Abstract
Quantifying target microbial populations in complex communities remains a barrier to studying species interactions in soil environments. Quantitative PCR (qPCR) assays were developed for quantifying pathogenic Streptomyces scabiei and antibiotic-producing Streptomyces lavendulae strains in complex soil communities. This assay will be useful for evaluating the competitive dynamics of streptomycetes in soil.
Streptomyces spp. are ubiquitous soil bacteria that are noted for their capacity to produce a vast array of bioactive compounds, including antibiotics (10). Antibiotic-mediated species interactions are believed to be important to Streptomyces fitness and plant disease biocontrol in soil, and yet quantitative data on Streptomyces interactions in soil are limited. Moreover, because the impacts of one species on another can be mediated through interactions with other microbes in the community, detecting these impacts requires a sensitive and accurate method for quantifying the target populations within a complex community. Here, we describe a sensitive and specific assay that targets a short hypervariable region of the 16S rRNA gene to distinguish among Streptomyces organisms in complex soil communities. Streptomyces strains DL93 (Streptomyces lavendulae, an antibiotic producer that is effective in plant disease biocontrol [9]) and DL87 (Streptomyces scabiei, a plant pathogen) were studied in the present work. This approach has significant potential to shed light on the diversity and complexity of Streptomyces species interactions in soil.
Primer selection.
A 324-bp segment of the 16S rRNA gene from over 400 Streptomyces strains from native prairie and agricultural soils in the Midwestern United States (reference 4 and unpublished data) and 14 additional representative Streptomyces type strains retrieved from GenBank was evaluated using ClustalW (11). The aligned sequences spanned a 326-bp region from 54 bp to 380 bp of the Streptomyces coelicolor 16S rRNA (Y00411 [1]), including the hypervariable γ region (5). Previously the γ region was found to account for approximately 30% of the variation among streptomycete 16S rRNA gene sequences (5). Nucleotide sequence variation was observed in species- and/or group-specific sequences (see Fig. S1 in the supplemental material), suggesting that the different species included in the alignment could be distinguished by PCR-based approaches in a species- and/or group-specific manner.
Primers were designed for detecting S. lavendulae strain DL93 and S. scabiei strain DL87. Primers for the SYBR green assay utilized species-specific forward primers and a single conserved reverse primer designed from the hypervariable region (Table 1; see also Fig. S1 in the supplemental material). Primers for the TaqMan assay utilized species-specific forward and reverse primers and a single TaqMan MGB probe for detecting amplicons from multiple strains (Table 1; see also Fig. S1). All primers and the probe were designed using Primer Express (Applied Biosystems, Foster City, CA). Primers were from Integrated DNA Technologies (Coralville, IA), and the probe was from Applied Biosystems.
TABLE 1.
Oligonucleotides used in this study
| Target organism | Oligonucleotide | Sequence (5′-3′) | Product size (bp) |
|---|---|---|---|
| S. scabiei DL87 | 87FSYBR | GGTCTAATACCGGATACGACACTCT | 164 |
| 87/93RSYBR | GCTACCCGTCGTCGCCT | ||
| 87taqF | CGGGCATCCGATGAGTGT | ||
| 87taqR | GAGCCGTTACCTCACCAACAA | 82 | |
| S. lavendulae DL93 | 93FSYBR | TCTAATACCGGATACCACTCCTG | 161 |
| 93taqF | GGATACCACTCCTGCCTGCAT | 89 | |
| 93taqR | ATTACCCCACCAACAAGCTGAT | ||
| Streptomyces sp. | Strep-probe | FAMa-CGGTGAAGGATGAGC-MGB |
FAM, 6-carboxyfluorescein.
Assay development.
Initial experiments utilized primers designed for a SYBR green-based assay. However, detection of strains in nonsterilized field soil was not achieved due to a large amount of nonspecific background amplification, which was assumed to be from indigenous streptomycetes. This problem of nonspecific amplification was eliminated by the TaqMan-based assay.
For testing the sensitivity and specificity of the TaqMan-based assay, DNA was extracted from cultures as described previously (3). Reaction mixtures (25 μl) consisted of 12.5 μl 2× TaqMan Universal PCR master mix (Applied Biosytems), 1 μl of each primer at 40 pmol/μl, 1.5 μl of the probe at 2.5 μM, and 5 μl DNA. Separate reactions were done with strain DL87 and DL93 primer pairs. Reaction conditions consisted of a single cycle of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min using the ABI Prism 7000 sequence detection system (Applied Biosystems). The lower limit of detection for both strains was 0.01 pg genomic DNA (see Fig. S2A and B in the supplemental material). Based on the genome size of streptomycetes sequenced to date (2, 6, 7), 0.01 pg represents approximately one genome equivalent, indicating that the sensitivity of the TaqMan assay is equivalent to a single bacterial cell. Standard curves generated for both strains showed a linear response from 0.01 pg to 10 ng DNA (see Fig. S2C and D). The sensitivity and range of the TaqMan assay are similar to the lower level and range of detection of pathogenic Streptomyces strains obtained in real-time quantitative PCR (qPCR) assays targeting the txtAB operon (8). Mixtures of each DNA were used in amplification reactions, and no interference in accurate measurement of DNA from either strain was detected (see Table S1 in the supplemental material).
