ end (i.e., the coding sequence for residues 1 to 47 of PyrG), followed by a 312-bp fragment carrying the terminator region of the aspnd1 gene from A. nidulans (9), which exhibits strong bidirectional transcriptional termination activity (unpublished data). Next to the aspnd1 terminator, we introduced a multiple cloning site (MCS) (EheI-BglII-XbaI-NcoI-KpnI-SmaI) followed by a 931-bp fragment containing the wild-type transcriptional terminator region of pyrG and the sequence downstream. Hence, any DNA fragment can be inserted at the MCS and introduced at the pyrG locus of A. fumigatus through a double homologous recombination event. In addition, the pyrG gene in pPYRGQ3 encodes a nonfunctional orotidine 5′-monophosphate decarboxylase that prevents selection of pyrG+ prototrophic strains not bearing the DNA fragment of interest. Additionally, the aspnd1 terminator prevents the transcription of the pyrG gene from being influenced by any opposite transcriptional activation activity potentially promoted by the inserted DNA.
To construct the pZRF19 plasmid, a SpeI-EcoRV fragment of 2.67 kb from plasmid pZRF1g was ligated into pPYRGQ3 digested with XbaI and SmaI.
The pZRF27 plasmid was constructed by inserting the NheI-HpaI fragment of 1.1 kb from plasmid pZHA2 into the pPYRGQ4 plasmid digested with XbaI and SmaI. To construct the pPYRGQ4 plasmid, a 1.26-kb fragment that contained the promoter region of zrfB was obtained by high-fidelity PCR (using genomic DNA from the A. fumigatus AF14 strain as the template and the oligonucleotides JA191 and JA192 as primers), digested with SfoI and BglII, and ligated to pPYRGQ3 digested with the same restriction enzymes.
To construct the pZRF313 plasmid, a BglII-HpaI fragment of 1.73 kb from plasmid pZRF39 was ligated to pPYRGQ5 digested with BglII and SmaI. A 1.75-kb fragment containing the coding sequence of zrfC was obtained by high-fidelity PCR, using genomic DNA from the A. fumigatus AF14 strain as the template and the oligonucleotides JA54 and JA60 as primers, and was cloned in pGEM-T Easy to generate the pZRF39 plasmid. The pPYRGQ5 plasmid was generated by inserting into pPYRGQ3 digested with SfoI and BglII a 0.91-kb fragment containing the zrfC-aspf2 promoter region, which was obtained by high-fidelity PCR using genomic DNA from the A. fumigatus AF14 strain as the template and the oligonucleotides JA194 and JA196 as primers and was digested with HpaI and BglII.
The pZRF317 plasmid was constructed by inserting the BglII-HpaI fragment of 1.12 kb from plasmid pZRF316 into pPYRGQ5 digested with BglII and SmaI. A 1.14-kb fragment containing the coding sequence of zrfCΔ13→622 was obtained by high-fidelity PCR, using genomic DNA from the A. fumigatus AF14 strain as the template and the oligonucleotides JA54 and JA165 as primers, and was cloned in pGEM-T Easy to generate the pZRF316 plasmid.
To obtain an aspf2Δ null mutant, the pASPF25 plasmid was constructed by replacing the XhoI-HpaI fragment containing the entire coding sequence of aspf2 by an SmaI-XhoI fragment of 3.64 kb from pPYRG4 containing the lacI-pyrG-lacI cassette. This plasmid carries a transforming DNA designed to delete a 1.2-kb BstZ17I-XhoI fragment containing the entire coding sequence of aspf2. To reintroduce the aspf2 gene at the pyrG1 locus of an auxotrophic aspf2Δ null mutant, plasmid pASPF26 was constructed by inserting a 1.87-kb HpaI-NheI fragment from pASPF24 into pPYRGQ3 digested with SmaI and XbaI.
The plasmids pZRF320 and pASPF361 were constructed by sequentially changing the PR sites of the zrfC-aspf2 promoter region in plasmids pZRF313 and pASPF26, respectively, by site-directed mutagenesis, as described above.
All DNA fragments obtained by PCR were confirmed by sequencing. The plasmids used to transform A. fumigatus were linearized by digestion with an appropriate restriction enzyme, extracted with phenol-chloroform-isoamyl alcohol, precipitated with acetate-isopropanol, washed in 70% ethanol, resuspended in STC buffer (1 M sorbitol, 10 mM CaCl2, 10 mM Tris-HCl, pH 7.5), and used for transformation as described below.
Preparation of protoplasts and transformation and verification of A. fumigatus mutants.
Protoplasts of A. fumigatus were prepared and transformation was performed as described previously (45). Plates were incubated at 37°C until pyrG-positive fungal transformants had grown. Several 10s of independent transformants were reisolated on AMMH agar medium. To identify the transformants that had undergone homologous recombination at the expected locus, genomic DNA obtained from conidia was analyzed by PCR using appropriate oligonucleotides as described previously (45). Finally, samples of genomic DNA from about 6 to 10 independent transformants for each fungal strain preselected by PCR were digested with at least two different combinations of restriction enzymes and analyzed by Southern blotting, using an appropriate probe. Spontaneous pyrG-negative fungal strains were selected on AMMH agar medium supplemented with 200 μM ZnSO4·7H2O, 0.05% uracil, 0.12% uridine, and 1.0 mg/ml 5-fluoroorotic acid.
Alphanumeric codes for all genes from A. fumigatus analyzed in this study.
The complete genomic sequences of two A. fumigatus strains (AF293 and A1163) are now available (14, 29). The genomic DNA of the AF293 strain was sequenced first, and it is considered a reference by many investigators (29). However, the A1163 strain is a CEA17 derivative (14), as is the AF14 strain (1, 45), which is the wild-type strain used in this study. Thus, to allow rapid access to the genomic DNA for the A. fumigatus genes from either of the strains (AF293 or A1163) used in this study, the alphanumeric identification codes for every gene in each strain are presented as follows: AFUA_1G01550 (in strain AF293) or AFUB_079250 (in A1163) for zrfA, AFUA_2G03860 (in AF293) or AFUB_020930 (in A1163) for zrfB, AFUA_4G09560 (in AF293) or AFUB_066680 (in A1163) for zrfC, AFUA_4G09580 (in AF293) or AFUB_066690 (in A1163) for aspf2, AFUA_1G10080 (in AF293) or AFUB_009490 (in A1163) for zafA, and AFUA 3G11970 (in AF293) or AFUB_037210 (in A1163) for pacC.
Nucleotide sequence accession numbers.
The cDNA sequence data for both zrfC and aspf2 are available in GenBank/EMBL/DDBJ under accession numbers GQ923786 and GQ923787, respectively.
RESULTS
The zrfC gene encodes a putative zinc transporter with potential zinc-binding motifs (ZBMs) at its N terminus.
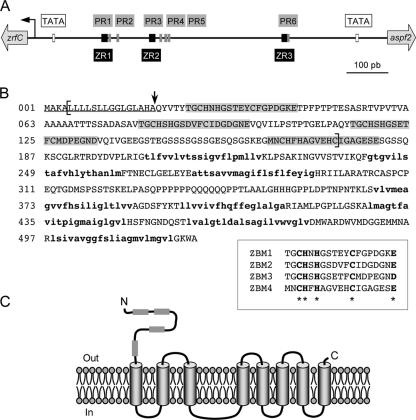
The zrfC gene of A. fumigatus has a discontinuous open reading frame of 1,720 bp with three introns of 48, 52, and 51 bp. The putative TATA box is located at position −68 (for numbering purposes, we assigned position −1 to the nucleotide preceding the A of the ATG start codon). The initiation transcription site (position +1 in the mRNA) was mapped by rapid amplification of cDNA ends at position −32. Interestingly, the ATG start codon of the aspf2 gene was located at position −885 (in the antiparallel strand). Therefore, the coding sequences of aspf2 and zrfC are divergent and separated by a DNA fragment of 884 bp. This DNA fragment includes the promoter region that drives the divergent transcription of zrfC and aspf2 and is henceforth abbreviated to zaP (for zrfC-aspf2 promoter) (Fig. 1A). In the zaP region, there are three 15-bp consensus sequences with a common 5′-CAAGGT-3′ core. These 15-bp sequences were designated ZR1 to ZR3 (for zinc response), since preliminary data obtained at our laboratory have indicated that they are cis-acting sequences required for regulating the expression of zrfC and aspf2 at the transcriptional level (unpublished data), most likely through binding of the zinc-responsive transcriptional activator ZafA (28). In addition, in the zaP region there are six PacC-like binding motifs (5′-GCCARG-3′) (42) designated PR1 to PR6 (for pH response).
Fig. 1.
Structural features of the zaP region and the ZrfC protein. (A) Schematic representation of the zaP region. The pH response sites (PR) and the putative zinc-response elements (ZR) are represented as gray- and black-shaded boxes, respectively. (B) Amino acid sequence of ZrfC. The predicted signal peptide and transmembrane domains are underlined and in bold lowercase, respectively. The four putative zinc-binding domains at the N terminus are shown separately for comparison. (C) Topological model of ZrfC insertion in the plasma membrane.
The coding sequence of zrfC encodes a 522-amino-acid protein with a predicted molecular mass of 54.3 kDa. Analysis of the amino acid sequence of the ZrfC protein performed using the dense alignment surface method (10) revealed that it contains nine putative hydrophobic transmembrane domains (Fig. 1B). The first hydrophobic domain, closer to the N terminus of ZrfC, matches a signal peptide that should be cleaved by the signal peptidase between residues A19 and Q20, as predicted by the SignalP method (version 3.0) (5). The N terminus of ZrfC extends approximately 200 residues toward the extracellular side, and it shows four repeats that define the CHXHX5CX6E/D consensus motif (Fig. 1B). These repeats were designated ZBM1 to ZBM4 (for zinc-binding motifs), since they resemble zinc-binding motifs found in other proteins (2). It is worth noting that the sequence CHFHAGVEHC within ZBM4 has remained unchanged throughout evolution in all ZrfC-like proteins deposited at the GenBank database to date, indicating a highly conserved function. The remaining transmembrane domains cross the membrane eight times, and, as predicted by the “positive-inside” rule (46), the mature form of ZrfC is most likely a membrane-embedded protein with both the N and C termini located on the outer surface of the membrane (Fig. 1C).
A search in the GenBank database performed using the BLASTP algorithm revealed that the ZrfC of A. fumigatus has the highest overall identity with EAW10413 from Aspergillus clavatus (71.6% identity), BAE55941 from A. oryzae (68.5%), and EAA64998 from A. nidulans (60.5%). Nevertheless, ZrfC also exhibits an overall identity of between 36% and 44% with other proteins from filamentous fungi (e.g., Neurospora crassa), polymorphic fungi (e.g., Candida albicans), and some yeasts (e.g., Debaryomyces hansenii). Interestingly, no orthologues to ZrfC have been found either in humans or in nonpathogenic yeasts such as S. cerevisiae, Schizosaccharomyces pombe, or Kluyveromyces lactis. In addition, a multiple alignment analysis performed with Clustal X (41) revealed that all ZrfC-like proteins found in the GenBank database are closely related to the ZIP-I subfamily of zinc transporters. However, in contrast to most proteins of the ZIP-I subfamily, ZrfC-like proteins have a signal peptide followed by a long N terminus, typically with four ZBMs (although they may contain three to seven ZBDs). Besides, a phylogenetic analysis revealed that all ZrfC-like proteins cluster close to ZIP-I proteins from fungi (e.g., Zrt2 and ZrfB) but are clearly separated from these (see Fig. S1 in the supplemental material). Thus, ZrfC-like proteins might represent a new subfamily of ZIP proteins, with members exclusively distributed among fungi.
The zrfC and aspf2 genes are expressed only in A. fumigatus growing in alkaline zinc-limiting media.
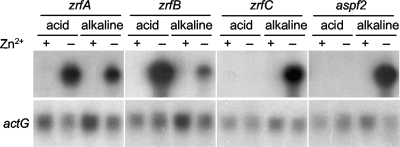
The presence of six PR motifs and three ZR sites arranged in a precise order in the zaP region suggested that expression of zrfC and that of aspf2 might be coregulated at the transcriptional level in response to pH and zinc availability. Indeed, Northern blot analysis revealed that both zrfC and aspf2 were expressed only under alkaline zinc-limiting conditions whereas zrfA and zrfB were expressed at a higher level in acidic zinc-limiting media (Fig. 2), as reported previously (45). Thus, the transcriptional profile of zrfC and aspf2 suggests that these genes might play a specific role in fungal growth under neutral or alkaline zinc-limiting conditions.
Fig. 2.
Analysis by Northern blotting of zrfA, zrfB, zrfC, and aspf2 transcription under both zinc-limiting and zinc-replete acidic and alkaline conditions. The AF14 strain was grown for 20 h at 37°C in acid SDA medium (pH 4.4) or alkaline SDN medium (pH 7.5) with a supplement of 100 μM Zn2+ (+) or without a supplement of Zn2+ (−), as indicated at the top of each lane.
ZrfC functions as a zinc transporter in yeast in alkaline zinc-limiting media but not under acidic growth conditions.
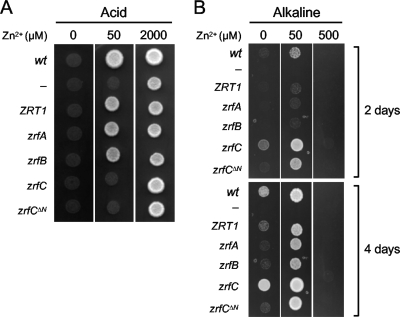
We speculate that the activity of ZrfC might be devoted to fully satisfying fungal zinc requirements under neutral or alkaline zinc-limiting conditions but not under acidic growth conditions. To test this hypothesis in a rapid and feasible way, a yeast complementation assay was performed in the genetic background provided by a zrt1Δzrt2Δ strain of S. cerevisiae (ZHY3). This yeast strain was not able to grow under acidic zinc-limiting conditions unless a large amount of zinc was added to the medium (57). The ability of ZHY3 expressing zrfC to grow was compared with that of ZHY3 expressing zrfA or zrfB from A. fumigatus or ZRT1 from S. cerevisiae. In addition, the most remarkable structural difference between ZrfC and the zinc transporters Zrt1, ZrfA, and ZrfB is that the former has an N-terminal tail with four putative ZBMs. Hence, to investigate the role of the ZrfC N terminus in the functionality of the ZrfC transporter, a deleted zrfC coding sequence (zrfCΔN) encoding a ZrfC protein (ZrfCΔ5→174) without its N terminus (the bracketed amino acid sequence in Fig. 1B) was also expressed in ZHY3. To prevent any misinterpretation of the data due to differences at the transcriptional level, each gene was expressed from a centromeric plasmid under the control of the ZRT1 promoter, as reported previously (28, 45). The acidic zinc-limiting medium (pH 4.4) was supplemented with 1 mM EDTA (SDAEB4) to further limit zinc availability. The ability of ZHY3 derivative strains expressing ZRT1, zrfA, zrfB, zrfC, or zrfCΔN to grow was tested with zinc-depleted SDAEB4 medium supplemented with increasing amounts of Zn2+ (from 0 to 2,000 μM). As shown, expression of either zrfC or zrfCΔN failed to restore the growth ability of ZHY3 in the acidic zinc-limiting medium (i.e., medium supplemented with 50 μM Zn2+) (Fig. 3A), which indicates that ZrfC does not exhibit zinc transport activity under these culture conditions. However, if ZrfC functions as a zinc transporter that operates only under alkaline conditions, it would be expected that expression of zrfC would restore the ability of the yeast to grow under alkaline zinc-limiting conditions. To test this hypothesis, all strains were cultured in a medium with the same composition of SDAEB4 except that it was buffered at pH 7.5 and EDTA was added at a final concentration of 0.1 mM. This concentration of EDTA was seen to be optimal for the control of zinc availability at pH 7.5 in the 0 to 100 μM Zn2+ range (a higher concentration of Zn2+ was toxic for yeast cells, even in the presence of 0.1 mM EDTA, whereas increasing the concentration to 1 mM EDTA in the medium inhibited yeast growth even in the presence of 2,000 μM Zn2+). As shown in Fig. 3B, expression of zrfC fully restored the growth ability of the ZHY3 strain in alkaline zinc-limiting media (i.e., medium without a supplement of Zn2+). Furthermore, this strain expressing zrfC grew even faster than the wild type, which carries both the ZRT1 and ZRT2 genes. In contrast, ZHY3 expressing only one of the zinc transporter-encoding genes (i.e., ZRT1, zrfA, or zrfB) grew less well than the wild type, whereas the ZHY3 strain transformed with the empty vector was unable to grow in alkaline zinc-replete medium. This indicates that both genes—ZRT1 and ZRT2—provide wild-type yeast cells with the full ability to take up the zinc that is required for them to grow optimally in alkaline media, an aspect of yeast physiology that, although expected, has not been reported previously. Finally, it was observed that the expression of zrfCΔN improved yeast growth to the wild-type level only with alkaline non-zinc-limiting medium (i.e., medium supplemented with 50 μM Zn2+), which indicates that the N terminus of ZrfC does enhance the zinc transport activity of the membrane-embedded part of ZrfC, particularly under extreme zinc-limiting conditions.