The specificity of the assay was tested with DNA from 15 Streptomyces strains commonly found in soil (4). Each reaction mixture contained 10 ng of template DNA. The DL93 primers amplified a product detected by the probe with DNA from S. lavendulae operational taxonomic unit (OTU) groups 1 and 5 (Table 2). Strain DL93 shares sequence identity with these strains in the targeted region of the 16S rRNA gene (see Fig. S1 in the supplemental material). The ability to detect multiple strains in two OTUs extends the capacity of the assay to follow the population dynamics of additional strains. Weak amplification occurred with Streptomcyes flavogriseus DNA (threshold cycle [CT] = 21), which shares 10/14 nucleotides with the forward primer and 17/18 nucleotides with the reverse primer. The primers designed for strain DL87 were highly specific and amplified DNA only from strain DL87.
TABLE 2.
Specificity of SYBR green and TaqMan assays against common Streptomyces species found in Cedar Creek Ecosystem Science Reserve soil
| Species and strain designation | Accession no. | OTU | Result for assay and strain primera |
|||
|---|---|---|---|---|---|---|
| SYBR green |
TaqMan |
|||||
| DL87 | DL93 | DL87 | DL93 | |||
| S. flavogriseus LK1234.1 | AY465284.1 | 14 | − | + | − | +/− |
| S. lavendulae | ||||||
| LK1312.4 | AY465295.1 | 1 | − | + | − | + |
| LK1132.3 | AY465223.1 | 1 | − | + | − | + |
| LK1334.1 | AY465333.1 | 1 | − | + | − | + |
| LK1223.4 | AY465263.1 | 5 | − | + | − | + |
| LK1231.3 | AY465271.1 | 5 | − | + | − | + |
| S. lydicus | ||||||
| LK1111.4 | AY465187.1 | 3 | − | − | − | − |
| LK1314.5 | AY465301.1 | 3 | − | − | − | − |
| LK1133.5 | AY465228.1 | 3 | − | − | − | − |
| LK1212.1 | AY465237.1 | 3 | − | − | − | − |
| S. olivochromogenes | ||||||
| LK1111.3 | AY465186.1 | 2 | − | − | − | − |
| LK1332.1 | AY465324.1 | 2 | − | − | − | − |
| LK1334.2 | AY465334.1 | 2 | − | − | − | − |
| S. rimosus | ||||||
| LK1124.1 | AY465212.1 | 4 | − | − | − | − |
| LK1324.5 | AY465318.1 | 4 | − | − | − | − |
| S. scabiei DL87 | AY277383.1 | + | − | + | − | |
| S. lavendulae DL93 | AY277380.1 | − | + | − | + | |
−, no product detected; +, specific amplicon detected; +/−, small amount of amplicon detected.
Field soil was inoculated with a 10-fold serial dilution series of strain DL87 or DL93 spores, and total soil DNA was isolated with the PowerSoil DNA kit (MoBio, Carlsbad, CA) according to the manufacturer's protocol after sonication for 10 min in extraction buffer and two rounds of homogenization using the FastPrep FP120 (MP Biomedicals, Solon, OH). The modification was needed for detecting low densities of DL93. PCRs were done as described above using 10 ng DNA. No amplification was detected in assays using DNA from noninoculated soil. Population estimates for both strains were linear from 102 to 106 CFU/g soil (see Fig. S3 in the supplemental material).
Quantifying Streptomyces colonization and competitive dynamics in complex soil communities.
We evaluated interactions of DL87 and DL93 by inoculating them alone and together to total combined densities of 106 CFU/g autoclaved soil or 5 × 105 and 5 × 103 CFU/g field soil. Soils were incubated in the dark at room temperature for 6 days. Each treatment was replicated three times. Culturable cell counts were determined by removing 1.0-g soil samples at 1 and 6 days postinoculation, shaking them in 10 ml sterile water for 1 h at 4°C, and dilution plating them onto oatmeal agar. Dilution plating was not conducted for the nonsterile field soil samples, as it is impossible to differentiate DL87 and DL93 from other Streptomyces strains in complex mixtures. Samples (0.25 g) were removed from sterilized and field soils at the same times for DNA extraction and used in qPCRs to provide corresponding estimates of DL87 and DL93 densities.