Fig. 3.
Functional analysis of zrfC in the S. cerevisiae background. The yeast strain ZHY3 was transformed with derivative pRS316 plasmids carrying, under the control of the ZRT1 promoter, the coding sequence of ZRT1 (pMC5-HSET), zrfA (pZHA1), zrfB (pZHA2), zrfC (pZSF30), or zrfCΔ13→622 (zrfCΔN) (pZSF310), as indicated on the left side of each picture. The DY1457 strain transformed with pRS316 is formally considered to be a wild type (wt). A total of 104 yeast cells were spotted onto SDA zinc-limiting agar plates buffered at pH 4.4 (A) or 7.5 (B) with 100 mM potassium phosphate and supplemented with Zn2+ at the specified concentrations (0 to 2,000 μM). Acid plates were supplemented with 1 mM EDTA and incubated for 2 days at 28°C. Alkaline plates were supplemented with 0.1 mM EDTA and incubated for 2 and 4 days at 28°C.
In sum, as suggested from data obtained using a yeast complementation assay, the zrfC gene of A. fumigatus encodes a protein involved in zinc uptake from alkaline media and the ZrfC zinc transport activity is enhanced by its N terminus, particularly under zinc-limiting conditions.
A. fumigatus requires zrfC to grow under alkaline and extreme zinc-limiting conditions.
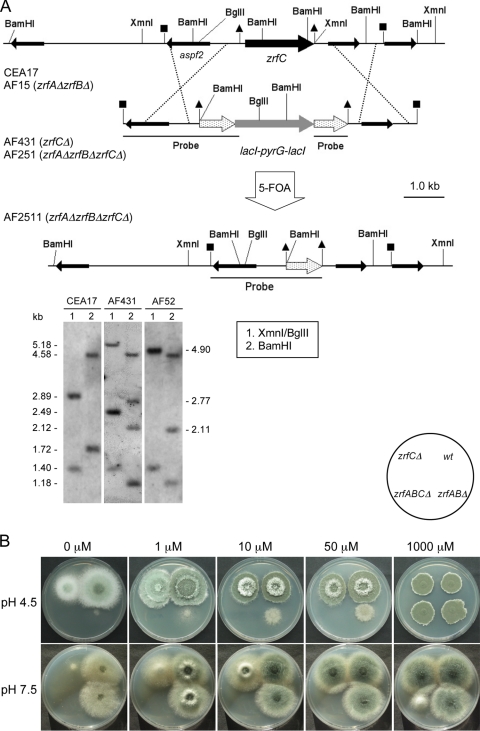
To investigate the role of zrfC in the growth of A. fumigatus, protoplasts of both the CEA17 and AF15 (zrfAΔzrfBΔ) uridine-uracil-auxotrophic strains were transformed to generate the prototrophic zrfCΔ (AF431) and zrfAΔzrfBΔzrfCΔ (AF251) null strains, respectively (Fig. 4A). The ability of these strains to grow was tested on acid and alkaline agar media under both zinc-limiting and zinc-replete conditions (Fig. 4B). The growth of the zrfCΔ null strain (AF431) was more impaired in alkaline than in acidic zinc-limiting medium. The growth ability of the zrfAΔzrfBΔzrfCΔ null strain (AF251) was abolished under both acidic and alkaline zinc-limiting conditions, and the strain hardly grew at all in the alkaline zinc-replete medium, whereas it grew at the wild-type level in the acidic zinc-replete medium. However, the zrfAΔzrfBΔ null mutant (AF10) grew poorly in acidic zinc-limiting medium but identically to the wild type in alkaline zinc-limiting medium, as reported previously (45). Therefore, the genes zrfA and zrfB are dispensable for fungal growth in alkaline, zinc-limiting medium in the presence of zrfC. In contrast, the zrfC gene was dispensable for fungal growth in acidic zinc-limiting medium in the presence of zrfA and zrfB. However, deleting zrfC is harmful to fungal growth, even under alkaline zinc-replete conditions in the absence of zrfA and zrfB, which indicates that either zrfA or zrfB (or both) might also contribute to zinc uptake in alkaline zinc-replete environments. Similarly, deleting both zrfA and zrfB is harmful to fungal growth under acidic and mild zinc-limiting conditions (i.e., media supplemented with ≤50 μM Zn2+) in the absence of zrfC, which indicates that zrfC must express in acidic zinc-limiting media at a very low level (not detectable by Northern blotting) and that ZrfC might also contribute to zinc uptake from acidic, mildly zinc-limiting media.
Fig. 4.
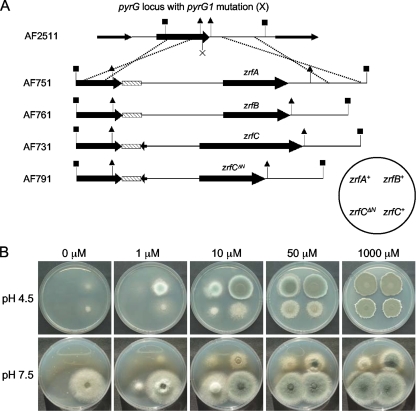
Construction and phenotypic analysis of zrfCΔ strains. (A) A 1.86-kb NheI-XmnI DNA genomic fragment containing the complete coding sequence of zrfC in the CEA17 and AF15 strains (delimited with triangles) was replaced by the lacI-pyrG-lacI cassette (gray arrow flanked by dotted arrows) in the zrfCΔ (AF431) and zrfAΔzrfBΔzrfCΔ (AF251) strains by the use of a 7.22-kb DNA fragment (delimited with closed squares) obtained from plasmid pZRF35 as transforming DNA. The thinner arrows indicate putative open reading frames surrounding the zrfC gene. The pyrG gene of AF251 was removed by spontaneous intrachromosomal recombination to generate the uridine-uracil-auxotrophic strain AF2511, in which the zrfC coding sequence had been replaced by only one lacI fragment (dotted arrow delimited with triangles). All strains harbored the correct integration event at the zrfC locus, as verified by Southern blotting analyses, using as a probe a DNA fragment obtained by PCR with the oligonucleotide pair JA8 and JA26 and plasmid pZRF35 as the template. Only relevant restriction sites are indicated. The source of the genomic DNA, the restriction enzymes used in the digestions, and the sizes of the fragments detected that match the expected sizes are specifically indicated in each panel. (B) Growth of A. fumigatus strains AF431 (zrfCΔ), AF10 (zrfAΔzrfBΔ), and AF251 (zrfAΔzrfBΔzrfCΔ) on both acid (SDAE; pH 4.5) and alkaline (SDNE; pH 7.5) zinc-limiting agar media not supplemented with Zn2+ or supplemented with 1 to 1,000 μM Zn2+, as indicated at the top of each panel.
To ascertain the role of zrfA, zrfB, or zrfC in the growth of A. fumigatus more precisely, each gene was reintroduced at the pyrG1 locus of the AF2511 strain (a spontaneous pyrG1 uridine-uracil-auxotrophic zrfAΔzrfBΔzrfCΔ mutant obtained from strain AF251 as shown in Fig. 4A) to generate the prototrophic strains zrfA+zrfBΔzrfCΔ (AF751), zrfAΔzrfΔB+zrfCΔ (AF761), and zrfAΔzrfBΔzrfC+ (AF731). In addition, to investigate whether the zinc transport activity of ZrfC was influenced by its N terminus, a deleted zrfC gene (zrfCΔ13→622) encoding a ZrfC protein without its N terminus (ZrfCΔ5→174) was also introduced at the pyrG1 locus of strain AF2511 to generate the prototrophic strain zrfAΔzrfBΔzrfCΔ13→622 (AF791) (Fig. 5A). The ability of these strains to grow was tested under acid and alkaline zinc-limiting and zinc-replete conditions (Fig. 5B). As expected, the zrfC+ strain (AF731) grew like strain zrfAΔzrfBΔ (AF10) under each set of conditions tested (compare Fig. 4B and 5B). Under all conditions tested, the zrfB+ strain (AF761) grew better than the zrfA+ strain (AF751), which did not grow at all under either alkaline or acidic, extreme zinc-limiting conditions (i.e., in medium supplemented with ≤1 μM Zn2+). In addition, the zrfB+ strain grew better than the zrfC+ strain in the acidic medium whereas it grew worse than zrfC+ in the alkaline medium. Therefore, the ability of A. fumigatus to grow in the alkaline zinc-limiting medium mainly depends on the zrfC gene and, to a lesser extent, on that of the zrfB gene. In contrast, the ability of A. fumigatus to grow in the acidic zinc-limiting medium primarily depends on the zrfB gene and secondarily on that of the zrfA gene. The zrfA and zrfC genes contribute minimally to fungal growth under alkaline and acidic zinc-limiting conditions, respectively. Additionally, it was observed that the N terminus of ZrfC is required for enhancement of fungal growth ability, particularly under extreme to mild zinc-limiting conditions (i.e., medium supplemented with ≤10 μM Zn2+).
Fig. 5.
Construction and phenotypic analyses of AF2511-derivative pyrG+ strains of A. fumigatus. (A) Construction of A. fumigatus strains that express zrfA (AF751), zrfB (AF761), zrfC (AF731), or zrfCΔ13→622 (zrfCΔN; AF791) at the pyrG locus. All strains harbored the correct integration event, as verified by Southern blotting analyses (not shown). The dashed boxes represent the terminator of the aspnd1 gene, as described in Materials and Methods. (B) Growth of the same strains on both acid (SDAE, pH 4.5) and alkaline (SDNE, pH 7.5) zinc-limiting agar media not supplemented with Zn2+ or supplemented with 1 to 1,000 μM Zn2+, as indicated at the top of each panel.
Expression of both zrfC and aspf2 is induced in zinc-limiting media by the ZafA zinc-responsive transcriptional activator.
The ZafA protein of A. fumigatus activates the transcription of the zrfA and zrfB genes under acidic zinc-limiting conditions (28). Therefore, it would be expected that the transcription of these genes as well as of others related to zinc homeostasis in A. fumigatus would also be regulated by ZafA under alkaline zinc-limiting conditions. To check this, the expression of the zrfC and aspf2 genes was analyzed using Northern blotting in a zafAΔ null strain grown in an alkaline zinc-replete and zinc-limiting medium (Fig. 6). Neither zrfC nor aspf2 was transcribed in a zafAΔ strain, which indicates that expression of these genes under alkaline zinc-limiting conditions is induced by the ZafA zinc-responsive transcriptional activator.
Fig. 6.
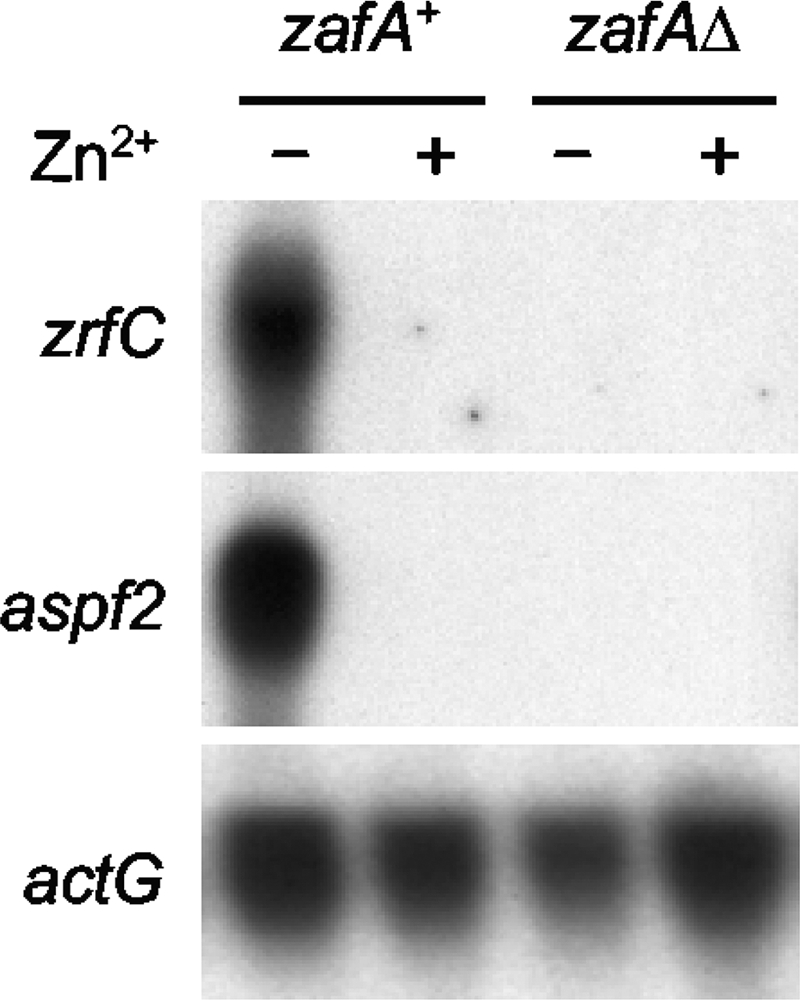
Transcription analysis by Northern blotting of zrfC and aspf2 in the A. fumigatus zafAΔ null (AF17) and zafA+ revertant (AF56R) strains grown for 20 h at 37°C in the alkaline SDN medium with a supplement of 100 μM Zn2+ (+) or without a supplement of Zn2+ (−), as indicated at the top of each lane.
Environmental pH influences the transcription of zrfC and aspf2 through the PacC transcription factor of A. fumigatus.
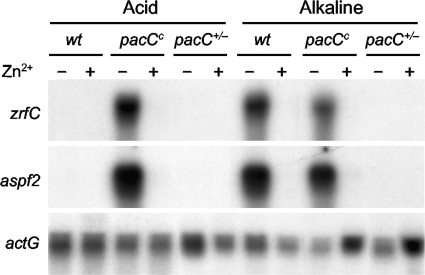
The presence of PR cis-acting sites in the zaP region suggested that the induction of the transcription of zrfC and aspf2 under alkaline zinc-limiting conditions probably depended on the PacC transcriptional regulator of A. fumigatus as well. To investigate this possibility, the transcription profile of these genes was analyzed by Northern blotting in acidity-mimicking pacC+/− (AF60) and alkalinity-mimicking pacCc (AF58) mutants (1). The expression profile displayed by the pacCc mutant for these genes under acidic and alkaline growth conditions was identical to that of a wild-type strain grown in the alkaline medium (Fig. 7). In contrast, the transcription profile of the zrfC and aspf2 genes displayed by the pacC+/− mutant under acidic and alkaline growth conditions was identical to that of a wild-type strain grown in acidic medium. Therefore, the pH-regulated transcription of the zrfC and aspf2 genes depends on PacC.
Fig. 7.