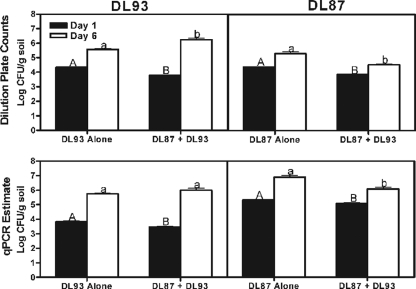
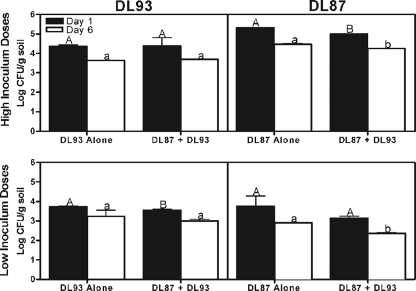
In autoclaved soil, DL93 and DL87 population estimates were similar whether measured by qPCR or by dilution plating. Densities of DL87 were negatively impacted by DL93 at both time points (Fig. 1; P < 0.05), reflecting the negative impacts of the antibiotic producer on the establishment and growth of DL87 in autoclaved soil. In contrast, after 6 days DL93 populations were larger when coinoculated with DL87 than when inoculated alone, suggesting a positive impact of DL87 on DL93 (Fig. 1). In field soil, DL93 also negatively impacted DL87 populations at each dose and time except after 1 day at the low-inoculum dose (Fig. 2). However, no positive impact of DL87 on DL93 was observed, and after 1 day, at the low-inoculum dose populations of DL93 were actually smallest in the presence of DL87.
FIG. 1.
Comparison of direct colony counts and qPCR for quantification of Streptomyces strains in sterile soil when inoculated alone and together at 1 and 6 days postinoculation. Different letters of the same case (uppercase or lowercase) within each figure indicate statistical significance at P < 0.05.
FIG. 2.
Quantification of Streptomyces strains in field soil when inoculated alone and together at 1 and 6 days postinoculation. Different letters of the same case (uppercase or lowercase) within each figure indicate statistical significance at P < 0.05.
These data suggest that significant competitive interactions occur between DL93 and DL87 in autoclaved soil and in the presence of a complex indigenous soil microbial community. However, the positive impact of DL87 on DL93 in autoclaved but not field soil indicates that the outcomes of microbial interactions in complex soil microbial communities may be different than those in sterilized soil. The qPCR assay offers a powerful tool for elucidating microbial population and competitive dynamics under “real-world” conditions.
Supplementary Material
Acknowledgments
This work was supported by award DEB-0543213 from the National Science Foundation and by USDA grant 2006-35319-17445.
This publication is a joint contribution from the USDA-ARS-Plant Science Research Unit and the Minnesota Agricultural Experiment Station.
Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the USDA, and does not imply its approval to the exclusion of other products and vendors that might also be suitable.
Footnotes
Published ahead of print on 15 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baylis, H. A., and M. J. Bibb. 1987. The nucleotide sequence of a 16S rRNA gene from Streptomyces coelicolor A3(2). Nucleic Acids Res. 15:7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, J. D. James, D. E. Harris, M. A. Quali, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Davelos, A. L., K. Xiao, J. M. Flor, and L. L. Kinkel. 2004. Genetic and phenotypic traits of streptomycetes used to characterize antibiotic activities of field-collected microbes. Can. J. Microbiol. 50:79-89. [DOI] [PubMed] [Google Scholar]
- 4.Davelos, A. L., K. Xiao, D. A. Samac, A. P. Martin, and L. L. Kinkel. 2004. Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microb. Ecol. 48:601-612. [DOI] [PubMed] [Google Scholar]
- 5.Kim, E., H. Kim, S. Hong, K. H. Kang, Y. H. Kho, and Y. Park. 1993. Gene organization and primary structure of a ribosomal RNA gene cluster from Streptomyces griseus subsp. griseus. Gene 132:21-31. [DOI] [PubMed] [Google Scholar]
- 6.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahaski, M. Shinose, Y. Takahaski, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu, X., L. A. Wanner, and B. J. Christ. 2008. Using the TxtAB operon to quantify pathogenic Streptomyces in potato tubers and soil. Phytopathology 98:405-412. [DOI] [PubMed] [Google Scholar]
- 9.Ryan, A. D., and L. L. Kinkel. 1997. Inoculum density and population dynamics of suppressive and pathogenic Streptomyces strains and their relationship to biological control of potato scab. Biol. Control 10:180-186. [Google Scholar]
- 10.Tanaka, Y., and S. Omura. 1990. Metabolism and products of actinomycetes: an introduction. Actinomycetologica 4:13-14. [Google Scholar]
- 11.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.