Transcription analysis by Northern blotting of zrfC and aspf2 in the pacCc (AF58) and pacC+/− (AF60) strains grown in either the acid SDA or alkaline SDN medium for 20 h at 37°C, both with a supplement of 100 μM Zn2+ (+) or without a supplement of Zn2+ (−), as indicated at the top of each lane.
The aspf2 gene is required for fungal growth in extreme zinc-limiting media, particularly under alkaline conditions.
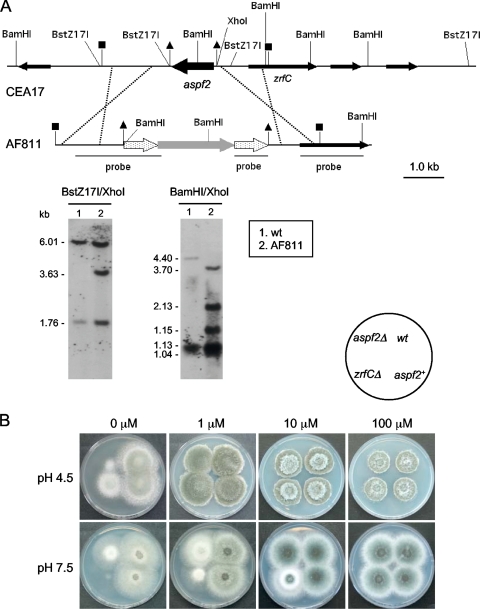
The zrfC and aspf2 genes exhibited identical transcription patterns under all conditions tested. Hence, if zrfC encodes a zinc transporter required for fungal growth in alkaline zinc-limiting media, it is likely that aspf2 would be also involved in maintaining zinc homeostasis under these environmental conditions. Accordingly, to investigate the role of Aspf2 in the biology of A. fumigatus, the aspf2 gene was deleted in the CEA17 strain to generate the prototrophic aspf2Δ null mutant (AF811) (Fig. 8A). To confirm that the phenotypic traits observed in the aspf2Δ mutant were specifically linked to the deletion of aspf2 rather than to any undesired mutation, the aspf2 gene was reintroduced at the pyrG1 locus of strain AF861, a spontaneous pyrG1 uridine-uracil-auxotrophic aspf2Δ mutant obtained from strain AF811, to generate the prototrophic AF881 aspf2+ revertant strain. The growth capacity of an aspf2Δ mutant was reduced under both acidic and alkaline extremely zinc-limiting conditions (Fig. 8B). In addition, it was consistently observed that the aspf2Δ mutant grew poorly under acidic and extreme zinc-limiting conditions compared to a zrfCΔ mutant. In contrast, a zrfCΔ mutant hardly grew under alkaline and extreme zinc-limiting conditions compared to an aspf2Δ strain. However, the growth ability of aspf2Δ could be restored to the wild-type level in the acidic medium supplemented with a minute amount of zinc (1 μM Zn2+) whereas an amount of zinc 10-fold higher was required for the aspf2Δ mutant to grow at the wild-type level in the alkaline medium. Therefore, expression of aspf2 is required at a very low level (not detectable by Northern blotting) for optimal fungal growth in acidic and extreme zinc-limiting media whereas it is required at a higher level for optimal fungal growth under alkaline zinc-limiting conditions, as for zrfC. In sum, the Aspf2 protein plays a role in zinc homeostasis when fungus grows in extreme zinc-limiting media, particularly under alkaline conditions.
Fig. 8.
Construction and phenotypic analysis of aspf2Δ strains. (A) A 1.2-kb BstZ17I-XhoI DNA genomic fragment containing the complete coding sequence of aspf2 in CEA17 (delimited with triangles) was replaced by the lacI-pyrG-lacI cassette (gray arrow flanked by dotted arrows) in the aspf2Δ (AF811) strain by the use of a 6.65-kb DNA fragment obtained from plasmid pASPF25 as transforming DNA. All strains harbored the correct integration event at the aspf2 locus, as verified by Southern blotting using as a probe a mixture of a DNA fragment obtained by PCR with the oligonucleotide pair JA187 and JA26 and plasmid pASPF25 as the template and a SmaI-BglII fragment obtained from plasmid pZRF39. Only relevant restriction sites are indicated. The source of the genomic DNA, the restriction enzymes used in the digestions, and the sizes of the fragments detected that match the expected sizes are specifically indicated in each panel. (B) Growth of A. fumigatus strains AF811 (aspf2Δ), AF881 (aspf2+), and AF431 (zrfCΔ) on both acid (SDAE; pH 4.5) and alkaline (SDNE; pH 7.5) zinc-limiting agar media not supplemented with Zn2+ or supplemented with 1 to 100 μM Zn2+, as indicated at the top of each panel.
PacC does not activate the transcription of either zrfC or aspf2 at alkaline pH but represses it at acidic pH.
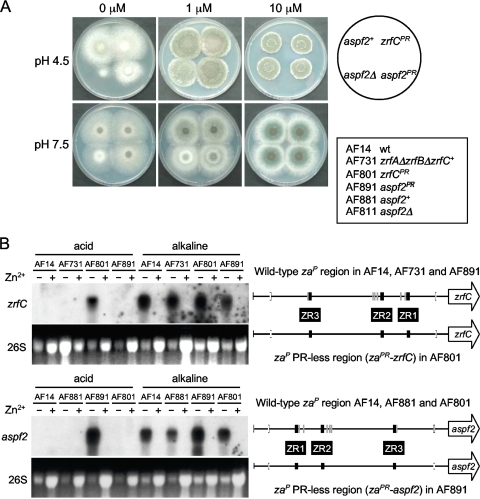
The transcription profiles of zrfA, zrfB, zrfC, and aspf2 indicated that ZafA must adopt an active conformation under zinc-limiting conditions, enabling it to induce gene expression regardless of the ambient pH. Similarly, the transcription profile of these genes under alkaline zinc-limiting conditions indicates that PacC apparently induces the transcription of zrfC and aspf2 whereas it represses the transcription of the zrfA and zrfB genes that are mainly expressed in acidic, zinc-limiting media (1). However, if ZafA is active under zinc-limiting conditions, why is it not able to induce the expression of zrfC and aspf2 under acidic zinc-limiting conditions? To answer this, we surmised that the arrangements and locations of most PR sites with respect to the ZR elements in the zaP region (PR1 and PR2 are respectively 2 bp and 22 bp from ZR1; PR3, PR4, and PR5 are located respectively 10 bp, 22 bp, and 30 bp from ZR2) (Fig. 1) could enable a physical interaction between ZafA and PacC when bound to their cognate sequences in the DNA, such that under acidic zinc-limiting conditions the transcriptional activation activity of ZafA might be negatively influenced by PacC, resulting in the repression of zrfC and aspf2 transcription. Accordingly, to investigate whether PacC binding to zaP really did mediate repression, the PacC binding capacity of this promoter was abolished by site-directed mutagenesis, taking into consideration the investigations carried out elsewhere (13). Thus, the six 5′-GCCARG-3′ PR cis-acting elements of the zaP region were converted into 5′-GCGTRG-3′ sequences that do not bind PacC (where the underlined sequence represents the conversion). Either the zrfC or the aspf2 coding sequence under the control of the PR-less zaP region (abbreviated to zaPR) was inserted at the pyrG locus of the AF52 (a spontaneous pyrG1 uridine-uracil-auxotrophic zrfCΔ mutant obtained from strain AF431) and AF861 (aspf2Δ) mutants to generate the prototrophic zrfC+ (AF801) and aspf2+ (AF891) strains, respectively. These strains were grown in an acidic or alkaline medium under either zinc-replete or zinc-limiting conditions, and the transcription of both the zrfC and aspf2 genes was analyzed by Northern blotting (Fig. 9). The AF801 strain grew at the wild-type level under all conditions tested, whereas the growth capacity of strain AF891 was consistently slightly reduced in an acidic and extreme zinc-limiting medium compared to the wild-type growth ability of the AF881 strain expressing aspf2 with a wild-type zaP region. This indicated that expression of aspf2 in acidic zinc-limiting media at the same level as in alkaline zinc-limiting media negatively influences fungal growth in the former (Fig. 9A). The transcriptional analysis of zrfC and aspf2 revealed that zrfC (in AF801) and aspf2 (in AF891) were expressed equally in acidic and alkaline zinc-limiting media whereas the expression of aspf2 (in AF801) and zrfC (in AF891) occurred at the wild-type strain level (Fig. 9B). This led us to conclude that the repression of these genes under acidic zinc-limiting conditions in strains with a wild-type zaP region must be mediated by PacC binding to their cognate PR sites, thereby negatively interfering with the ZafA activity. In addition, it also indicated that, under alkaline zinc-limiting conditions, the PacC protein does not influence, either positively or negatively, ZafA activity with respect to the transcription of either zrfC or aspf2 (i.e., PacC is irrelevant for zrfC-aspf2 transcription under alkaline zinc-limiting conditions).
Fig. 9.
Influence of zrfC or aspf2 expression on the growth ability in acidic media of the AF801 (zaPR → zrfC) and AF891 (zaPR → aspf2) strains, respectively, and transcription of zrfC and aspf2 in both strains. (A) Growth of AF801 and AF891 on both acid (SDAE; pH 4.5) and alkaline (SDNE; pH 7.5) zinc-limiting agar media not supplemented with Zn2+ or supplemented with 1 or 10 μM Zn2+, as indicated at the top of each panel. (B) Analysis by Northern blotting of the transcription of zrfC and aspf2 under the control of the zaPR region in the AF801 and AF891 strains. These strains were constructed following the same strategy used previously to construct prototrophic pyrG+ strains at the pyrG1 locus of A. fumigatus but using a fragment contained in either the pZRF320 (for zrfC) or pASPF361 (for aspf2) plasmid as transforming DNA. The expression levels of zrfC in the AF891 strain and aspf2 in AF801 were analyzed as endogenous controls for the correct functioning of the wild-type zaP regions carried by these strains. Expression of zrfC in the AF731 strain and that of aspf2 in AF881 were also analyzed as additional controls, since these strains also express at the pyrG loci zrfC and aspf2, respectively, but under the control of a wild-type zaP region. The zaPR region that drives transcription of the tested gene is depicted on the right side of each blot. All fungal strains were grown in either the acid SDA or alkaline SDN medium for 20 h at 37°C with a supplement of 100 μM Zn2+ (+) or without a supplement of Zn2+ (−), as indicated at the top of each lane.
DISCUSSION
Microorganisms adapted to grow in neutral or alkaline environments have to deal with problems related to both the availability and toxicity of essential cations, including Zn2+. Free Zn2+ ions tend to form insoluble metal oxides and phosphates as pH increases, resulting in decreased solution-phase Zn2+ concentrations (4, 24, 33). Therefore, microbial zinc availability is highly conditioned by the environmental pH, such that for a given zinc-limiting medium (e.g., a defined medium without a supplement of zinc), the amount of readily available zinc (i.e., free Zn2+) is much lower in the medium buffered at an alkaline pH than in medium buffered at an acidic pH. In addition, increasing the pH also raises the toxicity of zinc for microorganisms (17). The mechanism by which pH increases zinc toxicity has not been well established, but it may involve the adsorption of large amounts of zinc on the cell surface (under conditions of greater acidity, protons would compete with Zn2+ for binding sites) (33) and/or the formation of toxic species of soluble hydroxylated zinc (e.g., ZnOH+) (17). In this work, we focused on how A. fumigatus, a fungal pathogen that may readily adapt to grow in neutral or alkaline environments, obtains Zn2+ from alkaline zinc-limiting environments by means of ZrfC.
In contrast to most proteins of the ZIP-I subfamily, including ZrfA and ZrfB, whose overall similarity to ZrfC is rather low (∼19%), ZrfC and all ZrfC-like proteins have a signal peptide followed by a long N terminus that clearly differentiates them from typical ZIP-I transporters. Interestingly, these structural features are also present in the zinc transporters of the LZT subfamily (40). However, ZrfC-like proteins lack the metalloprotease signature (HEXPHE) that characterizes proteins of the LZT subfamily in their transmembrane V domain (40), whereas LZT proteins lack the CHFHAGVEHC motif that characterizes all ZrfC-like proteins. In light of this, we propose that the ZrfC-like proteins should be considered a new subfamily of ZIP transporters characterized by having as a signature the CHFHAGVEHC motif located at the N terminus, close to transmembrane I domain.
We speculate that, in neutral or alkaline environments, Zn2+ ions would bind to the ZBMs of the ZrfC N terminus with high affinity, thereby increasing by severalfold the zinc uptake capacity of the membrane-embedded part of ZrfC, which, as deduced from a yeast complementation assay, would exhibit zinc-uptake activity even in the absence of the N terminus. The binding of Zn2+ ions to the ZBMs could induce the proper folding of the N terminus either to interact with the membrane-embedded part of ZrfC responsible for zinc transport across the plasma membrane or to promote the formation of a Zn2+ bridge between ZrfC molecules, leading to the formation of homo-oligomers and thereby stimulating its endocytosis. Indeed, a similar mechanism has been described for proteins involved in neuronal copper and zinc homeostasis, such as the PrP prion protein (49), which is endocytosed upon the proper folding of the metal-binding domain present at its N terminus induced by binding to either Cu2+ or Zn2+ ions at neutral pH (19, 30, 53). Nevertheless, it is also possible that the endocytosis of ZrfC might be stimulated through a Zn2+-mediated interaction with the putative ZBM present in other proteins. In this regard, Aspf2 would be a good candidate, since it exhibits at its C terminus a CHTHXGX2HC motif (abbreviated to CH motif) (9) that resembles the CHFHAGVEHC signature within the ZBM4 of ZrfC. The CH motif is highly conserved in most Aspf2-like proteins, but it is absent in Zps1 (the S. cerevisiae Aspf2 orthologue). Thus, if the true function of the CH motif is to mediate the interaction with ZrfC through a Zn2+ bridge, it would not be surprising that Zps1 lacks the CH motif, since the yeast does not have a ZrfC orthologue.
Another question to be investigated is the mechanism underlying the regulation of zrfC-aspf2 expression by ZafA and PacC at the transcriptional level. It should be recalled that the arrangement of the ZR and PR elements in the zaP region is quite different from that seen in the promoter regions of zrfA or zrfB, in which there are several ZR elements clustered upstream from the TATA box but only one PR site (1, 45). In contrast to the zrfA and zrfB promoters, between the TATA boxes of zrfC and aspf2 there are two ZR-PR clusters, each formed by one ZR and two (or three) PR elements (Fig. 1). In addition, given the proximity between the ZR and PR sites in the zaP region, it is likely that a ZafA molecule bound to any ZR motif would interact physically with a PacC molecule bound to either adjacent PR site, in similarity to other transcriptional activators bound to neighboring DNA sites that interact with each other (26, 58). Indeed, a physical interaction between Zap1 and Rim101 (the yeast ZafA and PacC orthologues, respectively) may occur in vivo, as detected in a large-scale yeast two-hybrid screening (43). Nevertheless, the Zap1-Rim101 interaction detected by this means most likely does not reflect a genuine physiological interaction, since two-hybrid experiments have been performed on acidic zinc-replete media where neither Zap1 (7, 47) nor Rim101 (22, 52) was in an active conformation. Indeed, expression of ZPS1 preferentially occurs under alkaline zinc-limiting conditions and is induced directly by Zap1 but indirectly by the Rim101-processed form (i.e., the active form of Rim101) (20–23) and hence does not involve a Zap1-Rim101 interaction. However, if ZafA and PacC do interact under physiological conditions, the actual situation might be far more complex, considering the mode of action of the different forms of PacC, referred to as PacC72, PacC53, and PacC27 (31). In addition, in contrast to what has been described for other neutral and alkaline expressed genes, PacC does not activate zrfC-aspf2 expression under alkaline zinc-limiting conditions. Furthermore, under these environmental conditions PacC is not necessary for zrfC-aspf2 transcription. Therefore, this finding indicates that not all genes expressed in alkaline media are actually induced by PacC; at the same time, it raises a much more intriguing question concerning the repression of zrfC-aspf2 transcription under acidic growth conditions. In this regard, PacC53 could play a key role in zrfC repression in acidic media. Indeed, transcriptional repression is the main biological function of the Rim101-processed form (i.e., the analogous yeast form of PacC53) and may be exerted either indirectly (e.g., repression of ZPS1) or directly (e.g., repression of RIM8) (20). Thus, PacC53 might bind to PR sites to interact negatively with ZafA, thereby directly repressing zrfC-aspf2 transcription. In addition, as suggested for Rim101 (20), the repression by PacC53 induced by the presence of PR sites might also depend upon the context and orientation of ZR and PR elements within the zaP region, and a single PR site may not be sufficient to mediate PacC repression, which could explain why zrfA is not subject to PacC repression. In sum, study of the transcriptional regulation of zrfC and aspf2 has revealed that PacC appears to function as a repressor of alkaline-expressed genes under acidic growth conditions by influencing the transcriptional activation activity of ZafA; at the same time, it has provided a good framework for further investigation of the role of the PacC53 conformation in the physiology of Aspergillus species.
Readily available free Zn2+ ions are scarce in living tissue, since they are tightly bound to proteins (38). In addition, under physiological conditions the pH in normal tissues is 7.1 to 7.4 (37), which further contributes to restricting microbial zinc availability. Accordingly, the capacity of A. fumigatus to grow in lung tissue and cause tissue damage depends to a large extent on the presence of both the zinc-responsive transcriptional activator ZafA (28) and the pH-responsive transcriptional regulator PacC (6). Therefore, expression of downstream targets of these factors might be required for the pathogenicity of A. fumigatus. However, this does not indicate that all genes whose transcription is regulated by these factors are required for A. fumigatus virulence. For instance, transcription of both the zrfA and zrfB genes is regulated by ZafA and PacC. However, ZafA induces the transcription of these genes under zinc-limiting conditions regardless of ambient pH (28) whereas PacC partly represses it under alkaline zinc-limiting conditions (1). Thus, it would be expected that neither zrfA nor zrfB is required for fungal virulence, since the highest expression level of these genes occurs in acidic media whereas lung tissue provides a slightly alkaline environment. In contrast, other genes also regulated by ZafA and PacC such as zrfC and aspf2 could be required for fungal growth within host tissue, since their expression occurs only in an alkaline and extreme zinc-limiting environment closely mimicking the ambient conditions that the fungus presumably encounters when growing in the lung tissue of susceptible individuals. In this context, the ZrfC protein could be essential for obtaining zinc from host tissues and hence for sustained fungal growth within living tissue. Obviously, this would indicate that expression of zrfC must occur in fungus growing within host tissues, as is the case with aspf2. Indeed, A. fumigatus synthesizes large amounts of Aspf2 when invading tissue and is readily detected in the serum samples of most immunocompetent patients with aspergillosis (3, 9). Aspf2 shows very low (∼15%) identity to the metalloproteases of the aspzincin family (15, 39), but most cysteine residues and their spacing in both Aspf2-like proteins and aspzincin metalloproteases are highly conserved, which is indicative of a similar folding conformation based on the same pattern of disulfide-bridge formation previously described for this family of metalloproteases (39). In spite of these similarities, Aspf2 lacks the HEXXH signature that characterizes all metalloproteases, and no protease activity has been detected under any of the conditions tested to date (unpublished data). Nevertheless, in similarity to other proteins involved in sequential or closely related biochemical processes whose encoding genes transcribe divergently (54), Aspf2 might also contribute to zinc homeostasis, as does ZrfC. In this regard, we surmise that Aspf2 has evolved into a zinc-binding protein from an ancient zinc-requiring metalloprotease that lost its proteolytic activity while conserving its zinc-binding capacity. Furthermore, if Aspf2 is located in the fungal periplasm, like its orthologue Aspnd1 from A. nidulans (reference 9 and unpublished data), it could take up Zn2+ ions, forming an Aspf2-Zn2+ complex around fungal plasma membranes. This complex would increase the concentration of zinc around fungal cells in a bioavailable form that could be incorporated along with Aspf2, either directly or through an interaction with ZrfC. However, it is also possible that Zn2+ ions bound to Aspf2 could be more easily transferred to the ZBMs at the N terminus of ZrfC. Moreover, Aspf2 could also contribute to supplying Zn2+ in a more readily available form to zinc transporters operating in acidic and extreme zinc-limiting media (i.e., ZrfA and ZrfB). This would explain the severely reduced growth ability of the aspf2Δ mutant under these culture conditions (Fig. 8B). In contrast, expression of aspf2 in acidic zinc-limiting conditions at the same level as seen under alkaline zinc-limiting conditions could even more greatly improve the zinc uptake activity of the acidic transporters ZrfA and ZrfB. However, this would elicit a transient excess of intracellular zinc that could be sufficient to repress transcription of these transporters, particularly that of zrfA, whose expression is dramatically reduced when media are supplemented with an amount of Zn2+ as low as 0.5 μM (45), leading to a growth delay. Interestingly, this would explain the slightly reduced growth ability of the AF891 strain expressing aspf2 in acidic media (Fig. 9A). In sum, this report affords a basis for investigation of several important aspects of fungal biology, including the functioning of Aspf2 at the molecular level and the interrelationship that clearly exists among tissue pH, metal availability in host tissue, and fungal virulence.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Rodríguez for her excellent technical assistance.
We are also grateful for the support of the Spanish Ministry of Science and Technology and of the Regional Government of Castile & Leon (Spain) through grants BFU2007-66512 and SA080A06, respectively. J.A. and R.V. were recipients of fellowships from the Spanish Ministry of Science and Technology.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/cgi/content/full/9/3/424/DC1.
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Amich J., Leal F., Calera J. A. 2009. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int. Microbiol. 12:39–47 [PubMed] [Google Scholar]
- 2.Auld D. S. 2001. Zinc coordination sphere in biochemical zinc sites. Biometals 14:271–313 [DOI] [PubMed] [Google Scholar]
- 3.Banerjee B., Greenberger P. A., Fink J. N., Kurup V. P. 1998. Immunological characterization of Aspf2, a major allergen from Aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infect. Immun. 66:5175–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basta N. T., Ryan J. A., Chaney R. L. 2005. Trace element chemistry in residual-treated soil: key concepts and metal bioavailability. J. Environ. Qual. 34:49–63 [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 6.Bignell E., Negrete-Urtasun S., Calcagno A. M., Haynes K., Arst H. N., Jr., Rogers T. 2005. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 55:1072–1084 [DOI] [PubMed] [Google Scholar]
- 7.Bird A. J., McCall K., Kramer M., Blankman E., Winge D. R., Eide D. J. 2003. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 22:5137–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calera J. A., Hass H. 2009. Cations (Zn, Fe), p. 107–129InLatgé J. P., Steinbach W. J. (ed.), Aspergillus fumigatus and aspergillosis ASM Press, Washington, DC: [Google Scholar]
- 9.Calera J. A., Ovejero M. C., López-Medrano R., Segurado M., Puente P., Leal F. 1997. Characterization of the Aspergillus nidulans aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belong to the same family. Infect. Immun. 65:1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- 11.d'Enfert C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76–82 [DOI] [PubMed] [Google Scholar]
- 12.Eide D. J. 2006. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 1763:711–722 [DOI] [PubMed] [Google Scholar]
- 13.Espeso E. A., Tilburn J., Sanchez-Pulido L., Brown C. V., Valencia A., Arst H. N., Jr., Penalva M. A. 1997. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 274:466–480 [DOI] [PubMed] [Google Scholar]
- 14.Fedorova N. D., Khaldi N., Joardar V. S., Maiti R., Amedeo P., Anderson M. J., Crabtree J., Silva J. C., Badger J. H., Albarraq A., Angiuoli S., Bussey H., Bowyer P., Cotty P. J., Dyer P. S., Egan A., Galens K., Fraser-Liggett C. M., Haas B. J., Inman J. M., Kent R., Lemieux S., Malavazi I., Orvis J., Roemer T., Ronning C. M., Sundaram J. P., Sutton G., Turner G., Venter J. C., White O. R., Whitty B. R., Youngman P., Wolfe K. H., Goldman G. H., Wortman J. R., Jiang B., Denning D. W., Nierman W. C. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet 4e1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fushimi N., Ee C. E., Nakajima T., Ichishima E. 1999. Aspzincin, a family of metalloendopeptidases with a new zinc-binding motif. J. Biol. Chem. 274:24195–24201 [DOI] [PubMed] [Google Scholar]
- 16.Gitan R. S., Luo H., Rodgers J., Broderius M., Eide D. 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273:28617–28624 [DOI] [PubMed] [Google Scholar]
- 17.Hahne H. C. H., Kroontje W. 1973. Significance of pH and chloride concentration on behaviour of heavy metal pollutants: mercury (II), cadminum (II), zinc (II), and lead (II). J. Environ. Qual. 2:444–450 [Google Scholar]
- 18.Jensen L. T., Ajua-Alemanji M., Culotta V. C. 2003. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 278:42036–42040 [DOI] [PubMed] [Google Scholar]
- 19.Kenward A. G., Bartolotti L. J., Burns C. S. 2007. Copper and zinc promote interactions between membrane-anchored peptides of the metal binding domain of the prion protein. Biochemistry 46:4261–4271 [DOI] [PubMed] [Google Scholar]
- 20.Lamb T. M., Mitchell A. P. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb T. M., Xu W., Diamond A., Mitchell A. P. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850–1856 [DOI] [PubMed] [Google Scholar]
- 22.Li W., Mitchell A. P. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., Eide D. J. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97:7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez C. E., Motto H. L. 2000. Solubility of lead, zinc and copper added to mineral soils. Environ. Pollut. 107:153–158 [DOI] [PubMed] [Google Scholar]
- 25.Matthews T. M., Webb C. 1991. Culture systems, p. 249–289 InTuite M. F., Oliver S. G. (ed.), Saccharomyces, vol. 4 Plenum Press, New York, NY [Google Scholar]
- 26.Mitton K. P., Swain P. K., Chen S., Xu S., Zack D. J., Swaroop A. 2000. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 275:29794–29799 [DOI] [PubMed] [Google Scholar]
- 27.Moreno M. A., Amich J., Vicentefranqueira R., Leal F., Calera J. A. 2007. Culture conditions for zinc- and pH-regulated gene expression studies in Aspergillus fumigatus. Int. Microbiol. 10:187–192 [PubMed] [Google Scholar]
- 28.Moreno M. A., Ibrahim-Granet O., Vicentefranqueira R., Amich J., Ave P., Leal F., Latge J. P., Calera J. A. 2007. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol. Microbiol. 64:1182–1197 [DOI] [PubMed] [Google Scholar]
- 29.Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., Berriman M., Abe K., Archer D. B., Bermejo C., Bennett J., Bowyer P., Chen D., Collins M., Coulsen R., Davies R., Dyer P. S., Farman M., Fedorova N., Fedorova N., Feldblyum T. V., Fischer R., Fosker N., Fraser A., García J. L., García M. J., Goble A., Goldman G. H., Gomi K., Griffith-Jones S., Gwilliam R., Haas B., Haas H., Harris D., Horiuchi H., Huang J., Humphray S., Jiménez J., Keller N., Khouri H., Kitamoto K., Kobayashi T., Konzack S., Kulkarni R., Kumagai T., Lafon A., Latge J. P., Li W., Lord A., Lu C., Majoros W. H., May G. S., Miller B. L., Mohamoud Y., Molina M., Monod M., Mouyna I., Mulligan S., Murphy L., O'Neil S., Paulsen I., Peñalva M. A., Pertea M., Price C., Pritchard B. L., Quail M. A., Rabbinowitsch E., Rawlins N., Rajandream M. A., Reichard U., Renauld H., Robson G. D., Rodríguez de Córdoba S., Rodríguez-Peña J. M., Ronning C. M., Rutter S., Salzberg S. L., Sánchez M., Sánchez-Ferrero J. C., Saunders D., Seeger K., Squares R., Squares S., Takeuchi M., Tekaia F., Turner G., Vazquez de Aldana C. R., Weidman J., White O., Woodward J., Yu J. H., Fraser C., Galagan J. E., Asai K., Machida M., Hall N., Barrell B., Denning D. W. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 30.Pauly P. C., Harris D. A. 1998. Copper stimulates endocytosis of the prion protein. J. Biol. Chem. 273:33107–33110 [DOI] [PubMed] [Google Scholar]
- 31.Peñalva M. A., Arst H. N., Jr 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425–451 [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.Sandrin T. R., Maier R. M. 2003. Impact of metals on the biodegradation of organic pollutants. Environ. Health Perspect. 111:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal B. H. 2009. Aspergillosis. N. Engl. J. Med. 360:1870–1884 [DOI] [PubMed] [Google Scholar]
- 35.Serrano R., Bernal D., Simón E., Ariño J. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279:19698–19704 [DOI] [PubMed] [Google Scholar]
- 36.Serrano R., Ruíz A., Bernal D., Chambers J. R., Ariño J. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319–1333 [DOI] [PubMed] [Google Scholar]
- 37.Soller B. R., Khan T., Favreau J., Hsi C., Puyana J. C., Heard S. O. 2003. Investigation of muscle pH as an indicator of liver pH and injury from hemorrhagic shock. J. Surg. Res. 114:195–201 [DOI] [PubMed] [Google Scholar]
- 38.Tapiero H., Tew K. D. 2003. Trace elements in human physiology and pathology: zinc and metallothioneins. Biomed. Pharmacother. 57:399–411 [DOI] [PubMed] [Google Scholar]
- 39.Tatsumi H., Ikegaya K., Murakami S., Kawabe H., Nakano E., Motai H. 1994. Elucidation of the thermal stability of the neutral proteinase II from Aspergillus oryzae. Biochim. Biophys. Acta 1208:179–185 [DOI] [PubMed] [Google Scholar]
- 40.Taylor K. M., Nicholson R. I. 2003. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta 1611:16–30 [DOI] [PubMed] [Google Scholar]
- 41.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilburn J., Sarkar S., Widdick D. A., Espeso E. A., Orejas M., Mungroo J., Peñalva M. A., Arst H. N., Jr 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627 [DOI] [PubMed] [Google Scholar]
- 44.van der Rest M. E., Kamminga A. H., Nakano A., Anraku Y., Poolman B., Konings W. N. 1995. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol. Rev. 59:304–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicentefranqueira R., Moreno M. A., Leal F., Calera J. A. 2005. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot. Cell 4:837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 47.Wang Z., Feng L. S., Matskevich V., Venkataraman K., Parasuram P., Laity J. H. 2006. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J. Mol. Biol. 357:1167–1183 [DOI] [PubMed] [Google Scholar]
- 48.Waters B. M., Eide D. J. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277:33749–33757 [DOI] [PubMed] [Google Scholar]
- 49.Watt N. T., Hooper N. M. 2003. The prion protein and neuronal zinc homeostasis. Trends Biochem. Sci. 28:406–410 [DOI] [PubMed] [Google Scholar]
- 50.Weidner G., d'Enfert C., Koch A., Mol P. C., Brakhage A. A. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378–385 [DOI] [PubMed] [Google Scholar]
- 51.Wheeler K. A., Hurdman B. F., Pitt J. I. 1991. Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium. Int. J. Food Microbiol. 12:141–149 [DOI] [PubMed] [Google Scholar]
- 52.Xu W., Mitchell A. P. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahn R. 2003. The octapeptide repeats in mammalian prion protein constitute a pH-dependent folding and aggregation site. J. Mol. Biol. 334:477–488 [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Smith T. F. 1998. Yeast “operons.” Microb. Comp. Genomics 3:133–140 [DOI] [PubMed] [Google Scholar]
- 55.Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S., Eide D. 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273:28713–28720 [DOI] [PubMed] [Google Scholar]
- 56.Zhao H., Eide D. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93:2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H., Eide D. 1996. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271:23203–23210 [DOI] [PubMed] [Google Scholar]
- 58.Zhou Q., Gedrich R. W., Engel D. A. 1995. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J. Virol. 69:4323–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.